Identification of Reliable Reference Genes for the Expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ Sepal Color
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Isolation and cDNA Synthesis
2.3. Selection of Candidate Genes and Prime Design
2.4. Quantitative RT-PCR Analysis
2.5. Data Analysis
2.6. Assessment of Normalization
3. Results
3.1. Primer Quality Detection of Each Gene
3.2. geNorm
3.3. NormFinder
3.4. BestKeeper
3.5. RefFinder
3.6. Verifying Selected Reference Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kesumawati, E.; Kimata, T.; Uemachi, T.; Hosokaw, M.; Yazawa, S. Correlation of phytoplasma concentration in Hydrangea macrophylla with green-flowering stability. Sci. Hortic. 2006, 108, 74–78. [Google Scholar] [CrossRef]
- Ito, T.; Aoki, D.; Fukushima, K.; Yoshida, K. Direct mapping of Hydrangea blue-complex in sepal tissues of Hydrangea macrophylla. Sci. Rep. 2019, 9, 5450. [Google Scholar] [CrossRef]
- Galopin, G.; Codarin, S.; Viemont, J.D.; Morel, P. Architectural development of inflorescence in Hydrangea macrophylla cv. Hermann Dienemann. Horticulture 2008, 43, 361–365. [Google Scholar] [CrossRef]
- Schreiber, H.D. Curious chemistry guides Hydrangea colors. Am. Sci. 2014, 102, 444. [Google Scholar] [CrossRef]
- Liu, F.; Huang, L.L.; Li, Y.L.; Reinhoud, P.; Jongsma, M.A.; Wang, C.Y. Shoot organogenesis in leaf explants of Hydrangea macrophylla ‘Hyd1’ and assessing genetic stability of regenerants using ISSR markers. Plant Cell Tiss. Org. 2011, 104, 111–117. [Google Scholar] [CrossRef]
- Radonic, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven Golden Rules of Quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef]
- Remans, T.; Keunen, E.; Bex, G.J.; Smeets, K.; Vangronsveld, J.; Cuypersa, A. Reliable Gene Expression Analysis by Reverse Transcription-Quantitative PCR: Reporting and Minimizing the Uncertainty in Data Accuracy. Plant Cell 2014, 26, 3829–3837. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Chen, S.; Jiang, J.; Zhang, F.; Chen, F. Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci. Rep. 2015, 5, 8094. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference Gene Selection for RT-qPCR Analysis of Flower Development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Xin, H.; Chen, X.; Zhu, X.; Li, X. Reliable reference genes for normalization of gene expression data in tea plants (Camellia sinensis) exposed to metal stresses. PLoS ONE 2017, 12, e0175863. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Higgins, P.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Meng, Y.; Li, N.; Tian, J.; Gao, J.; Zhang, C. Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci. Hortic. 2013, 158, 16–21. [Google Scholar] [CrossRef]
- Xu, L.; Xu, H.; Cao, Y.; Yang, P.; Feng, Y.; Tang, Y.; Yuan, S.; Ming, J. Validation of Reference Genes for Quantitative Real-Time PCR during Bicolor Tepal Development in Asiatic Hybrid Lilies (Lilium spp.). Front. Plant Sci. 2017, 8, 669. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhou, Y.; Zhao, Y.; Liu, Y.; Ke, Y.; Jin, X.; Ma, H. Internal Reference gene selection for quantitative Real-Time RT-PCR normalization in potato tissues. Phyton 2020, 89, 329–344. [Google Scholar] [CrossRef]
- Jin, J.; Song, Z.; Zhao, B.; Zhang, Y.; Wang, R. Physiological and metabolomics responses of Hydrangea macrophylla (Thunb.) Ser. and Hydrangea strigosa Rehd. to lead exposure. Ecotox. Environ. Safe 2022, 243, 113960. [Google Scholar] [CrossRef]
- Chen, H.; Wang, D.; Zhu, Y.; Li, W.; Chen, J.; Li, Y. Integrative transcriptomics and proteomics elucidate the regulatory mechanism of Hydrangea macrophylla flower-color changes induced by exogenous aluminum. Agronomy 2022, 12, 969. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of realtime quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Peng, X.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 5–84. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pichon, M.; Courboun, I.; Beckert, M.; Boudet, A.M.; Grima-Pettenati, J. Cloning and characterization of two maize cDNAs encoding Cinnamoyl-CoA Reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 1998, 38, 671–676. [Google Scholar] [CrossRef]
- Piquemal, J.; Lapierre, C.; Myton, K.; Connell, A.O.; Boudet, A.M. Down-regulation of Cinnamoyl-CoA Reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 1998, 13, 71–83. [Google Scholar] [CrossRef]
- Bassil, E.; Blumwald, E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef]
- Yoshida, K.; Kondo, T.; Okazaki, Y.; Katou, K. Cause of blue petal colour. Nature 1995, 373, 291. [Google Scholar] [CrossRef]
- Zhang, H.; Du, C.; Wang, Y.; Wang, J.; Zheng, L.; Wang, Y. The Reaumuria trigyna leucoanthocyanidin dioxygenase (RtLDOX) gene complements anthocyanidin synthesis and increases the salt tolerance potential of a transgenic Arabidopsis LDOX mutant. Plant Physiol. Bioch. 2016, 106, 278–287. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Huggett, J.F.; Dheda, K.; Bustin, S.A.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef]
- Borrero, V.B.; Leidi, E.O.; Andrés, Z.; Rubio, L.; De Luca, A.; Fernández, J.A.; Cubero, B.; Pardo, J.M. Ion Exchangers NHX1 and NHX2 Mediate Active Potassium Uptake into Vacuoles to Regulate Cell Turgor and Stomatal Function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Chung, S.J.; Lee, G.J.; Lee, H.J.; Kim, J.B.; Kim, D.S.; Kang, S.Y. Isolation of a Leucoanthocyanidin Dioxygenase (LDOX) Gene from a Spray-type Chrysanthemum (Dendranthema × grandiflorum) and Its Colored Mutants. Korean J. Hortic. Sci. Technol. 2010, 28, 595–600. [Google Scholar] [CrossRef]
- Gollop, R.; Farhi, S.; Perl, A. Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci. 2001, 161, 579–588. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Y.; Zhao, L.; Liu, Y.N.; Zhao, M. Selection of reliable reference genes for RT-qPCR during methyl jasmonate, salicylic acid and hydrogen peroxide treatments in Ganoderma lucidum. J. Microbiol. Biotechn. 2018, 34, 92. [Google Scholar] [CrossRef]
- Jin, X.; Fu, J.; Dai, S.; Sun, Y.; Hong, Y. Reference gene selection for qPCR analysis in cineraria developing flowers. Sci. Hortic. 2013, 153, 64–70. [Google Scholar] [CrossRef]
- Wang, M.; Lu, S. Validation of Suitable Reference Genes for Quantitative Gene Expression Analysis in Panax ginseng. Front. Plant. Sci. 2016, 6, 696. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Chen, S.; Qi, X.; Feng, J.; Chen, H.; Wang, H.; Qin, Z.; Deng, Y. Selection and validation of reference genes for qRT-PCR gene expression analysis in Hydrangea macrophylla under aluminum treatment. Acta Agric. Boreali Sin. 2021, 36, 9–18. [Google Scholar] [CrossRef]

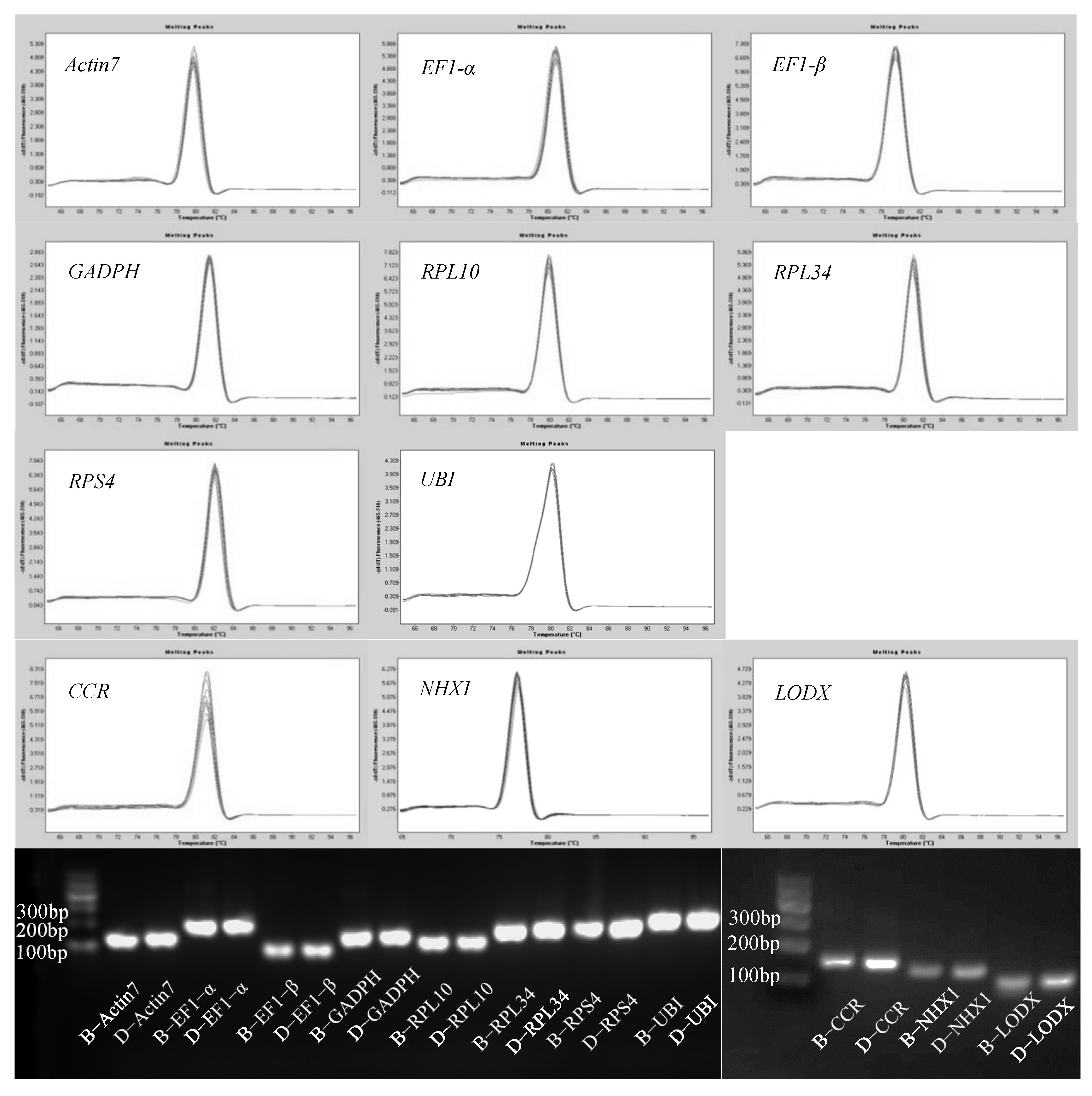
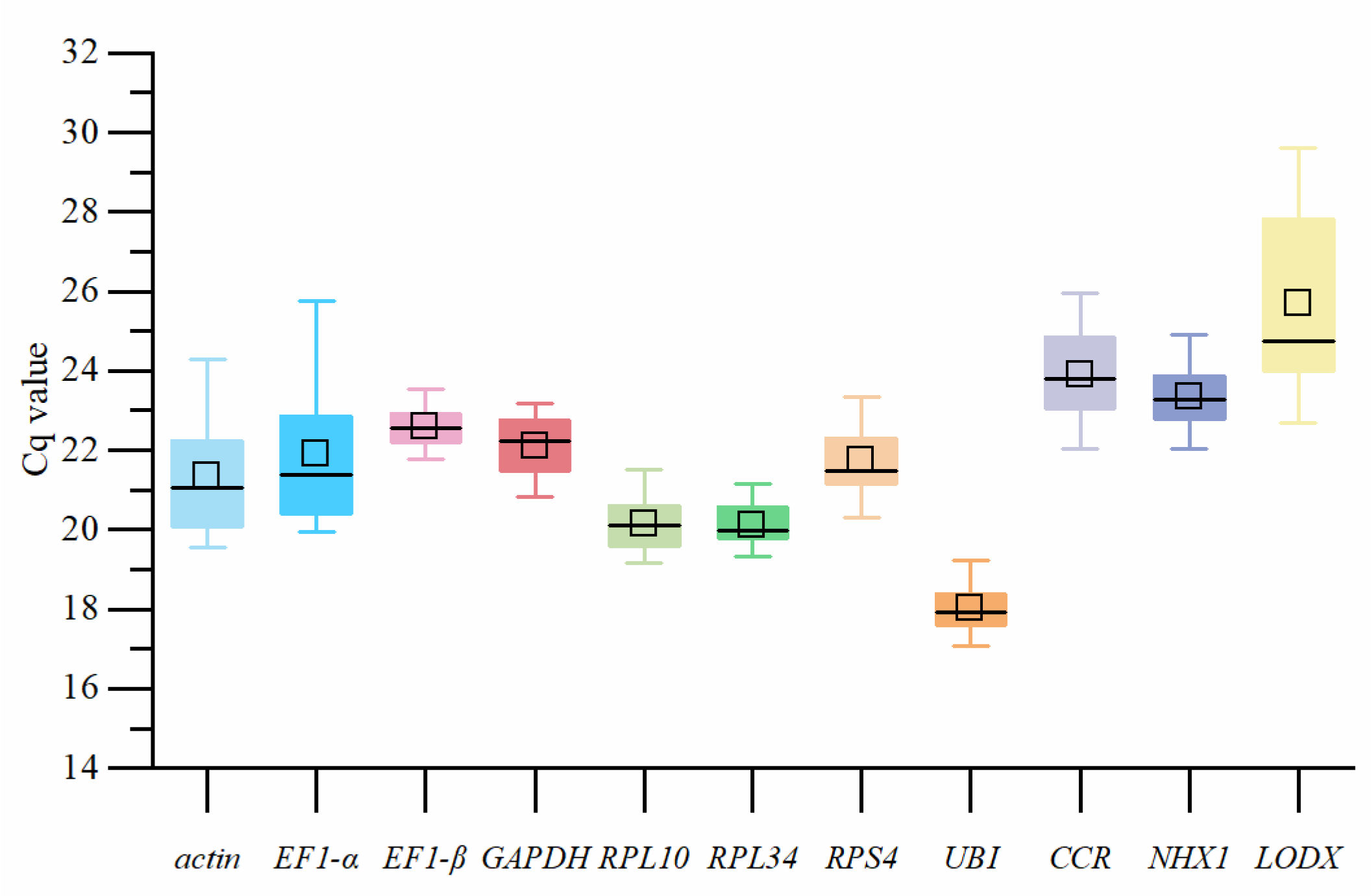
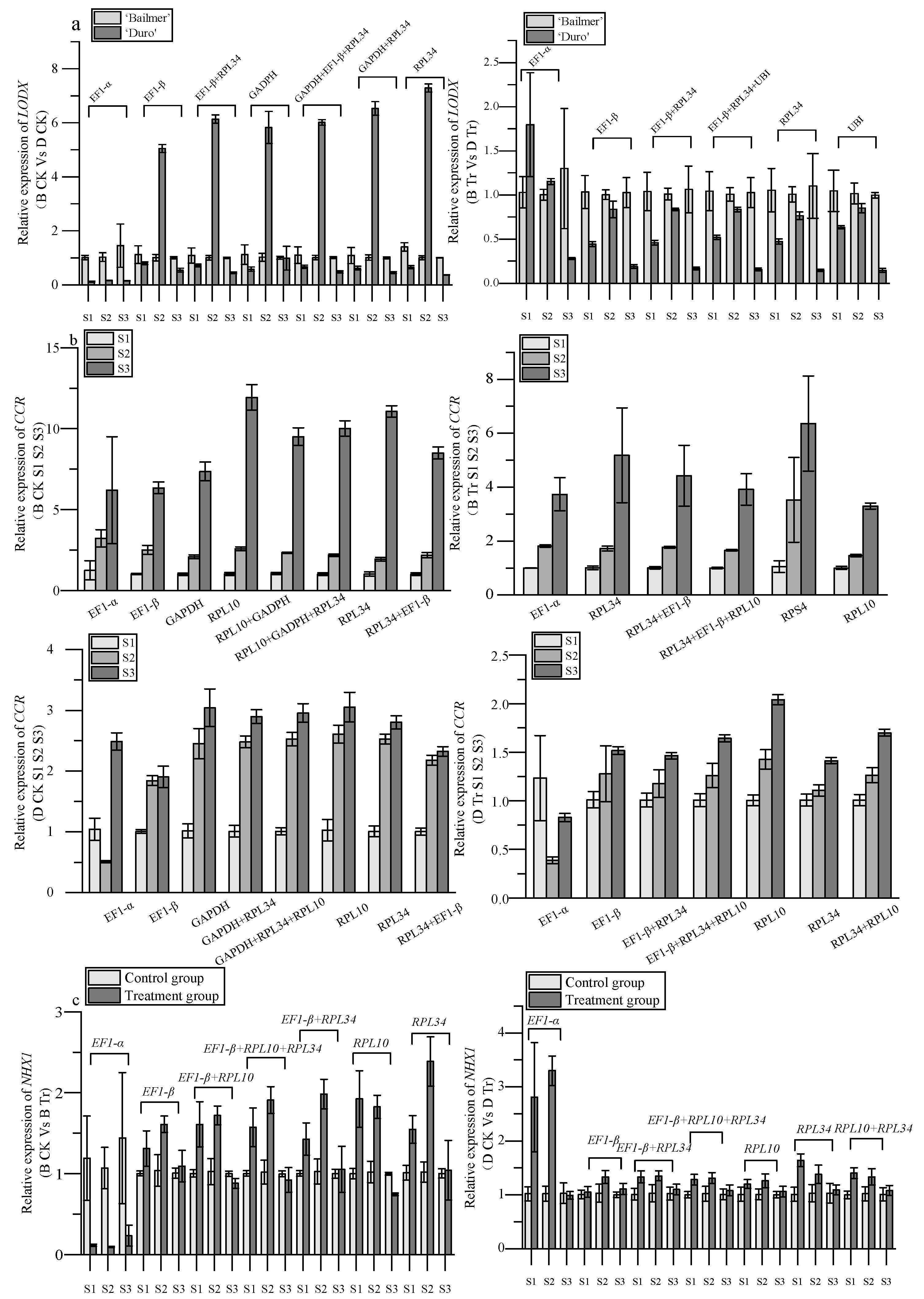
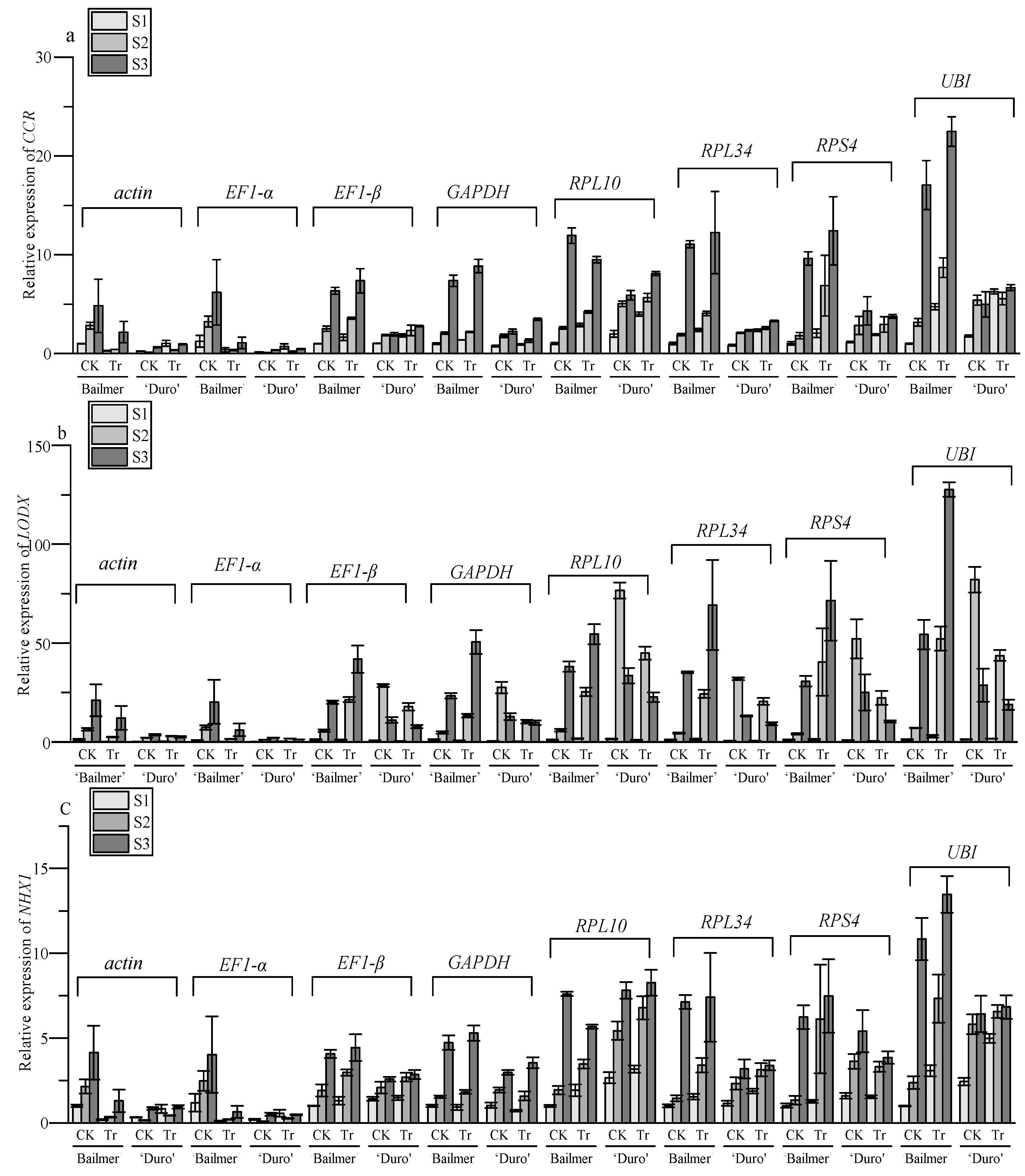
| Group | Subgroup |
|---|---|
| Comparison of different varieties | B CK vs. D CK |
| B Tr vs. D Tr | |
| Comparison of developmental stage | B CK S1 S2 S3 |
| B Tr S1 S2 S3 | |
| D CK S1 S2 S3 | |
| D Tr S1 S2 S3 | |
| Treatment or control group | B CK vs. B Tr |
| D CK vs. D Tr | |
| All materials | All materials |
| Gene Symbol | Gene Name | Primer Sequence (5’-3’) | Amplicon Length (bp) | Correlation Coefficients (R2) | Amplification Efficiency (E %) |
|---|---|---|---|---|---|
| ACT | Actin7 (Isoform0002522) | CCGAAGAGCACCCAGTTCTT | 131 | 0.999 | 109 |
| AGAAAGAACGGCCTGGATG | |||||
| EF1-α | elongation factor 1-Alpha (Isoform0002316) | ACCCGGTGACAATGTTGGTT | 180 | 0.988 | 86 |
| GACAATCGAGAACTGGGGCA | |||||
| EF1-β | elongation factor 1-beta (Isoform0062896) | CGCAGCTGTTTTAGGGAAGCC | 81 | 0.999 | 107 |
| GCGAGCTGCGAAGACACAGA | |||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase (Isoform0003402) | GGCTGAGACTGGAGCGGAAT | 134 | 0.999 | 104 |
| CAAACATGGGGGCGTCTTTGC | |||||
| RPL10 | 60S ribosomal protein L10 (Isoform0006092) | TGTCTCCAGCGAGGCTCTTGA | 104 | 0.958 | 126 |
| ATGGATGGACGCGCACTCTC | |||||
| RPL34 | 60S ribosomal protein L34 (Isoform0005815) | ACCCCCGGTGGAAAGCTAGT | 156 | 0.995 | 121 |
| ACGGTTCACAGTCCTCCGGT | |||||
| RPS4 | 40S ribosomal protein S4 (Isoform0004107) | GCTTCGAGACCATCCACGTC | 165 | 0.990 | 98.9 |
| GAGCGCTAAGCCTCTTCCTG | |||||
| UBI11 | Polyubiquitin (Isoform0002406) | TGTTGAGGCTGAGGGGAGGT | 190 | 0.980 | 99.5 |
| TGGAGGATGGAAGGACTCTTGC | |||||
| CCR | Cinnamoyl-CoA Reductase (Isoform0002511) | TTACTCCTTCGAGCCTCCTTCTGAC | 150 | 0.993 | 98 |
| ATGACAGCAGTGACAGAAGACGTGA | |||||
| NHX1 | sodium/hydrogen exchanger 1-like (Isoform0010678) | TGATGCCACATCAGTTGTGCTG | 128 | 0.992 | 106 |
| CACCCAGCAAAGTGCTTGTGAG | |||||
| LODX | Leucoanthocyanidin dioxygenase (Isoform0054522) | GAGTTGAACAAGGGCGCGAT | 101 | 0.998 | 105 |
| AACCCTTGTCCCGCAACCTT |
| Ranking Order | | B CK vs. D CK | B Tr vs. D Tr | B CK S1 S2 S3 | B Tr S1 S2 S3 | D CK S1 S2 S3 | D Tr S1 S2 S3 | B CK vs. B Tr | D CK vs. D Tr | All Materials |
|---|---|---|---|---|---|---|---|---|---|
| V2/3 | 0.121 | 0.091 | 0.105 | 0.093 | 0.095 | 0.077 | 0.127 | 0.094 | 0.149 |
| 1 | GAPDH | RPL34 | EF1-β | RPL34 | RPL34 | RPS4 | EF1-β | RPL10 | GAPDH | RPL34 | EF1-β | RPL34 | EF1-β | RPL10 | RPL10 | RPL34 | EF1-β | RPL34 |
| 2 | EF1-β | UBI | RPL10 | UBI | RPL10 | RPL10 | RPL34 | EF1-β | UBI |
| 3 | RPS4 | RPS4 | GAPDH | RPL34 | UBI | RPS4 | GAPDH | UBI | RPS4 |
| 4 | UBI | GAPDH | UBI | GAPDH | EF1-β | UBI | UBI | RPS4 | GAPDH |
| 5 | RPL10 | RPL10 | EF1-β | EF1-α | RPS4 | GAPDH | RPS4 | GAPDH | RPL10 |
| 6 | actin | actin | actin | actin | actin | actin | actin | actin | actin |
| 7 | EF1-α | EF1-α | EF1-α | RPS4 | EF1-α | EF1-α | EF1-α | EF1-α | EF1-α |
| Gene Name | B CK vs. D CK | B Tr vs. D Tr | B CK S1 S2 S3 | B Tr S1 S2 S3 | D CK S1 S2 S3 | D Tr S1 S2 S3 | B CK vs. B Tr | D CK vs. D Tr | All Materials |
|---|---|---|---|---|---|---|---|---|---|
| actin | 0.963 | 0.467 | 0.488 | 0.333 | 0.607 | 0.467 | 0.908 | 0.579 | 0.796 |
| EF1-α | 1.327 | 0.506 | 0.765 | 0.302 | 0.710 | 0.565 | 1.256 | 0.670 | 1.079 |
| EF1-β | 0.174 | 0.203 | 0.201 | 0.173 | 0.222 | 0.208 | 0.211 | 0.208 | 0.189 |
| GADPH | 0.097 | 0.413 | 0.194 | 0.243 | 0.153 | 0.476 | 0.110 | 0.400 | 0.276 |
| RPL10 | 0.572 | 0.352 | 0.187 | 0.306 | 0.197 | 0.197 | 0.296 | 0.186 | 0.473 |
| RPL34 | 0.167 | 0.158 | 0.255 | 0.019 | 0.173 | 0.102 | 0.302 | 0.123 | 0.208 |
| RPS4 | 0.477 | 0.417 | 0.268 | 0.481 | 0.334 | 0.255 | 0.518 | 0.357 | 0.444 |
| UBI | 0.510 | 0.180 | 0.397 | 0.174 | 0.323 | 0.170 | 0.614 | 0.246 | 0.480 |
| Gene Name | B CK vs. D CK | B Tr vs. D Tr | B CK S1 S2 S3 | B Tr S1 S2 S3 | D CK S1 S2 S3 | D Tr S1 S2 S3 | B CK vs. B Tr | D CK vs. D Tr | All Materials | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SD | CV | SD | CV | SD | CV | SD | CV | SD | CV | SD | CV | SD | CV | SD | CV | SD | CV | |
| actin | 1.30 | 5.87 | 0.69 | 3.37 | 0.43 | 1.84 | 0.50 | 2.49 | 0.64 | 3.07 | 0.58 | 2.76 | 1.70 | 7.82 | 0.61 | 2.89 | 1.22 | 5.71 |
| EF1-α | 1.68 | 7.33 | 0.66 | 3.14 | 0.84 | 3.43 | 0.41 | 1.99 | 0.71 | 3.34 | 0.69 | 3.23 | 2.03 | 9.03 | 0.69 | 3.25 | 1.45 | 6.63 |
| EF1-β | 0.24 | 1.05 | 0.21 | 0.94 | 0.10 | 0.44 | 0.19 | 0.85 | 0.38 | 1.68 | 0.22 | 1.00 | 0.36 | 1.59 | 0.46 | 2.04 | 0.41 | 1.83 |
| GADPH | 0.32 | 1.43 | 0.46 | 2.15 | 0.21 | 0.93 | 0.50 | 2.28 | 0.41 | 1.83 | 0.47 | 2.19 | 0.57 | 2.57 | 0.61 | 2.77 | 0.62 | 2.79 |
| RPL10 | 0.53 | 2.61 | 0.41 | 2.09 | 0.41 | 2.05 | 0.12 | 0.64 | 0.32 | 1.55 | 0.17 | 0.85 | 0.35 | 1.78 | 0.40 | 1.93 | 0.54 | 2.66 |
| RPL34 | 0.45 | 2.21 | 0.24 | 1.23 | 0.48 | 2.35 | 0.29 | 1.46 | 0.43 | 2.10 | 0.19 | 0.99 | 0.46 | 2.26 | 0.40 | 2.02 | 0.43 | 2.12 |
| RPS4 | 0.57 | 2.59 | 0.41 | 1.93 | 0.46 | 2.12 | 0.63 | 2.92 | 0.59 | 2.63 | 0.17 | 0.80 | 0.56 | 2.59 | 0.65 | 2.99 | 0.61 | 2.79 |
| UBI | 0.63 | 3.50 | 0.28 | 1.58 | 0.57 | 3.17 | 0.29 | 1.61 | 0.56 | 3.07 | 0.28 | 1.54 | 0.44 | 2.48 | 0.45 | 2.50 | 0.46 | 2.56 |
| Ranking Order | B CK vs. D CK | B Tr vs. D Tr | B CK S1 S2 S3 | B Tr S1 S2 S3 | D CK S1 S2 S3 | D Tr S1 S2 S3 | B CK vs. B Tr | D CK vs. D Tr | All Materials |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GAPDH | RPL34 | RPL10 | EF1-β | GAPDH | RPL34 | EF1-β | RPL34 | EF1-β |
| 2 | RPL34 | EF1-β | GAPDH | RPL34 | RPL34 | RPL10 | RPL10 | RPL10 | RPL34 |
| 3 | EF1-β | UBI | RPL34 | RPL10 | RPL10 | EF1-β | RPL34 | EF1-β | GAPDH |
| 4 | RPS4 | RPL10 | EF1-β | UBI | EF1-β | RPS4 | GAPDH | UBI | UBI |
| 5 | UBI | RPS4 | RPS4 | GAPDH | UBI | UBI | UBI | RPS4 | RPS4 |
| 6 | RPL10 | GAPDH | UBI | EF1-α | RPS4 | actin | RPS4 | GAPDH | RPL10 |
| 7 | actin | actin | actin | actin | actin | GAPDH | actin | actin | actin |
| 8 | EF1-α | EF1-α | EF1-α | RPS4 | EF1-α | EF1-α | EF1-α | EF1-α | EF1-α |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Yuan, S.; Qi, H.; Chu, Z.; Liu, C. Identification of Reliable Reference Genes for the Expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ Sepal Color. Horticulturae 2022, 8, 835. https://doi.org/10.3390/horticulturae8090835
Zhang G, Yuan S, Qi H, Chu Z, Liu C. Identification of Reliable Reference Genes for the Expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ Sepal Color. Horticulturae. 2022; 8(9):835. https://doi.org/10.3390/horticulturae8090835
Chicago/Turabian StyleZhang, Gaitian, Suxia Yuan, Hui Qi, Zhiyun Chu, and Chun Liu. 2022. "Identification of Reliable Reference Genes for the Expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ Sepal Color" Horticulturae 8, no. 9: 835. https://doi.org/10.3390/horticulturae8090835
APA StyleZhang, G., Yuan, S., Qi, H., Chu, Z., & Liu, C. (2022). Identification of Reliable Reference Genes for the Expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ Sepal Color. Horticulturae, 8(9), 835. https://doi.org/10.3390/horticulturae8090835





