A MADS-Box Gene-Based InDel Marker Discriminating Sex in Actinidia arguta
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. Resequencing and Polymorphic InDel Detection
2.3. Detection of Candidate InDels Discriminating Sex
2.4. Development of a Sex-Discriminating InDel Marker
3. Results
3.1. Resequencing and InDel Detection
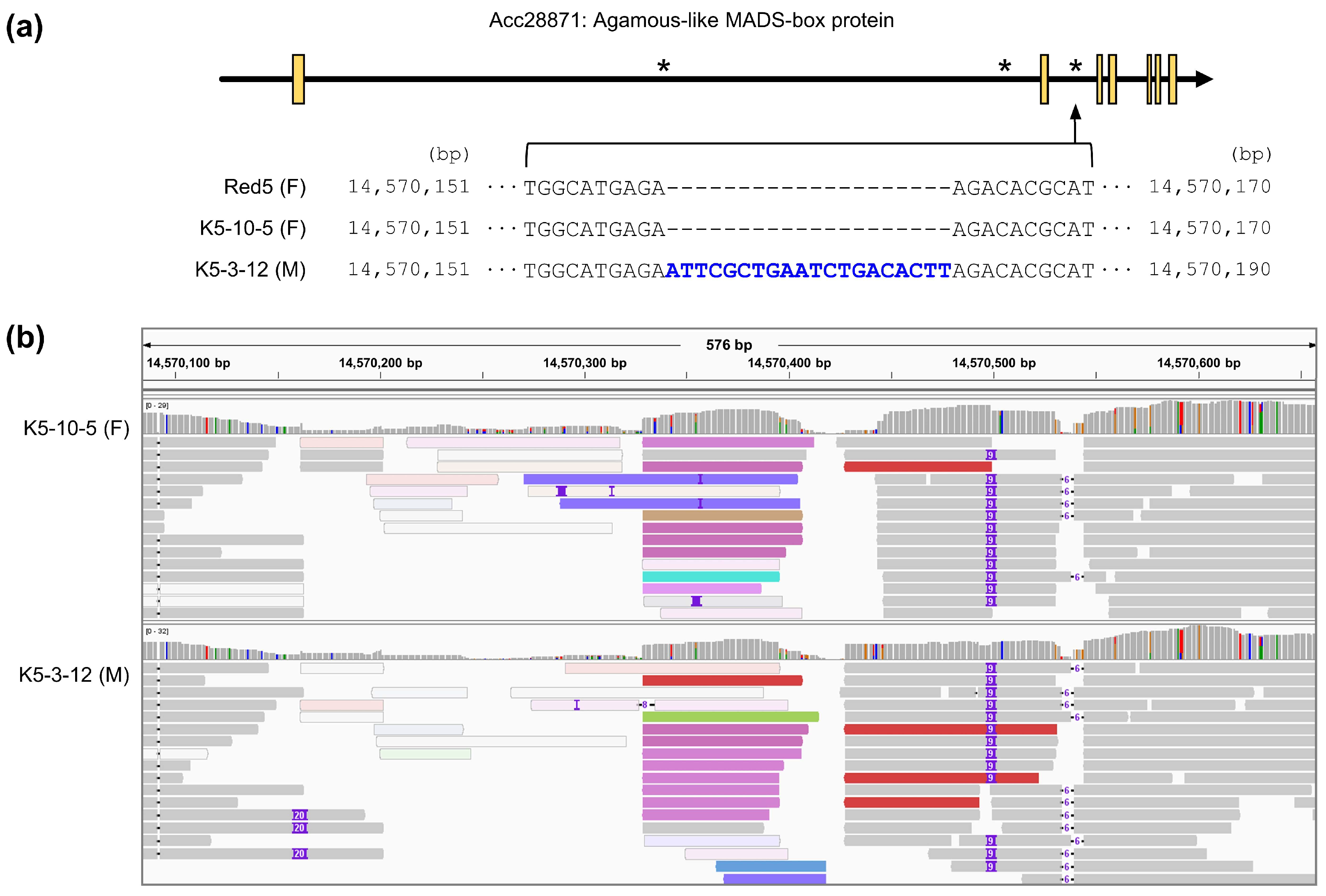
3.2. Selection of Polymorphic InDels Occurring in Sex-Related Genes
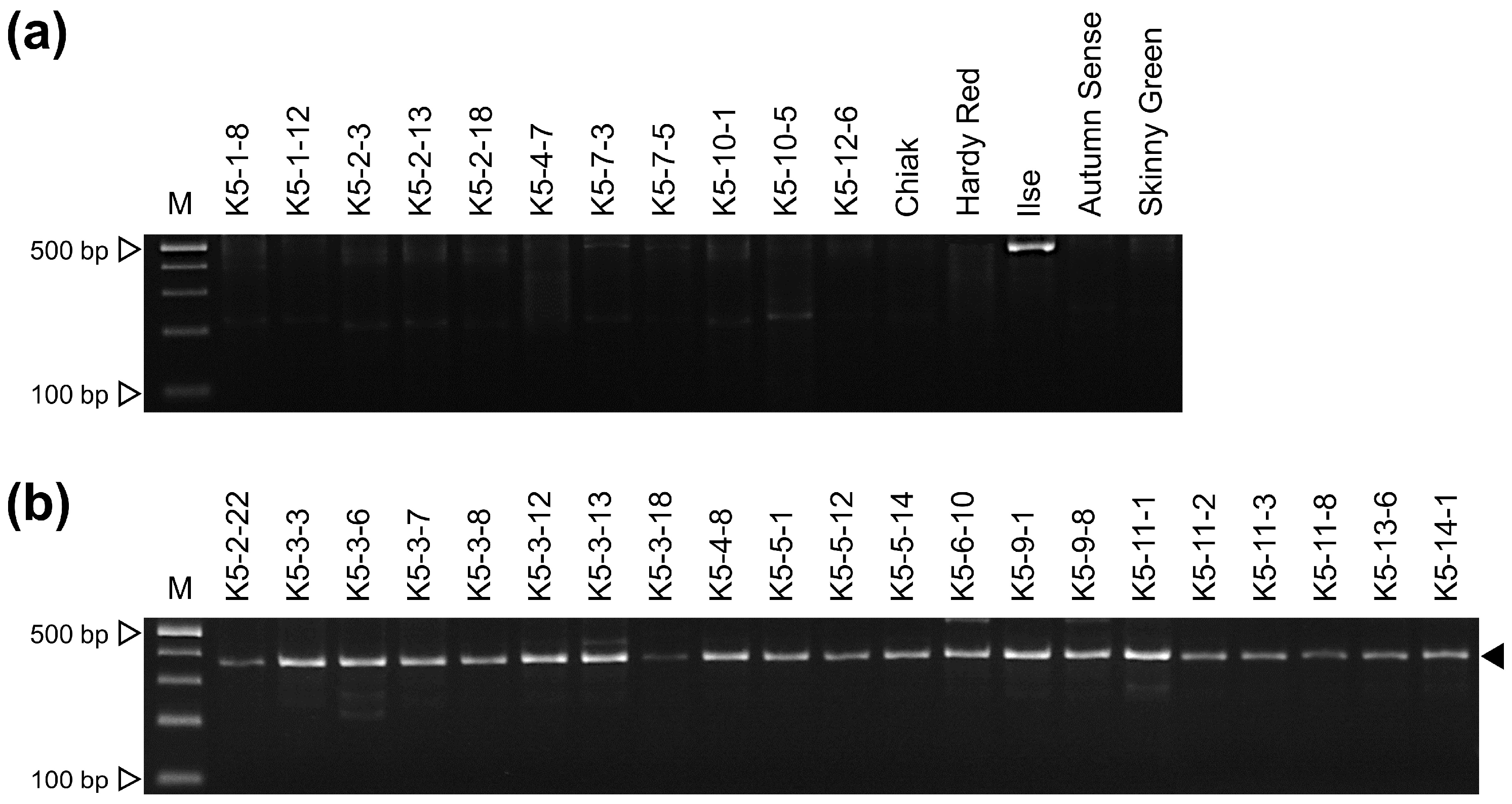
3.3. Development of a Sex-Discriminating InDel Marker
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wellmer, F.; Riechmann, J.L.; Alves-Ferreira, M.; Meyerowitz, E.M. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 2004, 15, 1314–1326. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, B.; Zhang, Q.; Altman, N.; Ma, H. Genome-wide expression profiling and identification of gene activities during early flower development in Arabidopsis. Plant Mol. Biol. 2005, 58, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Bendahmane, A.; Renner, S.S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011, 62, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Ishii, K.; Aonuma, W.; Ikeda, T.; Kawamoto, H.; Koizumi, A.; Filatov, D.A.; Chibalina, M.; Bergero, R.; Charlesworth, D.; et al. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 2016, 6, 18917. [Google Scholar] [CrossRef] [PubMed]
- Harkess, A.; Zhou, J.; Xu, C.; Bowers, J.E.; Ven der Hulst, R.; Ayyampalayam, S.; Mercati, F.; Riccardi, P.; McKain, M.R.; Kakrana, A.; et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 2017, 8, 1279. [Google Scholar] [CrossRef]
- Akagi, T.; Henry, I.M.; Tao, R.; Comai, L. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 2014, 346, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Henry, I.M.; Kawai, T.; Comai, L.; Tao, R. Epigenetic regulation of the sex-determination gene MeGI in polyploid persimmon. Plant Cell 2016, 28, 2905–2915. [Google Scholar] [CrossRef]
- Liao, Q.; Du, R.; Gou, J.; Shen, H.; Liu, H.; Nguyen, J.K.; Ming, R.; Yin, T.; Huang, S.; Yan, J. The genomic architecture of the sex-determining region and sex-related metabolic variation in Ginkgobiloba. Plant J. 2020, 104, 1399–1499. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.F.; Mathew, L.S.; Ahmed, I.; Al-Azwani, I.K.; Krueger, R.; Rivera-Nuñez, D.; Mohamoud, Y.A.; Clark, A.G.; Suhre, K.; Malek, J.A. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 2018, 9, 3969. [Google Scholar] [CrossRef]
- Okazaki, Y.; Takahata, S.; Hirakawa, H.; Suzuki, Y.; Onodera, Y. Molecular evidence for recent divergence of X- and Y-linked gene pairs in Spinacia oleracea L. PLoS ONE 2019, 14, e0214949. [Google Scholar] [CrossRef]
- Akagi, T.; Henry, I.M.; Ohtani, H.; Morimoto, T.; Beppu, K.; Kataoka, I.; Tao, R. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 2018, 30, 780–795. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Pilkington, S.M.; Varkonyi-Gasic, E.; Henry, I.M.; Sugano, S.S.; Sonoda, M.; Firl, A.; McNeilage, M.A.; Douglas, M.J.; Wang, T.; et al. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plant 2019, 5, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.F.; Fraser, L.G.; Gill, G.P. Sex determination in Actinidia. Acta Hortic. 1997, 444, 85–88. [Google Scholar] [CrossRef]
- Gill, G.P.; Harvey, C.F.; Gardner, R.C.; Fraser, L.G. Development of sex-linked PCR markers for gender identification in Actinidia. Theor. Appl. Genet. 1998, 97, 439–445. [Google Scholar] [CrossRef]
- Shirkot, P.; Sharma, D.R.; Mohapatra, T. Molecular identification of sex in Actinidia deliciosa var. deliciosa by RAPD markers. Sci. Hortic. 2022, 94, 33–39. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, C.; Liu, Y.; VanBuren, R.; Yao, X.; Zhong, C.; Huang, H. High-density interspecific genetic maps of kiwifruit and the identification of sex-specific markers. DNA Res. 2015, 22, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Hale, I.; Melo, A.T.O.; Gustafson, H. Sex-linked molecular markers for two cold-hardy kiwifruit species, Actinidia arguta and A. kolomikta. Eur. J. Hortic. Sci. 2018, 83, 236–246. [Google Scholar] [CrossRef]
- Kim, J.; Oh, S.; Kim, Y.; Lee, M.; Kwack, Y.B.; Kim, D. Development of a CAPS marker for discriminating sex in Actinidia arguta. Hortic. Sci. Technol. 2022, 40, 233–241. [Google Scholar] [CrossRef]
- Ye, L.X.; Luo, M.M.; Wang, Z.; Bai, F.X.; Luo, X.; Gao, L.; Peng, L.; Chen, Q.H.; Zhang, L. Genome-wide analysis of MADS-box gene family in kiwifruit (Actinidia chinensis var. chinensis) and their potential role in floral sex differentiation. Front. Genet. 2022, 13, 1043178. [Google Scholar] [CrossRef]
- Oh, S.; Lee, M.; Kim, K.; Han, H.; Won, K.; Kwack, Y.B.; Shin, H.; Kim, D. Genetic diversity of kiwifruit (Actinidia spp.), including Korean native A. arguta, using single nucleotide polymorphisms derived from genotyping-by-sequencing. Hortic. Environ. Biotehcnol. 2019, 60, 105–114. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. Solexa QA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 257. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Harkess, A.; Huang, K.; ven der Hulst, R.; Tissen, B.; Caplan, J.L.; Kloppula, A.; Batish, M.; Meyers, B.C.; Leebens-Mack, J. Sex determination by two Y-linked genes in garden asparagus. Plant Cell 2020, 32, 1790–1796. [Google Scholar] [CrossRef]
- Cegan, R.; Marais, G.A.B.; Kubekova, H.; Blavet, N.; Widmer, A.; Vyskot, B.; Doležel, J.; Šafář, J.; Hobza, R. Structure and evolution of Apetala3, a sex-linked gene in Silene latifolia. BMC Plant Biol. 2010, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Guttman, D.S.; Charlesworth, D. An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature 1998, 393, 263–266. [Google Scholar] [CrossRef]
- Delichère, C.; Veuskens, J.; Hernould, M.; Barbacar, N.; Nouras, A.; Negrutiu, I.; Monéger, F. SlY1, the first active gene cloned from a plant Y chromosome, encodes a WD-repeat protein. EMBO J. 1999, 18, 4169–4179. [Google Scholar] [CrossRef]
- Atanassov, I.; Delichère, C.; Filatov, D.A.; Charlesworth, D.; Negrutiu, I.; Monéger, F. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol. Biol. Evol. 2001, 18, 2162–2168. [Google Scholar] [CrossRef]
- Moore, R.C.; Kozyreva, O.; Lebel-Hardenack, S.; Siroky, J.; Hobza, R.; Vyskot, B.; Grant, S.R. Genetic and functional analysis of DD44, a sex-linked gene from the dioecious plant Silene latifolia, provides clues to early events in sex chromosome evolution. Genetics 2003, 163, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Bergero, R.; Forrest, A.; Kaiser, V.B.; Charlesworth, D. Nucleotide diversity in Silene latifolia autosomal and sex-linked genes. Proc. R. Soc. B 2010, 227, 3283–3290. [Google Scholar] [CrossRef] [PubMed]
- Muir, G.; Bergero, R.; Charlesworth, D.; Filatov, D.A. Does local adaptation cause high population differentiation of Silene latifolia Y chromosomes? Evolution 2011, 62, 3368–3380. [Google Scholar] [CrossRef]
- Weingartner, L.A.; Delph, L.F. Neo-sex chromosome inheritance across species in Silene hybrids. J. Evol. Biol. 2014, 27, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Chanderbali, A.S.; Yoo, M.J.; Zahn, L.M.; Brockington, S.F.; Wall, P.K.; Gitzendanner, M.A.; Albert, V.A.; Leebens-Mack, J.; Altman, N.S.; Ma, H.; et al. Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Biol. Sci. 2010, 107, 22570–22575. [Google Scholar] [CrossRef] [PubMed]
- Men, Y.; Li, J.R.; Shen, H.L.; Yang, Y.M.; Fan, S.T.; Li, K.; Guo, Y.S.; Lin, H.; Liu, Z.D.; Guo, X.W. VaAPRT3 gene is associated with sex determination in Vitis amurensis. Front. Genet. 2022, 12, 727260. [Google Scholar] [CrossRef] [PubMed]
- Khanum, P.; Khan, A.A.; Khan, I.A.; Chaffar, A.; Khan, Z. TPD1-like gene as a suitable marker for early sex determination in date palm (Phoenix dactylifera L.). Genes 2023, 14, 907. [Google Scholar] [CrossRef]
- Kong, X.; Wang, F.; Geng, S.; Guan, J.; Tao, S.; Jia, M.; Sun, G.; Wang, Z.; Wang, K.; Ye, X.; et al. The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 2021, 20, 75–88. [Google Scholar] [CrossRef]
- Shen, G.; Yang, C.H.; Shen, C.Y.; Huang, K.S. Origination and selection of ABCDE and AGL6 subfamily MADS-box genes in gymnosperms and angiosperms. Biol. Res. 2019, 52, 25. [Google Scholar] [CrossRef]
- Varkony-Gasic, E.; Moss, M.S.; Voogd, G.; Wu, R.; Lough, R.H.; Wnag, Y.Y.; Hellens, R.P. Identification and characterization of flowering genes in kiwifruit: Sequence conservation and role in kiwifruit flower development. BMC Plant Biol. 2011, 11, 72. [Google Scholar] [CrossRef]
- De Mori, G.; Testolin, R.; Cipriani, G. A molecular protocol for early sex discrimination (ESD) in Actinidia spp. J. Berry Res. 2022, 12, 249–266. [Google Scholar] [CrossRef]
| Sex | Species | Accession | |||
|---|---|---|---|---|---|
| Female | Actinidia arguta | K5-1-8 | K5-1-12 | K5-2-3 | K5-2-13 |
| K5-2-18 | K5-4-7 | K5-7-3 | K5-7-5 | ||
| K5-10-1 | K5-10-5 | K5-12-6 | Chiak | ||
| Hardy Red | Ilse | Autumn Sense | |||
| (A. arguta × A. deliciosa) × A. arguta | Skinny Green | ||||
| Male | A. arguta | K5-2-22 | K5-3-3 | K5-3-6 | K5-3-7 |
| K5-3-8 | K5-3-12 | K5-3-13 | K5-3-18 | ||
| K5-4-8 | K5-5-1 | K5-5-12 | K5-5-14 | ||
| K5-6-10 | K5-9-1 | K5-9-8 | K5-11-1 | ||
| K5-11-2 | K5-11-3 | K5-11-8 | K5-13-6 | ||
| K5-14-1 | |||||
| Sex-Related Gene | Gene ID 1 | Description | Identity (%) | E-Value |
|---|---|---|---|---|
| GbMADS1 | Acc28871 | Agamous-like MADS-box protein | 68.33 | 1.08 × 10−41 |
| GbMADS2 | ||||
| GbMADS4 | ||||
| GbMADS18 | ||||
| CYP703 | Acc28333 | Cytochrome P450 CYP736A12 like | 58.84 | 1.85 × 10−75 |
| TDF1 | Acc28900 | Hypothetical protein | 78.63 | 7.05 × 10−57 |
| Acc28821 | Transcription factor like | 73.85 | 1.77 × 10−49 | |
| Acc28639 | Transcription factor like | 68.64 | 2.73 × 10−39 | |
| Acc28638 | Transcription factor like | 68.64 | 2.73 × 10−39 | |
| Acc28429 | Myb-related protein | 73.26 | 1.98 × 10−30 | |
| GDSL esterase/lipase | Acc28776 | GDSL esterase/lipase, partial | 74.78 | 9.52 × 10−145 |
| AP3Y | Acc28719 | Agamous-like MADS-box protein | 69.49 | 5.06 × 10−12 |
| Acc28717 | Agamous-like MADS-box protein | 62.22 | 2.03 × 10−22 |
| Variation | Position (bp) | InDel (Reference/Alternative) | Gene ID 1 | Region | Description | |
|---|---|---|---|---|---|---|
| Female | Male | |||||
| Insertion | 12,919,896 | -/AT | -/- | Acc28719 | Exon | Agamous-like MADS-box protein |
| 14,557,224 | -/AG | -/- | Acc28871 | Intron | Agamous-like MADS-box protein | |
| 14,559,075 | -/- | -/T | Acc28871 | Intron | Agamous-like MADS-box protein | |
| 14,570,161 | -/- | -/ATTCGGTGAATCTGACACTT | Acc28871 | Intron | Agamous-like MADS-box protein | |
| Deletion | 12,896,671 | TGAT/- | TGAT/TGAT | Acc28717 | Exon | Agamous-like MADS-box protein |
| 14,814,902 | A/A | A/- | Acc28900 | Intron | Hypothetical protein | |
| 14,815,837 | -/- | -/T | Acc28900 | Intron | Hypothetical protein | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, S.; Kim, J.; Kim, Y.; Lee, M.; Kim, D. A MADS-Box Gene-Based InDel Marker Discriminating Sex in Actinidia arguta. Horticulturae 2023, 9, 1310. https://doi.org/10.3390/horticulturae9121310
Oh S, Kim J, Kim Y, Lee M, Kim D. A MADS-Box Gene-Based InDel Marker Discriminating Sex in Actinidia arguta. Horticulturae. 2023; 9(12):1310. https://doi.org/10.3390/horticulturae9121310
Chicago/Turabian StyleOh, Sewon, Jung Kim, Yumi Kim, Mockhee Lee, and Daeil Kim. 2023. "A MADS-Box Gene-Based InDel Marker Discriminating Sex in Actinidia arguta" Horticulturae 9, no. 12: 1310. https://doi.org/10.3390/horticulturae9121310





