Abstract
The double-petal varieties of ornamental pomegranate have higher ornamental value and garden development potential than the single-petal varieties but there has been no study on the genomic variation between them. This study aimed to determine the genomic variation between the two kinds of varieties and the relationship between the variation and phenotype by identifying the DNA variation of three single-petal varieties and three double-petal varieties using re-sequencing technology. The results showed that the variation number of each variety was in the order of single nucleotide polymorphisms (SNPs) > insertions and deletions (InDels) > structural variations (SVs) > copy number variations (CNVs). The number of SNPs and InDels in the double-petal varieties was significantly higher than that in the single-petal varieties, and there was no significant difference in the number of SVs and CNVs. The number of non-synonymous SNPs in the coding region (Nonsyn_CDS_SNPs) and InDels with a 3X length in the coding region (3X_shiftMutation_CDS_InDel) was significantly higher in the double-petal varieties than that in the single-petal varieties. The number of the two variants was strongly positively correlated with each morphological index that was related to the phenotypic difference between the two varieties. Nonsyn_CDS_SNPs and 3X_shiftMutation_CDS_InDel were enriched in the cell membrane system, cell periphery, and signal transduction, from which 15 candidate genes were screened. Our results provide genomic data for the study of the formation mechanism of the double-petal flower and lay a theoretical foundation for new variety breeding of ornamental pomegranate.
1. Introduction
Pomegranate has rich germplasm resources. At present, the research on the phenotypic and genetic diversity of pomegranate varieties mainly focuses on the shape, quality, and related molecular markers of fruit [1,2,3], while the research on the main ornamental character—the flower—has received less attention. The single-petal varieties of ornamental pomegranate (Punica granatum L.) have a narrow flower shape and single-petal layer, while the double-petal varieties have a full flower shape, more petaloid stamens, and numerous petals [4]. Thus, the double-petal varieties have higher ornamental value and garden development potential. The phenotypic differences between the two kinds of varieties are mainly manifested in the petalization of the stamens and the growth of the petal transitional form. Currently, research on the mechanism of double-petal flower formation mainly focuses on the regulation of transcription factors, while there is little research on DNA variation [5,6,7]. For example, scholars have established the ABCDE flower development model to explain how transcription factors regulate flower organ morphogenesis [8,9,10,11]. In recent years, second-generation sequencing technology, which is fast, efficient, and low cost, has realized the whole genome sequencing of many plants, and, thus, provided technical support for genomic research on flower types [12,13,14,15,16,17]. For instance, Xing [18] screened out flowering-related genetic variation by comparing the genomes of two apple varieties using re-sequencing technology. Then, Huang [13] screened out genes that are related to lip petal development by comparing the genomes of two Phalaenopsis aphrodite varieties and the transcriptomes of 21 tissues, and Wu [14] screened out single nucleotide polymorphism (SNP) variations that are related to sterile flowers and continuous flowering in Hydrangea macrophylla using a genome-wide association study of 82 bigleaf hydrangea cultivars. Currently, the whole genome sequences of three pomegranate varieties, ‘Taishanhong’ [19], ‘Dabenzi’ [20], and ‘Tunisia’ [21], have been published. However, pomegranate genome-based research has mainly focused on the fruit character [21], stress resistance [22], and the chloroplast genome [23], and a comparative study on the genomic variation between the single- and double-petal varieties of ornamental pomegranate has yet to be carried out.
Using re-sequencing technology, this study determined the genomic variation between single and double-petal varieties and the relationship between the variation and phenotype, thus contributing to the molecular mechanism of pomegranate petalization and further providing a reference for new breeding varieties of pomegranate.
2. Materials and Methods
2.1. Plant Material
Six ornamental pomegranate varieties with similar plant types and ecological habits were collected from the Chinese pomegranate germplasm resource nursery (Yicheng; 34°49′49.195″ N, 117°21′18.701″ E) for genome re-sequencing. The six ornamental pomegranate varieties included three single-petal varieties (‘Taiansanbaitian’, ‘Yichengdanbanfenhongtian’, and ‘Zipitian’) and three double-petal varieties (‘Luoyangbaimasi’, ‘Yichengfenhongmudan’, and ‘Taianhongmudan’; Table 1). Among them, ‘Taiansanbaitian’ and ‘Luoyangbaimasi’ are white flower varieties, ‘Yichengdanbanfenhongtian’ and ‘Yichengfenhongmudan’ are pink flower varieties, and ‘Zipitian’ and ‘Taianhongmudan’ are red flower varieties.

Table 1.
Six ornamental pomegranate varieties.
2.2. Collection of the Morphological Parameters of Six Varieties
Three plants were randomly selected from each variety for the investigation of morphological parameters. The morphological parameters included: the flower length, flower width, flower width/flower length, calyx length, calyx width, calyx width/calyx length, sepal number, petal number, and petaloid stamen number. A ruler or vernier caliper was used to measure the length, and two significant figures were retained after the decimal point.
2.3. Sample Collection, Library Establishment, and Genome Re-Sequencing
A one-leaf sample from mixed shoots for each variety were collected and immediately frozen using liquid nitrogen and then stored in an ultra-low temperature refrigerator at −80 °C. The improved cetyltrimethylammonium bromide (CTAB) method was used to extract the total DNA of the samples [24]. Then, the DNA samples were randomly fragmented by Covaris and the fragments were collected by magnetic beads. Adenine was added to 3′ end of end-repaired DNA fragments before adaptor ligation. The ligation products were then cyclized and then amplified by linear isothermal Rolling-Circle Replication and DNA NanoBall technology. Then, agarose gel electrophoresis was used to screen the size of the fragments. A 200–300 bp small fragment library was established using polymerase chain reaction amplification. The qualified library was sequenced on the BGISEQ platform.
2.4. Data Filtering and Mapping
We used SOAPnuke (v1.4.0) to obtain clean data. The BWA [25] software was used to match the clean reads to the reference genome of ‘Dabenzi’. Then, Picard tools (v1.118; http://broadinstitute.github.io/picard/, accessed on 5 March 2020) were used to sort the SAM files according to the reference genome and convert them into BAM files. The BAM files were used to detect genomic variation after repairing the mate-pair information, adding the read group information, and labeling the repetitive reads.
2.5. Detection of the Single Nucleotide Polymorphisms, Insertions and Deletions, Structural Variation, and Copy Number Variation Polymorphisms
The GATK [26] software was used to detect the SNPs and insertions and deletions (InDels). The SNP filtering parameters were: “QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0”. The InDel filtering parameters were: “QD < 2.0 || FS > 200.0 || ReadPosRankSum < −20.0”. Additionally, Breakdancer [27] was used to detect the structural variations (SVs) using the parameters “–m 100 –x 1,000,000 –s 30 –d 5”. Moreover, SOAPcnv [28] was used to detect the copy number variations (CNVs) using the parameters “–u 2 –z”.
2.6. Data Processing and Bioinformatics Analysis
Microsoft Excel 2020 was used for the basic statistics and mapping of the morphological parameters and the number of SNPs, InDels, SVs, and CNVs. The SPSS 24.0 software (IBM, Armonk, New York, NY, USA) was used to conduct an analysis of variance (ANOVA) by Duncan’s multiple-range test and Pearson correlation analyses. The differences between the means were considered statistically significant at both p < 0.05 and p < 0.01. Non-synonymous SNPs in the coding (CDS) region and InDels causing a frameshift mutation with a length of 3X in the CDS region between ‘Taiansanbaitian’ and ‘Luoyangbaimasi’, ‘Yichengdanbanfenhongtian’ and ‘Yichengfenhongmudan’, and ‘Zipidian’ and ‘Taianhongmudan’ were compared. The common genes in the three comparison groups with important genetic variation between the single- and double-petal varieties were identified, and ggVennDiagram in the R software (http://cran.r-project.org/web/packages/cluster/, accessed on 10 February 2021) was used to make a Venn diagram. Online software (https://www.omicshare.com/tools, accessed on 4 December 2022) was used to carry out Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) classification and enrichment analysis.
3. Results
3.1. Flower Morphological Parameters for the Two Types of Varieties
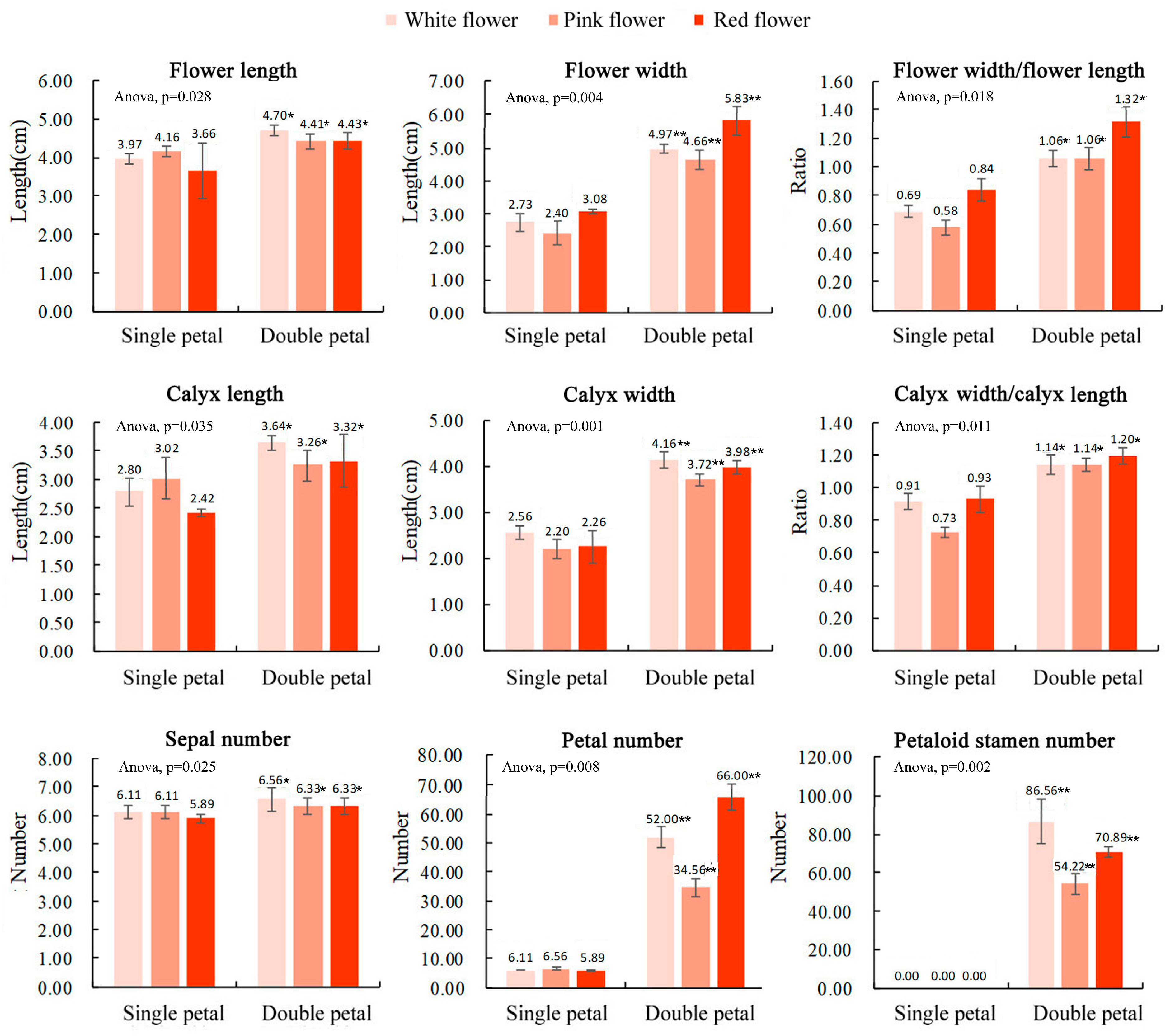
The results of the flower morphological parameters for the single and double-petal varieties of ornamental pomegranate are shown in Figure 1. The results of the variance analysis showed that the variety had significant effects on the flower length, flower width, flower width/flower length, calyx length, calyx width, calyx width/calyx length, sepal number, petal number, and petaloid stamen number. The flower length, flower width, flower width/flower length, calyx length, calyx width, calyx width/calyx length, sepal number, petal number, and petaloid stamen number of the double-petal varieties were significantly higher than those of the single-petal varieties. Flower color had no significant effect on the flower morphological parameters.
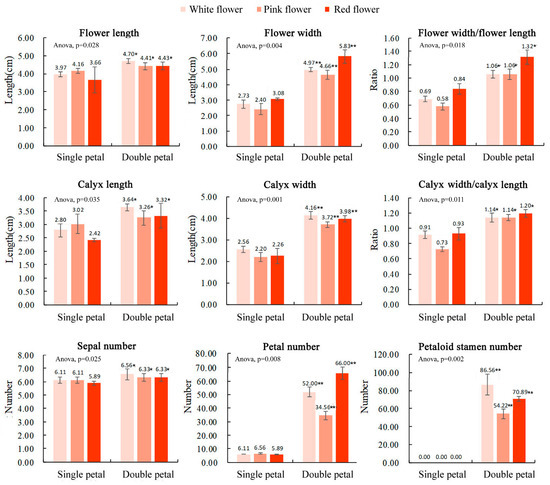
Figure 1.
Morphological flower parameters of 6 ornamental pomegranate varieties. One asterisk and two asterisks indicated significant differences at p < 0.05 and p < 0.01, respectively, between the two types of flowers (single- and double-petals) according to the Duncan test. The standard deviation was also indicated.
3.2. Quality Evaluation and Mapping of the Sequencing Data
The re-sequencing data of the pomegranate varieties were filtered and quality control was conducted, and the results are shown in Table 2. A total of 98.40 GB of original sequencing data were obtained from the six ornamental pomegranate varieties, and the average ratio of the clean data to the original sequencing data was 92.72%. In addition, the average rate of high-quality (Q30) bases was 91.27%, and the average GC content was 40.96%. Moreover, 95.69% of the clean reads were aligned to the reference genome, with a mean sequencing depth of 51.38-fold and a mean coverage of 95.24%.

Table 2.
Summary of the sequencing data of 6 ornamental pomegranate varieties.
3.3. Basic Analysis of the Four Variation Types
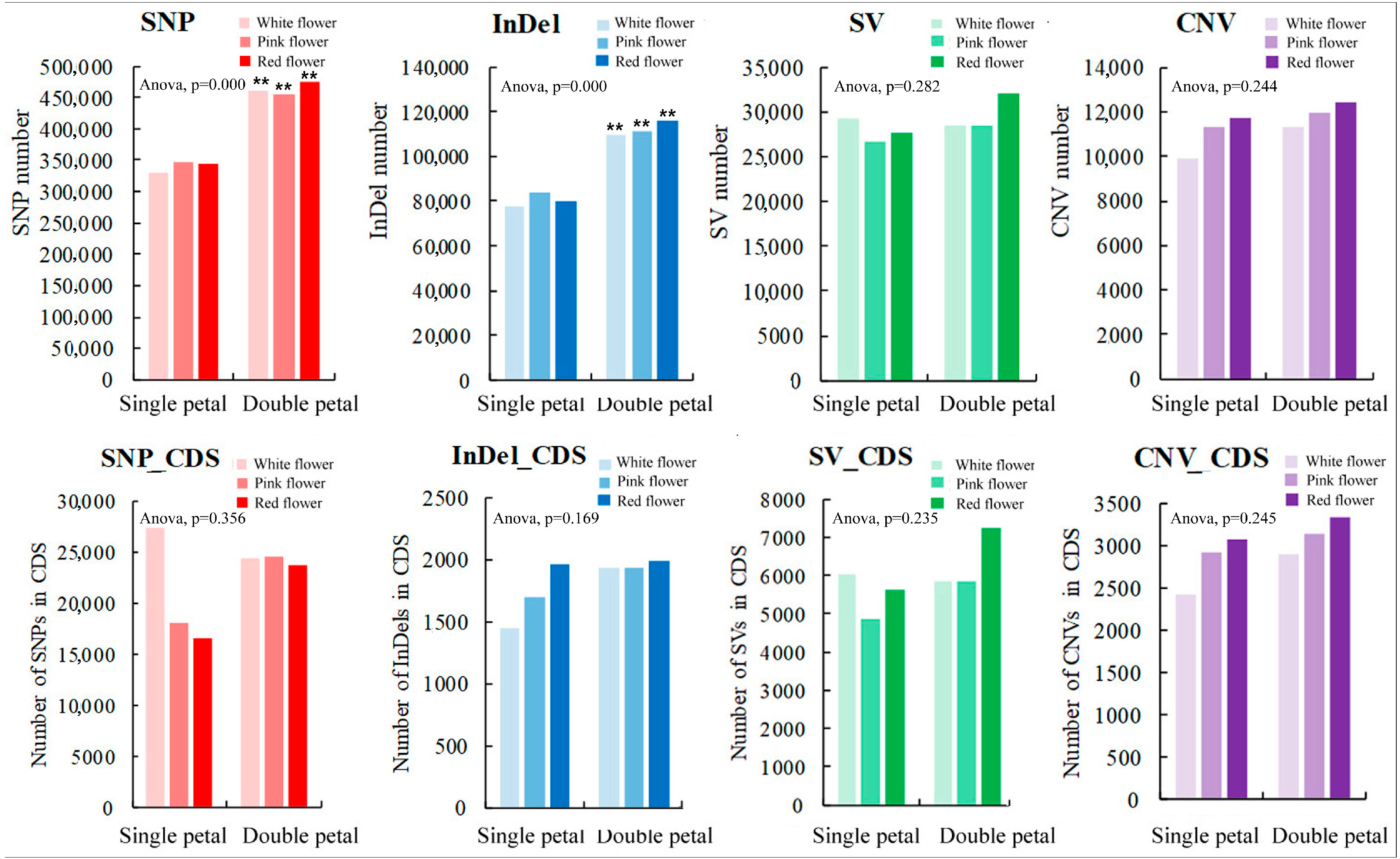
In this study, the SNPs, InDels, SVs, and CNVs of the ornamental pomegranate varieties were identified and counted. The results are shown in Figure 2. The average SNP number (463,840) in the double-petal varieties was significantly higher than that in the single-petal varieties (339,904). Furthermore, the average InDel number (111,995) in the double-petal varieties was significantly higher than that in the single-petal varieties (80,288). In addition, the average SV number in the single and double-petal varieties was 27,835 and 29,606, respectively, whereas the average CNV number in the single and double-petal varieties was 10,974 and 11,864, respectively. The variety had no significant effect on the SV and CNV numbers. The trend of the various types in the genome of the six ornamental pomegranate varieties was in the order of SNP > InDel > SV > CNV. Additionally, the variety had no significant effect on the number of SNPs, InDels, SVs, and CNVs. The trend of the various types in the coding sequences of the six ornamental pomegranate varieties was in the order of SNP_CDS > SV_CDS > CNV_CDS > InDel_CDS. On the other hand, there was no significant difference in the variable number of SNP, InDel, SV, CNV, SNP_CDS, InDel_CDS, SV_CDS, and CNV_CDS among varieties of different colors.
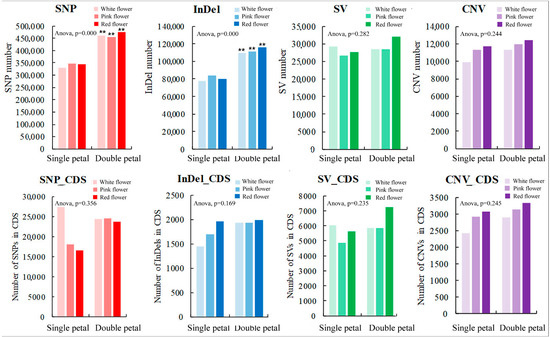
Figure 2.
Quantitative statistics of 4 variation types of six ornamental pomegranate varieties. Two asterisks indicated significant differences at p < 0.01, between the two types of flowers (single- and double-petals) according to the Duncan test.
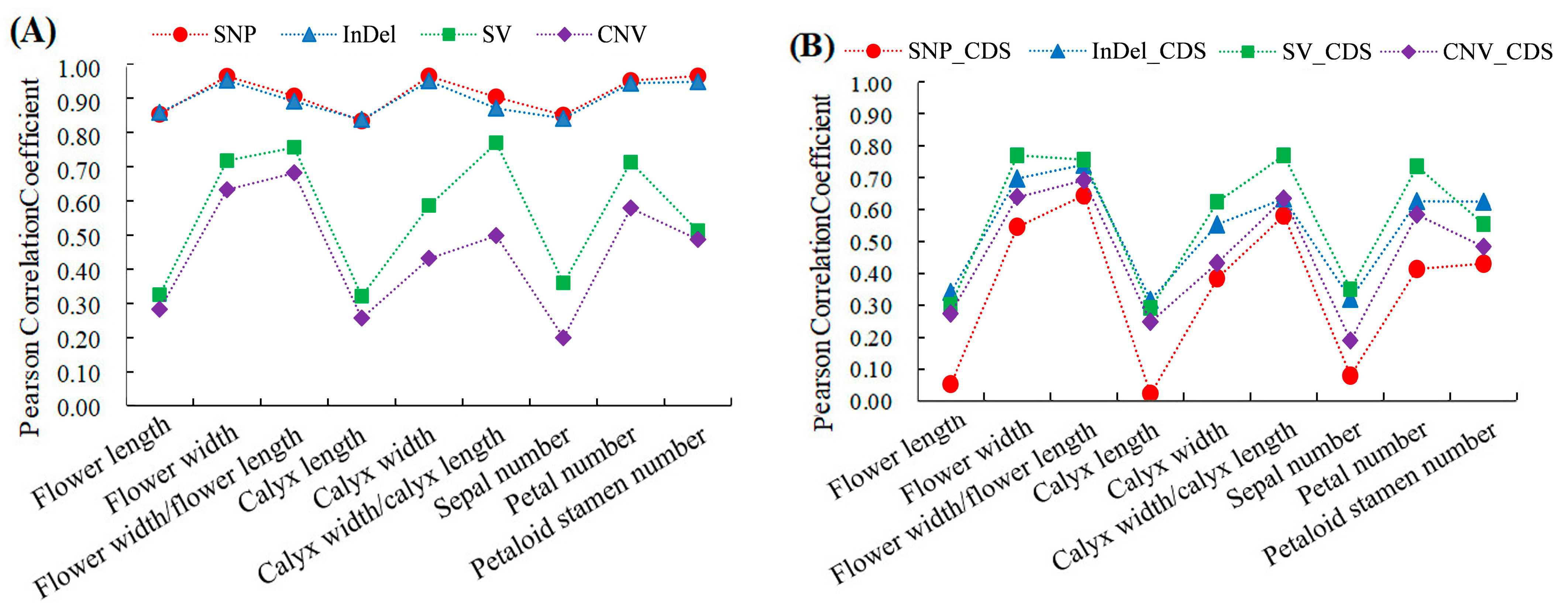
We conducted a Pearson correlation analysis between the number of different variation types identified in six varieties of genomes and the morphological parameters of 6 varieties in order to show the relationship between the variation and phenotype (Figure 3A). The SNP and InDel numbers were positively correlated with each morphological index, and the correlation coefficients ranged from 0.83 to 0.96 and 0.84 to 0.95, respectively. The correlation coefficients between the number of SNPs and InDels and the flower width, calyx width, petal number, and petaloid stamen number were higher than 0.90. The SV and CNV numbers were positively correlated with each morphological index, and the correlation coefficients ranged from 0.32 to 0.77 and 0.20 to 0.68, respectively. The trend of the correlation between the SNP number and each morphological index was similar to that between the InDel number and each morphological index, and the trend of the correlation between the SV number and each morphological index was similar to that between the CNV number and each morphological index.
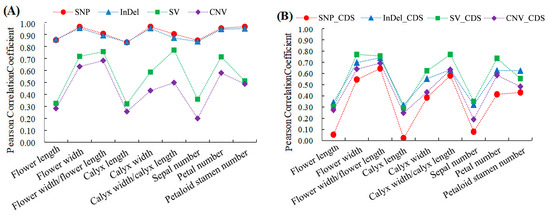
Figure 3.
Correlation analysis between four variations of number and phenotypic parameters of ornamental pomegranate. (A) Correlation between the number of four types of variations in genome region and phenotypic parameters; (B) Correlation between the number of four types of variations in CDS region and phenotypic parameters.
The relationship between the number of the four variation types ©n the CDS and the morphological parameters was also analyzed, and the results are shown in Figure 3B. The SNP_CDS, InDel_CDS, SV_CDS, and CNV_CDS numbers were all positively correlated with each morphological index, and the correlation coefficients between them and the flower width, flower width/flower length, calyx width, calyx width/calyx length, petal number, and petaloid stamen number were higher than those between them and the flower length, calyx length, and sepal number. The correlation coefficients between the number of the four variation types in the intergenic region and the morphological indicators were also calculated (Table S1). The correlation coefficients between the SNP number in the intergenic region and the morphological parameters and between the InDel number in the intergenic region and the morphological parameters were much higher than those in the CDS region, indicating that a large number of SNPs and InDels that are closely related to the morphological parameters occur in the intergenic region.
3.4. Annotation Analysis of the Single Nucleotide Polymorphisms
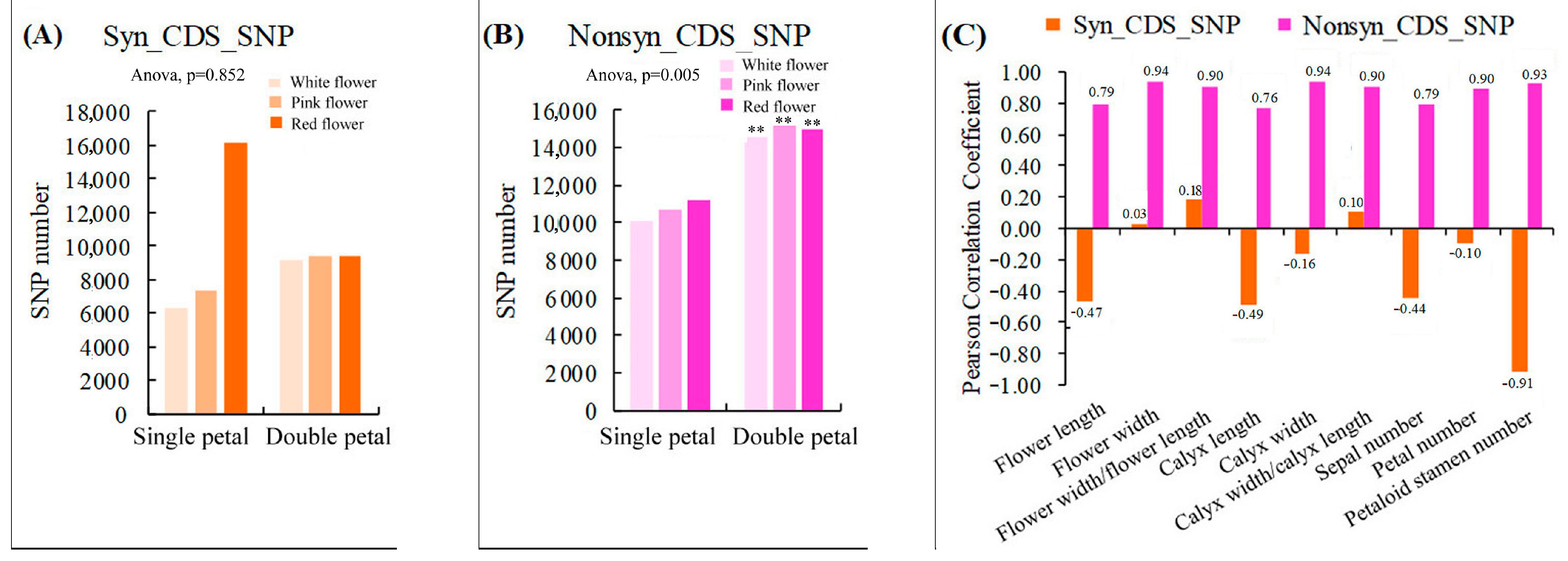
Our results showed that the SNP heterozygosity of the single and double-petal varieties of ornamental pomegranate ranged from 55.36% to 75.38% and 67.46% to 72.38%, respectively (Table 3). The single and double-petal varieties had no significant effect on the SNP heterozygosity. Furthermore, the ranges of the number of synonymous SNPs in the CDS region (Syn_CDS_SNP) of the single- and double-petal varieties were 6340–16,142 and 9140–9416 respectively, and the variety had no significant influence on the Syn_CDS_SNP number (Figure 4A). The ranges of the number of non-synonymous SNPs in the CDS region (Nonsyn_CDS_SNP) of the single and double petal varieties were 10,118–11,254 and 14,566–15,162, respectively. The single-petal varieties had a very significant impact on the Nonsyn_CDS_SNP number (p < 0.01; Figure 4B). Moreover, the correlation analysis showed that the Syn_CDS_SNP number was negatively correlated with most of the morphological parameters, while the Nonsyn_CDS_SNP number was strongly positively correlated with all the morphological parameters (Figure 4C), indicating that the Nonsyn_CDS_SNP were closely related to all the morphological parameters and had a great impact on the phenotypic traits. Additionally, the variety had extremely significant effects on the SNP variations that involved the loss of a start codon, acquisition of a start codon, loss of a stop codon, and acquisition of a stop codon and the SNP variations that were located at the splicing site, in the region within 5K upstream/downstream of the gene, and in the gene region (p < 0.01). There were also positive correlations between these SNPs and each of the morphological indicators (Tables S2 and S3).

Table 3.
SNPs statistics of six ornamental pomegranate varieties.
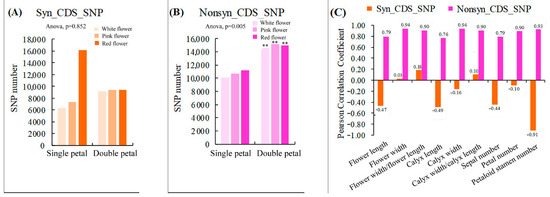
Figure 4.
Statistics of synonymous and nonsynonymous SNPs in coding regions and their correlation analysis with morphological parameters. (A) Statistics of Syn_CDS_SNP number in six varieties; (B) Statistics of Nonsyn_CDS_SNP number in six varieties; (C) Correlation analysis between the number of Syn_CDS_SNP/Nonsyn_CDS_SNP and morphological indicators. Two asterisks indicated significant differences at p < 0.01, between the two types of flowers (single- and double-petals) according to the Duncan test.
3.5. Annotation Analysis of the Insertions and Deletions
The annotation statistics of the InDels in the six varieties are shown in Table 4. The results of the analysis of variance showed that the variety had a very significant impact on the number of InDels in the whole genome (p < 0.01) but there was no significant impact on the number of InDels in the CDS region. Additionally, the variety had a very significant effect on the InDels that were located in the region within 5K upstream/downstream of the gene, gene region, exon region, intron region, and pseudogene region (p < 0.01; Table S4), and there were positive correlations between these InDels and the morphological parameters (Table S5).

Table 4.
Annotation statistics of InDels in six varieties.
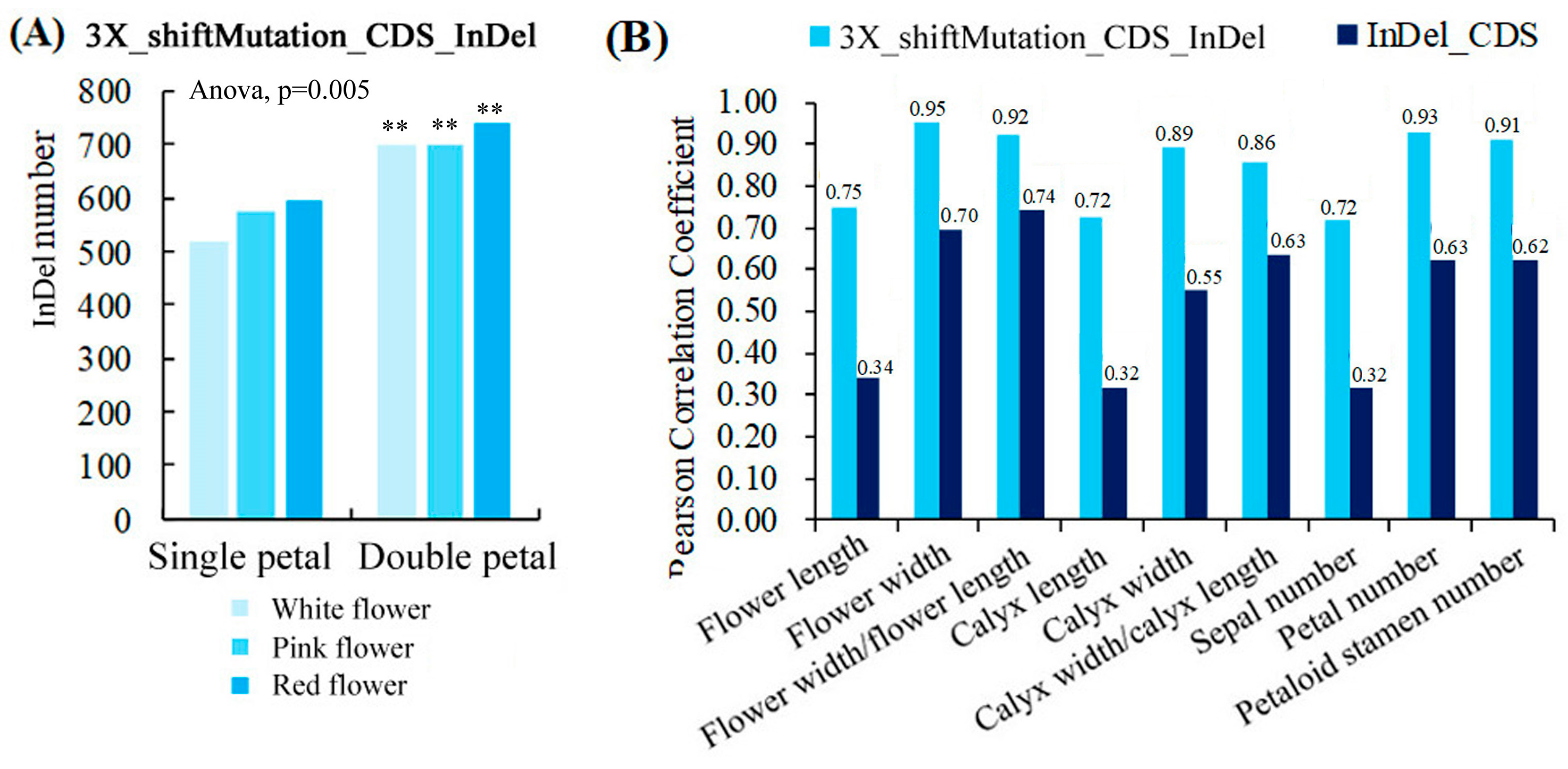
Figure 5A shows that the ranges of the number of InDels with a 3X length that caused a frameshift mutation in the CDS region (3X_shiftMutation_CDS_InDel) of the single and double-petal varieties were 517–593 and 697–738, respectively. The variety had a very significant impact on the number of 3X_shiftMutation_CDS_InDel (p < 0.01). In addition, the correlation analysis results showed that the number of 3X_shiftMutation_CDS_InDel was strongly positively correlated with the morphological parameters, and the correlation coefficients (0.72–0.95) between them were higher than those (0.32–0.74) between the InDels in the CDS region and the morphological parameters (Figure 5B), indicating that the 3X_shiftMutation_CDS_InDel had a great influence on the phenotypic characters of the ornamental pomegranates.
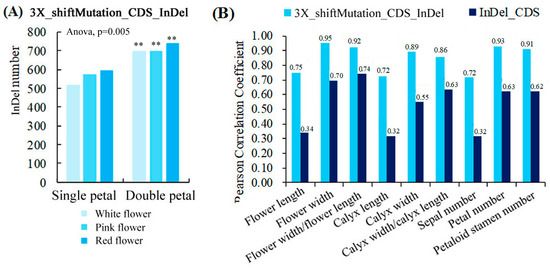
Figure 5.
Quantitative statistics of 3X_shiftMutation_CDS_InDels and its correlation with phenotypic parameters. (A) Quantitative statistics of 3X_shiftMutation_CDS_InDels; (B) Correlation analysis between two types of InDels and morphological parameters. Two asterisks indicated significant differences at p < 0.01, between the two types of flowers (single- and double-petals) according to the Duncan test.
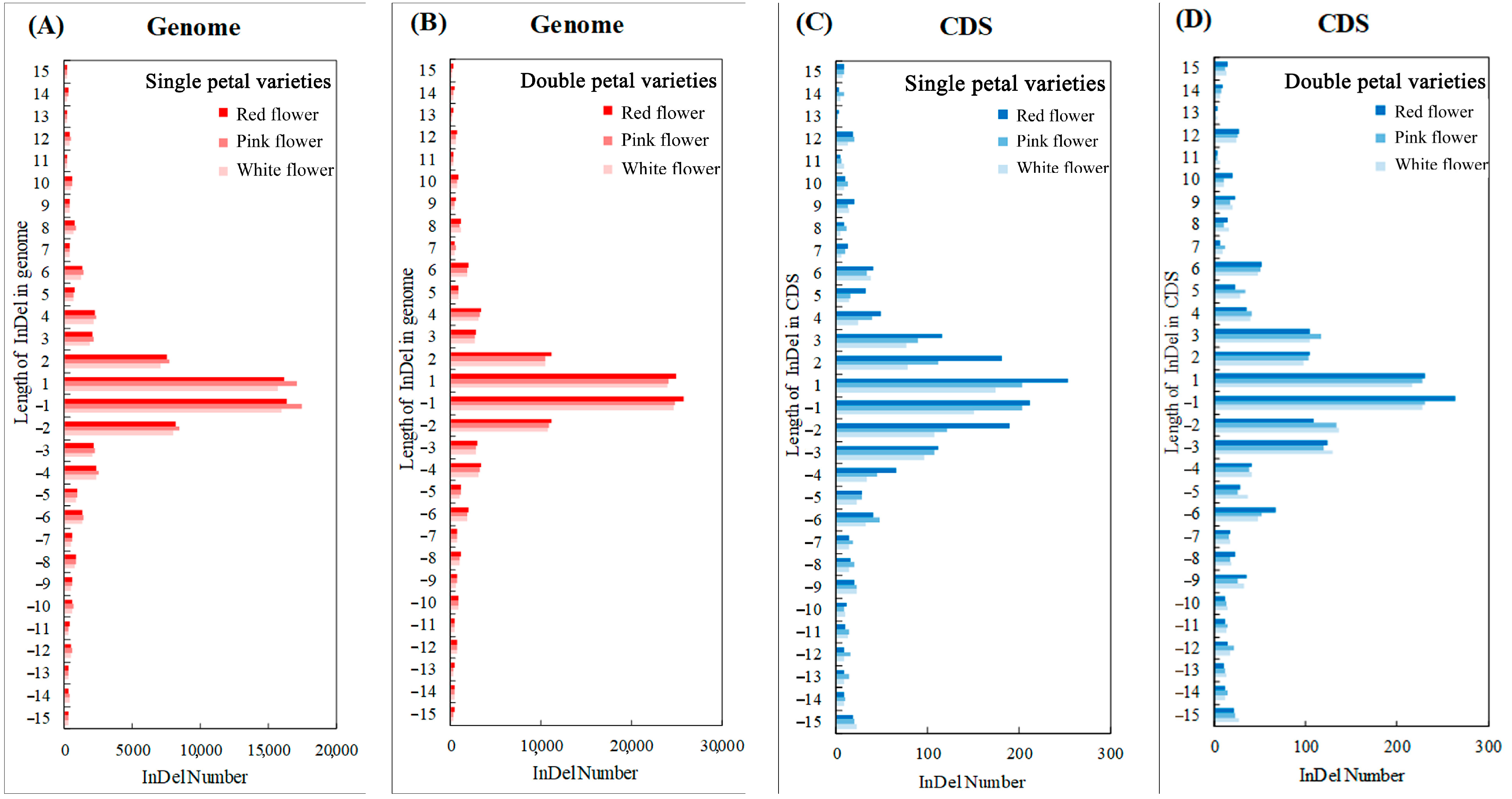
Figure 6 shows that the number of InDels of different lengths in the genome and CDS conforms to the normal distribution, with the largest number of one-base insertions or deletions. The analysis of variance showed that the variety had a significant impact on the number of InDels of different lengths in the genome (p < 0.05), while the variety had no significant impact in the CDS region. The number of InDels of each length in the single-petal varieties was significantly lower than that of the double-petal varieties. As shown in Figure 6A,B, within 15 bases that were inserted or deleted in the genome, the number of InDels of an even length was more than the number of adjacent InDels of an odd length (except for the one base InDels). As shown in Figure 6C,D, in the CDS region, the number of InDels with a length of a multiple of three was more than the number of adjacent InDels with a length that was not a multiple of three.
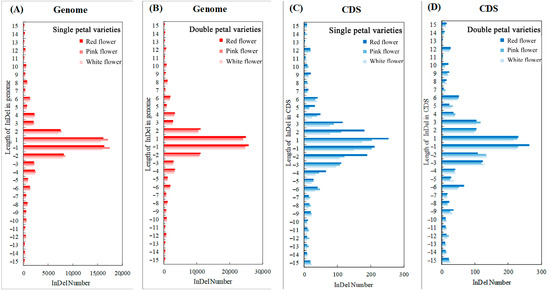
Figure 6.
Length distribution statistics of insertion or deletion. (A) Length distribution map of insertion or deletion in genome region of single-petal varieties; (B) Length distribution map of insertion or deletion in genome region of double-petal varieties; (C) Length distribution map of insertion or deletion in CDS region of single-petal varieties; (D) Length distribution map of insertion or deletion in CDS region of double-petal varieties.
3.6. Annotation Analysis of the Structural Variations
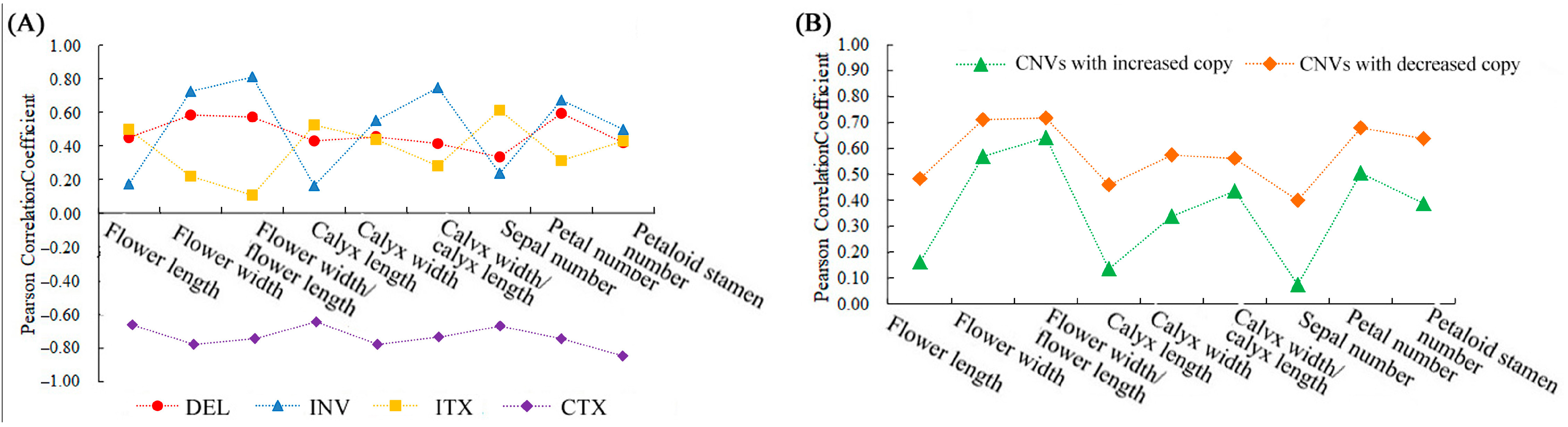
A total of 172,324 SVs were identified in the six ornamental pomegranate varieties (Table S6). The type with the largest number was inter-chromosomal translocation (CTX), followed by inversion (INV), deletion (DEL), and intra-chromosomal translocation (ITX), and no insertions were detected. The variety had no significant effect on the total SVs, DELs, INVs, and ITXs but it had a significant effect on the CTXs (p < 0.05). The CTX number in the single-petal varieties was significantly higher than that in the double-petal varieties. Moreover, the correlation analysis results showed that each morphological index was positively correlated with the number of DELs, INVs, and ITXs, while each morphological index was negatively correlated with the number of CTXs (Figure 7A).
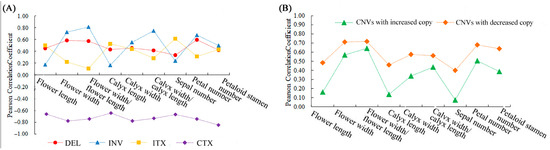
Figure 7.
The correlation coefficient between SV/CNV types and morphological parameters. (A) The correlation coefficient between SV types and morphological parameters; (B) the correlation coefficient between CNV types and morphological parameters.
3.7. Annotation Analysis of the Copy Number Variations
A total of 68,517 CNVs were identified in the six ornamental pomegranate varieties, of which the number of CNVs with an increased copy number was lower than that with a decreased copy number (Table S7). The analysis of variance showed that the variety had no significant effect on the number of total CNVs, CNVs with an increased copy number, and CNVs with a decreased copy number. Additionally, the correlation analysis results showed that the correlation coefficients between the morphological parameters and the number of CNVs with an increased copy number were lower than those between the morphological parameters and the number of CNVs with a decreased copy number (Figure 7B). In addition, the number of CNVs with a decreased copy number was positively correlated with the flower width, flower width/flower length, calyx width, calyx width/calyx length, petal number, and petaloid stamen number (R > 0.56).
3.8. Variation Analysis between the Single- and Double-Petal Varieties
According to the above results, the variety had a significant effect on the number of Nonsyn_CDS_SNP and 3X_shiftMutation_CDS_InDel, so we compared these two variations in three groups and screened out the genes that were important for determining the single- or double-petal variety of ornamental pomegranate. Among them, there were 548 variations (belonging to 228 genes) between the genomes of ‘Taiansanbaitian’ and ‘Luoyangbaimasi’, 792 variations (belonging to 345 genes) between the genomes of ‘Yichengdanbanfenhongtian’ and ‘Yichengfenhongmudan’, and 314 variations (belonging to 181 genes) between the genomes of ‘Zipidian’ and ‘Taianhongmudan’. In total, 37 common variants (belonging to 15 genes) were finally obtained from the three groups of genes. The GO enrichment analysis found (Figures S1–S9) that the enrichment of the variant genes among the three groups was similar. In terms of the cellular component GO terms, the three groups of mutant genes were enriched in the cell membrane system and cell periphery. Then, in terms of the biological process GO term, the three groups of mutant genes were enriched in signal transduction, and in terms of the molecular function GO terms, the three groups of mutant genes were enriched in transferase activity and GTP hydrolase activity. Finally, 15 mutant genes were screened from the three groups of comparison, and they were mainly involved in hormone pathways and stress responses, transcription and post-transcription regulation, translation, and post-translation regulation, and purine metabolism (Table 5).

Table 5.
Classification of variant genes between single- and double-petal pomegranate.
4. Discussion
The rapid development of bioinformatics and sequencing technology makes it possible to sequence the whole genome of many ornamental plants, which provides a starting point for revealing genetic variation at the genome level and exploring the relationship between genetic variation and phenotypic diversity [12,29,30]. When compared with the single-petal varieties, the double-petal varieties of ornamental pomegranates have higher ornamental value and wider garden application potential. Therefore, the genetic variation characteristics between the two varieties of ornamental pomegranates deserved further study. In this study, we found that the number of SNPs and InDels in the double-petal varieties was significantly higher than that in the single-petal varieties using whole genome re-sequencing technology, but there was no significant difference in the number of SNPs and InDels between varieties of different flower colors, indicating that the genetic variations among varieties of different flower types of ornamental pomegranates were more abundant than that among varieties of different flower colors. This also supported the previous researchers’ finding that the single-petal varieties and the double-petal varieties were clustered in different branches through quantitative classification and molecular markers. In cluster analysis and principal component analysis, pomegranates were first classified according to flower type, and flower color was the second classification standard [18]. We also found that the number of genomic SNPs and InDels had a strong positive correlation with the morphological parameters, and the correlation coefficient was higher than that of SNPs and InDels located in the coding regions. These results indicated that the genetic differences between the two kinds of varieties were closely related to the flower type phenotype in the whole genome regions, but the variations that were related to the flower type phenotype might be only a small part of the differences in the coding regions. Further research also showed that SNPs and InDels closely related to flower phenotypic traits were mainly located in the intergenic region. This result supported the previous findings that miRNAs targeting transcription factors and hormone-related regulatory factors involved in pomegranate fruit development were located in the intergenic region [31], indicating that the intergenic region has an important role in pomegranate development. In addition, we found that the variety had no significant effect on the number of CNVs in the genome or coding region, but the numbers of CNVs in the red flower varieties were the largest, followed by the pink flower varieties, and the white flower varieties were the least, indicating that CNVs affecting gene expressions by disturbing gene activities and changing gene dosages might not participate in the flower development, but in the anthocyanin accumulation process.
Single nucleotide polymorphism heterozygosity is related to the abundance of parental resources, and SNP heterozygosity in citrus, soybean, and other plants is significantly related to phenotypic characteristics [18,32,33]. However, this study found that the single- or double-petal variety had no significant effect on the SNP heterozygosity in ornamental pomegranate, indicating that ornamental pomegranate varieties may have undergone a complex natural selection and artificial breeding. Further analysis showed that the variety had no significant impact on the number of synonymous SNPs in the coding region but had a very significant impact on the number of non-synonymous SNPs in the coding region. Furthermore, it was found that Nonsyn_CDS_SNP were strongly positively correlated with the morphological parameters, indicating that Nonsyn_CDS_SNP was important to distinguish between single- and double petal-varieties.
According to genome variation studies of tomato, rice, apple, and other plants, it was found that the InDels number was generally less than the SNP number [24,34,35], which was consistent with our results. As with the SNPs, this study found that single- or double-petal variety had a very significant impact on the InDel number in the whole genome but had no significant impact on the InDels number in the CDS region. With further investigation, we found that the variety had a significant impact on the 3X_shiftMutation_CDS_InDel number, and the 3X_shiftMutation_CDS_InDel number was strongly positively correlated with the phenotypic parameters, indicating that 3X_shiftMutation_CDS_InDel were important for distinguishing between the single-petal and double-petal varieties. Interestingly, within 15 bases that were inserted or deleted in the genome, the number of InDels of an even length was more than the number of InDels that were adjacent to them of an odd length. However, the law between the length and number of InDels in apples, millet, and other plants is different from this study. They generally follow the law that the longer the length is, the less the number of InDels [36,37]. Therefore, it is speculated that the findings in this study may be specific to pomegranates and very different from species such as grapes [38]. Additionally, we also found that the number of InDels with a length of three or a multiple of three was more than the number of adjacent InDels with a length that was not a multiple of three. This is because DNA mutations with lengths of three or multiple of three will not cause a frameshift, thus, avoiding fatal damage.
This study screened 15 candidate genes that were related to petalization, most of which were reported to be involved in reproductive development. Auxin affects stamen development and petal growth and YUC1 encodes an important rate-limiting enzyme in the auxin synthesis pathway [39,40]. Yan [41] found that the overexpression of YUC1 led to the overproduction of auxin and the poor development of the stamens, while variation in YUC1 led to serious defects in the flower type development [42]. The cystathionine β-synthase domain proteins have the function of maintaining the balance of the redox reaction in the cells. In mutant plants, the scavenging capacity of active oxygen is reduced, the anthers are short and white, and there are no pollen grains [43]. Then, the phosphorus transporter gene, PHO1, can regulate grain filling and phosphorus distribution in crops [44,45] but there have been no studies on flower development. Additionally, the receptor-like protein kinase gene, RLK, regulates a series of biological processes, such as plant development, stress resistance, and hormone perception. It also plays an important role in the development of petunia pollen [46]. Moreover, the CCCH-type zinc finger protein is a transcription factor with a typical zinc finger structure. Liu [47] found that the C3H gene had the highest expression in the stamens of Chimonanthus praecox, and transgenic Arabidopsis thaliana had early flowering and abnormal stamen. Then, the pentatricopeptide repeat protein is an RNA-binding protein that participates in many post-transcriptional regulatory processes, such as splicing, editing, stabilization, and translation, and it plays a key role in cytoplasmic male sterility [48,49]. Furthermore, the ribosomal protein L23 is involved in the secretion and folding of new proteins. Moreover, the L23 gene is expressed in inflorescences and other tissues in Arabidopsis thaliana, and the reproductive organs in mutant plants are deformed [50]. Additionally, STPK catalyzes the phosphorylation of serine and threonine residues on proteins, which is negatively regulated by the flower development gene AGAMOUS in Arabidopsis thaliana [51] and determines the number of female flowers and spike length in maize [52]. Moreover, AGAMOUS, APETALA1, and APETALA2 resulted in more expression in brebas than in the main crop as reported in a recent investigation on Ficus carica [53]. Palmitoyltransferase catalyzes the palmitoylation modification of proteins [54], and pollen tube growth is defective in mutant plants [55]. The effect of these mutant genes on the petalization of ornamental pomegranate is worthy of further study.
5. Conclusions
In this paper, the genomic variation between single- and double-petal varieties was identified. The results showed that the number of SNPs and InDels caused by the mutation was larger than the number of SVs and CNVs caused by the recombination. The number of SNPs and InDels in the double-petal varieties was significantly higher than that in the single-petal varieties, and there was no significant difference in the number of SNPs and InDels between varieties of different flower colors, supporting the previous classification of pomegranate according to flower type. The variety had no significant effect on the SV and CNV numbers. In addition, the number of Nonsyn_CDS_SNP and 3X_shiftMutation_CDS_InDel was strongly positively correlated with the morphological parameters, showing that these two kinds of variants have an important influence on the phenotypic difference between the single- and double-petal varieties. Lastly, fifteen mutant genes were screened out from Nonsyn_CDS_SNPs and 3X_shiftMutation_CDS_InDels among the three groups of varieties and they were mainly involved in hormone pathways and stress responses, transcription and post-transcription regulation, translation, and post-translation regulation, and purine metabolism. This paper provides genomic mutation data between the single- and double-petal varieties and lays a theoretical foundation for double-flower molecular breeding in ornamental pomegranate. Further functional verification of mutation genes will provide insight and enable a deeper understanding of genetic involvement in the regulation of floral organ development.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9030361/s1, Table S1: Correlation coefficient between SNP, InDel, SV, CNV and morphological parameters; Table S2: SNP annotations of six ornamental pomegranate varieties; Table S3: Correlation coefficient between different SNP types and morphological parameters; Table S4: InDel annotations of six ornamental pomegranate varieties; Table S5: Correlation coefficient between different InDel types and morphological parameters; Table S6: Statistics of SV type in six ornamental pomegranate varieties; Table S7: Statistics of CNV type in six ornamental pomegranate varieties; Figure S1: Enrichment map of GO cell components of variant genes between ‘Taiansanbaitian’ and ‘Luoyangbaimasi’; Figure S2: Enrichment map of GO biological process of variant genes between ‘Taiansanbaitian’ and ‘Luoyangbaimasi’; Figure S3: Enrichment map of GO molecular function of variant genes between ‘Taiansanbaitian’ and ‘Luoyangbaimasi’; Figure S4: Enrichment map of GO cell components of variant genes between ‘Yichengdanbanfenhongtian’ and ‘Yichengfenhongmudan’; Figure S5: Enrichment map of GO biological process of variant genes between ‘Yichengdanbanfenhongtian’ and ‘Yichengfenhongmudan’; Figure S6: Enrichment map of GO molecular function of variant genes between ‘Yichengdanbanfenhongtian’ and ‘Yichengfenhongmudan’; Figure S7: Enrichment map of GO cell components of variant genes between ‘Zipitian’ and ‘Taianhongmudan’; Figure S8: Enrichment map of GO biological process of variant genes between ‘Zipitian’ and ‘Taianhongmudan’; Figure S9: Enrichment map of GO molecular function of variant genes between ‘Zipitian’ and ‘Taianhongmudan’.
Author Contributions
Conceptualization, Y.H., Z.Y. and Z.Z.; methodology, Y.H.; software, Y.H. and H.Y.; validation, Y.H.; formal analysis, Y.H.; investigation, Y.H.; resources, Z.Y.; data curation, Y.H.; writing—original draft preparation, Y.H.; writing—review and editing, Y.H., H.Y. and W.D.; visualization, Y.H. and H.Y.; supervision, Z.Y. and Z.Z.; project administration, Z.Y. and Z.Z.; funding acquisition, Z.Y. and Z.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32101582, 31770752), the Natural Science Foundation of Jiangsu Province of China (BK20210613), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (21KJB220008), the Initiative Project for Talents of Nanjing Forestry University (GXL2014070), the Priority Academic Program Development of Jiangsu High Education Institutions (PAPD), China Scholarship Council (CSC, 202008320482).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Patil, P.G.; Jamma, S.M.; Singh, N.V.; Bohra, A.; Parashuram, S.; Injal, A.S.; Gargade, V.A.; Chakranarayan, M.G.; Salutgi, U.D.; Dhinesh, B.K.; et al. Assessment of genetic diversity and population structure in pomegranate (Punica granatum L.) using hypervariable SSR markers. Physiol. Mol. Biol. Plants 2020, 26, 1249–1261. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, A.; Frabboni, L.; Mazzeo, A.; Ferrara, G.; Disciglio, G. Comparative evaluation of yield and fruit physico-chemical characteristics of five commercial cultivars of pomegranate grown in southeastern Italy in two consecutive years. Horticulturae 2022, 8, 497. [Google Scholar] [CrossRef]
- Giancaspro, A.; Mazzeo, A.; Giove, S.L.; Zito, D.; Marcotuli, A.; Gallotta, P.; Colasuonno, D.; Nigro, A.; Blanco, M.; Aradhya, A.; et al. Exploiting DNA-based molecular tools to assess genetic diversity in pomegranate (Punica granatum L.) selections and cultivars. Fruits 2017, 72, 292–305. [Google Scholar] [CrossRef]
- Ferrara, G.; Porfido, C.; Terzano, R.; Sarkhosh, A.; Mazzeo, A. A Study on the Characteristics of Buds and Flowers in Pomegranate: Differences among Cultivars. Horticulturae 2023, 9, 117. [Google Scholar] [CrossRef]
- Jing, D.; Chen, W.; Hu, R.; Zhang, Y.; Xia, Y.; Wang, S.; He, Q.; Guo, Q.; Liang, G. An integrative analysis of transcriptome, proteome and hormones reveals key differentially expressed genes and metabolic pathways involved in flower development in Loquat. Int. J. Mol. Sci. 2020, 21, 5107. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Chen, L.; Han, J.; Deng, X.; Tan, S.; Li, L.; Li, L.; Zhou, J.; Peng, H.; Yang, G.; He, G.; et al. Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon. Sci. Rep. 2016, 6, 21623. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Kim, S.; Koh, J.; Yoo, M.J.; Kong, H.; Hu, Y.; Ma, H.; Soltis, P.S.; Soltis, D.E. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 2005, 43, 724–744. [Google Scholar] [CrossRef]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Muioj, M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Saint-Oyant, L.H.; Ruttink, T.; Hamama, L. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 2018, 4, 473–484. [Google Scholar] [CrossRef]
- Huang, J.Z.; Lin, C.P.; Cheng, T.C.; Huang, Y.W.; Tsai, Y.J.; Cheng, S.Y.; Chen, Y.W.; Lee, C.P.; Chung, W.C.; Chang, B.C.; et al. The genome and transcriptome of Phalaenopsis yield insights into floral organ development and flowering regulation. PeerJ 2016, 4, e2017. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Alexander, L.W. Genome-wide association studies for inflorescence type and remontancy in Hydrangea macrophylla. Hortic. Res. 2020, 7, 27. [Google Scholar] [CrossRef]
- Mariette, S.; Wong Jun Tai, F.; Roch, G.; Barre, A.; Chague, A.; Decroocq, S.; Groppi, A.; Laizet, Y.; Lambert, P.; Tricon, D.; et al. Genome-wide association links candidate genes to resistance to Plum Pox Virus in apricot (Prunus armeniaca). New Phytol. 2016, 209, 773–784. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Jin, X.; Liu, L.; Dai, X.; Liu, Y.; Zhao, L.; Zheng, P.; Wang, X.; Liu, Y.; et al. Floral transcriptomes reveal gene networks in pineapple floral growth and fruit development. Commun. Biol. 2020, 3, 500. [Google Scholar] [CrossRef]
- Fan, L.; Chen, M.; Dong, B.; Wang, N.; Yu, Q.; Wang, X.; Xuan, L.; Wang, Y.; Zhang, S.; Shen, Y. Transcriptomic analysis of flower bud differentiation in Magnolia sinostellata. Genes 2018, 9, 212. [Google Scholar] [CrossRef]
- Wang, X.F. Studies on the Cultivar Classifieation of Punica granatum L. Ph.D. Thesis, Nanjing Forestry University, Nanjing, China, June 2007. [Google Scholar]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef]
- Qin, G.H.; Xu, C.Y.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D.; et al. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft- and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, J.; Munjal, V.; Sakthivel, K.; Thalor, S.K.; Mondal, K.K.; Chinchure, S.; Gharate, R. Polyphasic phenotypic and genetic analysis reveals clonal nature of Xanthomonas axonopodis pv. punicae causing pomegranate bacterial blight. Plant Pathol. 2020, 69, 347–359. [Google Scholar] [CrossRef]
- Yan, M.; Zhao, X.; Zhou, J.; Huo, Y.; Ding, Y.; Yuan, Z. The complete chloroplast genomes of Punica granatum and a comparison with other species in Lythraceae. Int. J. Mol. Sci. 2019, 20, 2886. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987, 19, 11–15. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Chen, K.; Wallis, J.W.; McLellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; McGrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef]
- Chalvin, C.; Drevensek, S.; Chollet, C.; Gilard, F.; Šolić, E.M.; Dron, M.; Bendahmane, A.; Boualem, A.; Cornille, A. Study of the genetic and phenotypic variation among wild and cultivated clary sages provides interesting avenues for breeding programs of a perfume, medicinal and aromatic plant. PLoS ONE 2021, 16, e0248954. [Google Scholar] [CrossRef]
- Anderson, N.O.; Kávová, T.; Daa, B.; Urn, V.; Květ, J. Phenotypic and genotypic variation in Czech forage, ornamental and wild populations of reed Canarygrass. Crop. Sci. 2016, 56, 2421–2435. [Google Scholar] [CrossRef]
- Saminathan, T.; Bodunrin, A.; Singh, N.V.; Devarajan, R.; Nimmakayala, P.; Jeff, M.; Aradhya, M.; Reddy, U.K. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 2016, 16, 122. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.J.; Li, Y.P.; Zhou, J.J.; Hu, C.G.; Zhang, J.Z. Genome-wide genetic variation and comparison of fruit-associated traits between kumquat (Citrus japonica) and Clementine mandarin (Citrus clementina). Plant Mol. Biol. 2018, 96, 493–507. [Google Scholar] [CrossRef]
- Guo, D.D.; Yuan, F.J.; Yu, X.M. Genome-wide variation analysis of grain and vegetable soybeans based on re-sequencing. Mol. Plant Breed. 2019, 17, 7306–7312. [Google Scholar] [CrossRef]
- Hideki, H.; Kenta, S.; Akio, O.; Hiroyuki, F.; Koh, A.; Christophe, R.; Shusei, S.; Sachiko, I.; Satoshi, T. Genome-Wide SNP genotyping to infer the effects on gene functions in tomato. DNA Res. 2013, 20, 221–233. [Google Scholar] [CrossRef]
- Parida, S.K.; Mukerji, M.M.; Singh, A.K.; Singh, N.K.; Mohapatra, T. SNPs in stress-responsive rice genes: Validation, genotyping, functional relevance and population structure. BMC Genom. 2012, 13, 26–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, W.; Xin, L.; Gao, Z.; Hou, Y.; Yu, X.; Zhang, Z.; Qu, S. Genomic variants of genes associated with three horticultural traits in apple revealed by genome re-sequencing. Hortic. Res. 2014, 1, 14045. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Cao, Y.; Quan, J.; Dong, L.; Li, Z.; Zhu, Y.; Zhu, L.; Dong, Z.; Li, D. Identifying the genome-wide sequence variations and developing new molecular narkers for genetics research by re-sequencing a landrace cultivar of Foxtail Millet. PLoS ONE 2013, 8, e73514. [Google Scholar] [CrossRef]
- Fanizza, G.; Colonna, G.; Resta, P.; Ferrara, G. The effect of the number of RAPD markers on the evaluation of genotypic distances in Vitis vinifera. Euphytica 1999, 107, 45–50. [Google Scholar] [CrossRef]
- Robert, H.S.; Crhak, K.L.; Mroue, S.; Benková, E. The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J. Exp. Bot. 2015, 66, 5029–5042. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Ding, L.; Song, A.; Shen, F.; Jiang, J.; Chen, S.; Chen, F. Transcriptomic and hormone analyses reveal mechanisms underlying petal elongation in Chrysanthemum morifolium ‘Jinba’. Plant Mol. Biol. 2017, 93, 593–606. [Google Scholar] [CrossRef]
- Yan, S.; Che, G.; Ding, L.; Chen, Z.; Liu, X.; Wang, H.; Zhao, W.; Ning, K.; Zhao, J.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Zafar, S.A.; Patil, S.B.; Uzair, M.; Fang, J.; Zhao, J.; Guo, T.; Yuan, S.; Uzair, M.; Luo, Q.; Shi, J.; et al. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 2020, 225, 356–375. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, L.; Gao, Q.; Wang, J.; Li, X.; Wang, H.; Liu, Y.; Lin, H.; Liu, J.; Wang, X.; et al. A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals. Nat. Genet. 2021, 53, 906–915. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, J.; Satheesh, V.; Meng, F.; Gao, W.; Dong, J.; Zheng, Z.; An, G.Y.; Nussaume, L.; Liu, D.; et al. SHORT-ROOT stabilizes PHOSPHATE1 to regulate phosphate allocation in Arabidopsis. Nat. Plants 2022, 8, 1074–1081. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Bauer, Z.; Boller, T. Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 2001, 13, 1155–1163. [Google Scholar] [CrossRef]
- Liu, H.; Huang, R.; Jing, M.; Sui, S.; Guo, Y.; Liu, D.; Li, Z.; Lin, Y.; Li, M. Two C3H type zinc finger protein genes, CpCZF1 and CpCZF2, from Chimonanthus praecox affect stamen development in Arabidopsis. Genes 2017, 8, 199. [Google Scholar] [CrossRef]
- Igarashi, K.; Kazama, T.; Toriyama, K. A gene encoding pentatricopeptide repeat protein partially restores fertility in RT98-Type cytoplasmic male-sterile rice. Plant Cell Physiol. 2016, 57, 2187. [Google Scholar] [CrossRef] [PubMed]
- Bentolila, S.; Bentolila, S.; Alfonso, A.A.; Hanson, M.R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 2002, 99, 10887–10892. [Google Scholar] [CrossRef] [PubMed]
- Mcintosh, K.B.; Bonham-Smith, P.C. The two ribosomal protein L23A genes are differentially transcribed in Arabidopsis thaliana. Genome 2005, 48, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Takahashi, N.; Shimura, Y.; Okada, K. A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 1997, 38, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, M.; Li, W.; Liu, L.; Jian, Y.; Yang, Z.; Shen, X.; Ning, Q.; Du, Y.; Zhao, R.; et al. A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat. Commun. 2020, 11, 988. [Google Scholar] [CrossRef] [PubMed]
- Marcotuli, I.; Mazzeo, A.; Colasuonno, P.; Terzano, R.; Nigro, D.; Porfido, C.; Tarantino, A.; Aiese Cigliano, R.; Sanseverino, W.; Gadaleta, A.; et al. Fruit development in Ficus carica L.: Morphological and genetic pproaches to fig buds for an evolution from monoecy toward dioecy. Front. Plant Sci. 2020, 11, 1208. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Hemsley, P.A.; Kemp, A.C.; Grierson, C.S. The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell. 2005, 17, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).