QTL Mapping of Leaf-Related Traits Using a High-Density Bin Map in Brassica rapa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Phenotype Evaluation
2.2. Isolation of Genomic DNA and Re-Sequencing
2.3. Genotyping and Bin-Map Construction
2.4. QTL Analysis of Morphological Traits
3. Results
3.1. Variation of Leaf Morphological Traits in the F2 Population
3.2. Construction of Bin Map Using Low-Coverage Sequencing
3.3. QTL Analysis
3.4. Co-Localization of QTLs and Candidate Gene Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiao, D.; Wang, H.; Basnet, R.K.; Zhao, J.; Lin, K.; Hou, X.; Bonnema, G. Genetic dissection of leaf development in Brassica rapa using a genetical genomics approach. Plant Physiol. 2014, 164, 1309–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.J.; Wang, X.; Deng, B.; Lou, P.; Wu, J.; Sun, R.; Xu, Z.; Vroman, J.; Koornneef, M.; Bonnema, G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005, 110, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yang, X.; Wang, H.; Qiu, N.; Chen, Y.; Wang, F.; Zhang, Y.; Li, H.; Li, J.; Gao, J. Construction of an Intragenic SSR-Based Linkage Map and QTL Mapping for Agronomic Traits in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 2022, 8, 165. [Google Scholar] [CrossRef]
- Kapoor, R.; Banga, S.S.; Banga, S.K. A microsatellite (SSR) based linkage map of Brassica rapa. New Biotechnol. 2009, 26, 239–243. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Kondo, M.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Hirai, M.; Matsumoto, S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics 2006, 173, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Sun, S.; Liu, B.; Wang, H.; Deng, J.; Liao, Y.; Wang, Q.; Cheng, F.; Wang, X.; Wu, J. A sequence-based genetic linkage map as a reference for Brassica rapa pseudochromosome assembly. BMC Genom. 2011, 12, 239. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, Y.; Xing, J.; Liu, Z.; Feng, H. Mapping quantitative trait loci for yield-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Euphytica 2013, 193, 221–234. [Google Scholar] [CrossRef]
- Choi, S.R.; Yu, X.; Dhandapani, V.; Li, X.; Wang, Z.; Lee, S.Y.; Heon Oh, S.; Pang, W.; Ramchiary, N.; Hong, C.; et al. Integrated analysis of leaf morphological and color traits in different populations of Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2017, 130, 1617–1634. [Google Scholar] [CrossRef]
- Kim, J.S.; Chung, T.Y.; King, G.J.; Jin, M.; Yang, T.J.; Jin, Y.M.; Kim, H.I.; Park, B.S. A sequence-tagged linkage map of Brassica rapa. Genetics 2006, 174, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Panigrahi, J.; Patnaik, A.; Kole, P.; Koleb, C. Addition of restriction fragment length polymorphism markers to the genetic linkage map of Brassica rapa L. (syn. campestris). Z. Naturforsch C J. Biosci. 2009, 64, 882–890. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.; Cao, J.; Yu, X.; Xiang, X.; Chen, H. Mapping QTLs for root morphological traits in Brassica rapa L. based on AFLP and RAPD markers. J. Appl. Genet. 2008, 49, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Soengas, P.; Hand, P.; Vicente, J.G.; Pole, J.M.; Pink, D.A. Identification of quantitative trait loci for resistance to Xanthomonas campestris pv. campestris in Brassica rapa. Theor. Appl. Genet. 2007, 114, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Kubo, N.; Saito, M.; Tsukazaki, H.; Kondo, T.; Matsumoto, S.; Hirai, M. Detection of quantitative trait loci controlling morphological traits in Brassica rapa L. Breed. Sci. 2010, 60, 164–171. [Google Scholar] [CrossRef] [Green Version]
- Bagheri, H.; El-Soda, M.; van Oorschot, I.; Hanhart, C.; Bonnema, G.; Jansen-van den Bosch, T.; Mank, R.; Keurentjes, J.J.; Meng, L.; Wu, J. Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Front. Plant Sci. 2012, 3, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Kitashiba, H.; Inaba, K.; Nishio, T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009, 16, 311–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Wang, H.; Zhong, W.; Bai, J.; Liu, P.; He, Y. QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 2013, 8, e76059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, X.; Su, T.; Xin, X.; Li, P.; Wang, W.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, J.; Sun, L. The Adaxial/Abaxial Patterning of Auxin and Auxin Gene in Leaf Veins Functions in Leafy Head Formation of Chinese Cabbage. Front. Plant Sci. 2022, 13, 918112. [Google Scholar] [CrossRef]
- Li, X.; Ramchiary, N.; Choi, S.R.; Van Nguyen, D.; Hossain, M.J.; Yang, H.K.; Lim, Y.P. Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 2010, 53, 939–947. [Google Scholar] [CrossRef] [Green Version]
- Li, G.H.; Chen, H.C.; Liu, J.L.; Luo, W.L.; Xie, D.S.; Luo, S.B.; Wu, T.Q.; Akram, W.; Zhong, Y.J. A high-density genetic map developed by specific-locus amplified fragment (SLAF) sequencing and identification of a locus controlling anthocyanin pigmentation in stalk of Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). BMC Genom. 2019, 20, 343. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Wang, R.; Zhang, Z.; Li, Q.; Wang, L.; Wang, Y.; Zhao, Z. High-resolution mapping of quantitative trait loci con-trolling main floral stalk length in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2019, 20, 437. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Wang, B.; Dong, X.; Liu, H.; Ren, L.; Chen, J.; Hauck, A.; Song, W.; Lai, J. An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F(2) maize population. BMC Genom. 2014, 15, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, G.; Zhai, G.; Feng, Q.; Yan, S.; Wang, A.; Zhao, Q.; Shao, J.; Zhang, Z.; Zou, J.; Han, B. Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J. Exp. Bot 2012, 63, 5451–5462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.; Jeong, H.J.; Yang, H.B.; Kang, S.M.; Kwon, J.K.; Kim, S.; Choi, D.; Kang, B.C. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 2016, 23, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.; Ogden, E.L.; Bostan, H.; Sargent, D.J.; Ward, J.; Gilbert, J.; Iorizzo, M.; Rowland, L.J. High-Density Linkage Map Construction and QTL Identification in a Diploid Blueberry Mapping Population. Front. Plant Sci. 2021, 12, 692628. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Sun, T.; Liu, X.; Li, C.; Li, M.; Wang, X.; Ren, H.; Zhao, Z.; Zhuang, F. Detection of Chromosomal Segments Introgressed from Wild Species of Carrot into Cultivars: Quantitative Trait Loci Mapping for Morphological Features in Backcross Inbred Lines. Plants 2022, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Ray, R.; Li, P.; Xu, J.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A.; Yang, X. Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genom. 2015, 290, 1457–1470. [Google Scholar] [CrossRef]
- Sun, X.; Luo, S.; Luo, L.; Wang, X.; Chen, X.; Lu, Y.; Shen, S.; Zhao, J.; Bonnema, G. Genetic Analysis of Chinese Cabbage Reveals Correlation Between Rosette Leaf and Leafy Head Variation. Front. Plant Sci. 2018, 9, 1455. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef]
- Rizzo, D.; Da Lio, D.; Bartolini, L.; Francia, C.; Aronadio, A.; Luchi, N.; Campigli, S.; Marchi, G.; Rossi, E. DNA Extraction Methods to Obtain High DNA Quality from Different Plant Tissues. Methods Mol. Biol. 2022, 2536, 91–101. [Google Scholar]
- Fu, L.; Cai, C.; Cui, Y.; Wu, J.; Liang, J.; Cheng, F.; Wang, X. Pooled mapping: An efficient method of calling variations for population samples with low-depth resequencing data. Mol. Breed. 2016, 36, 48. [Google Scholar] [CrossRef]
- Larose, D.T.; Larose, C.D. Discovering Knowledge in Data: An Introduction to Data Mining; Wiley-Interscience: New York, NY, USA, 2004. [Google Scholar]
- Ooijen, J. JoinMap 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Population; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Van Ooijen, J.W.; Boer, M.P.; Jansen, R.C.; Maliepaard, C.A. MapQTL 4.0: Software for the Calculation of QTL Positions on Genetic Maps (User Manual); Plant Research International: Wageningen, The Netherlands, 2000. [Google Scholar]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z. Mapping quantitative trait loci for leaf and heading-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinesis). Hortic. Environ. Biotechnol. 2011, 52, 494–501. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, B.H. Growth-regulating factor4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 2006, 49, 463–468. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [Green Version]
- Krogan, N.T.; Berleth, T. A dominant mutation reveals asymmetry in MP/ARF5 function along the adaxial-abaxial axis of shoot lateral organs. Plant Signal. Behav. 2012, 7, 940–943. [Google Scholar] [CrossRef] [Green Version]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Beltramino, M.; Ercoli, M.F.; Debernardi, J.M.; Goldy, C.; Rojas, A.M.L.; Nota, F.; Alvarez, M.E.; Vercruyssen, L.; Inze, D.; Palatnik, J.F. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 2018, 8, 13447. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Wu, F.; Yu, X.; Bai, J.; Zhong, W.; He, Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014, 164, 710–720. [Google Scholar] [CrossRef] [Green Version]
- He, Y.K.; Xue, W.X.; Sun, Y.D.; Yu, X.H.; Liu, P.L. Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 2000, 10, 151–160. [Google Scholar] [CrossRef]
- Guo, X.; Liang, J.; Lin, R.; Zhang, L.; Wu, J.; Wang, X. Series-Spatial Transcriptome Profiling of Leafy Head Reveals the Key Transition Leaves for Head Formation in Chinese Cabbage. Front. Plant Sci. 2021, 12, 787826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, J.; Cai, X.; Chen, H.; Wu, J.; Lin, R.; Cheng, F.; Wang, X. Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic. Res. 2021, 8, 68. [Google Scholar] [CrossRef]
- Liang, J.; Liu, B.; Wu, J.; Cheng, F.; Wang, X. Genetic Variation and Divergence of Genes Involved in Leaf Adaxial-Abaxial Polarity Establishment in Brassica rapa. Front. Plant Sci. 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinon, V.; Etchells, J.P.; Rossignol, P.; Collier, S.A.; Arroyo, J.M.; Martienssen, R.A.; Byrne, M.E. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 2008, 135, 1315–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Ling, Q.; Wang, H.; Huang, H. Ribosomal proteins promote leaf adaxial identity. Development 2008, 135, 1325–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Liang, J.; Lin, R.; Zhang, L.; Zhang, Z.; Wu, J.; Wang, X. Single-cell transcriptome reveals differentiation between adaxial and abaxial mesophyll cells in Brassica rapa. Plant Biotechnol. J. 2022, 20, 2233–2235. [Google Scholar] [CrossRef]
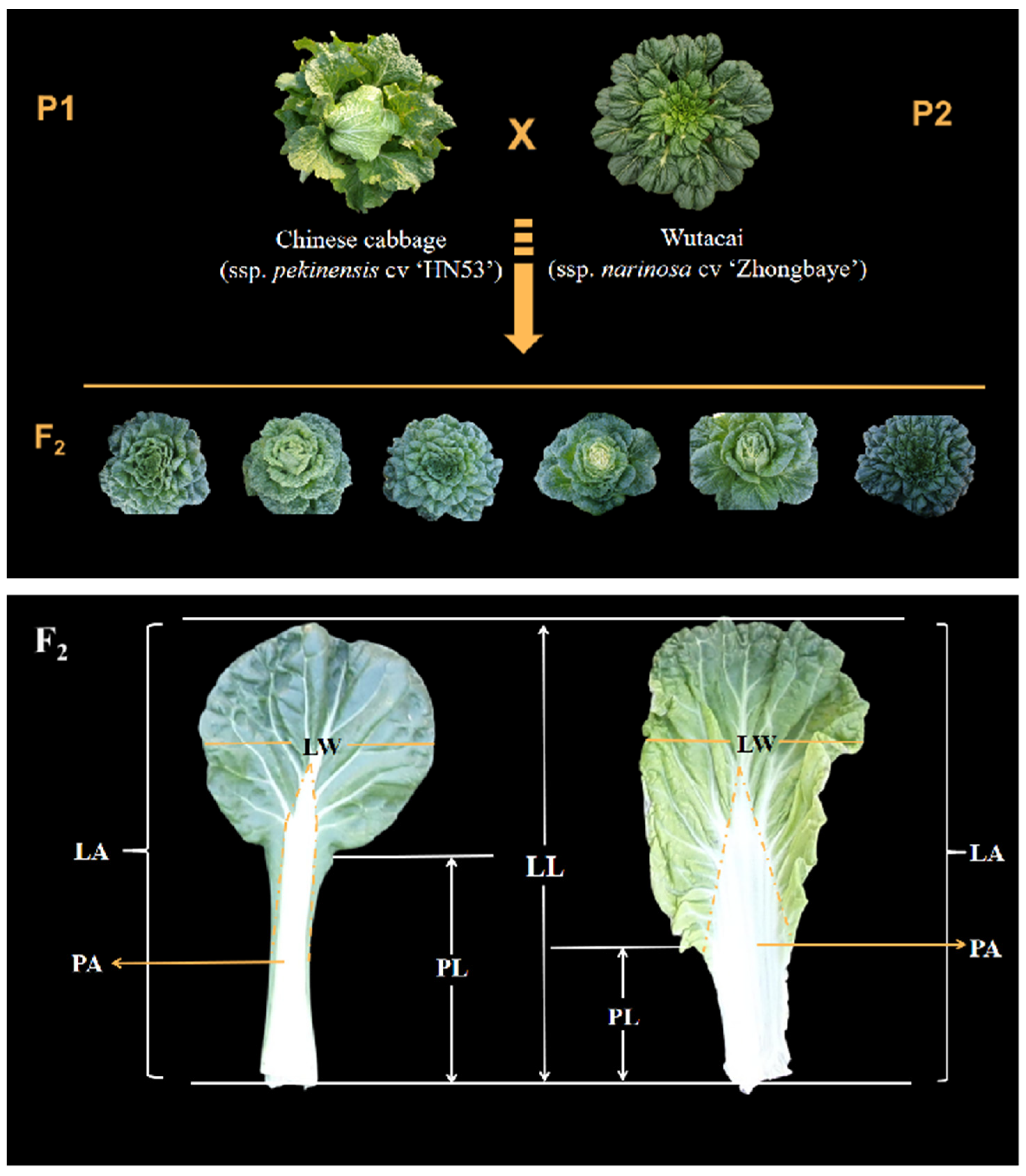




| Trait Name | Trait Description | Units | |
|---|---|---|---|
| LWT | Leaf weight | Total leaf weight | g |
| PWT | Petiole weight | Weight from the base of the petiole to the bottom of the lamina | g |
| PLTI | Ratio of total leaf weight to petiole weight | Measured by dividing LWT by PWT (LWT/PWT) | |
| LL | Leaf length | Length from the base of the petiole to the tip of the lamina | cm |
| LW | Leaf width | Width of leaves at the widest point | cm |
| LA | Leaf area | Total leaf surface area | cm2 |
| PA | Petiole area | Total petiole surface area | cm2 |
| PAI | Ratio of total leaf surface area to petiole surface area | Measured by dividing LA by PA (LA/PA) | |
| PL | Petiole length | Length from the base of the petiole to the bottom of the lamina | cm |
| PI | Ratio of total leaf surface area to petiole surface area | Measured by dividing LL by PL (LL/PL) | |
| LI | Index of the leaf | Measured by dividing LL by LW (LL/LW) |
| P1 | P2 | F2 Means | F2 Range | |
|---|---|---|---|---|
| LWT (g) | 110.89 ± 2.32 | 14.4 ± 0.44 | 45.62 ± 16.82 | 15.10–93.63 |
| PWT (g) | 51.12 ± 0.23 | 9.75 ± 0.58 | 28.85 ± 10.24 | 6.33–60.40 |
| PLTI | 2.17 ± 0.05 | 1.48 ± 0.05 | 1.58 ± 0.14 | 1.29–2.38 |
| LL (cm) | 44.09 ± 0.95 | 19.22 ± 0.53 | 33.79 ± 4.70 | 21.77–43.48 |
| LW (cm) | 25.04 ± 0.71 | 8.32 ± 0.08 | 17.01 ± 3.08 | 10.33–26.18 |
| LI | 1.76 ± 0.05 | 2.31 ± 0.06 | 2.02 ± 0.26 | 1.45–2.71 |
| PL (cm) | 25.15 ± 0.93 | 13.8 ± 0.20 | 20.92 ± 3.24 | 13.92–29.98 |
| PI | 1.75 ± 0.04 | 1.39 ± 0.02 | 1.63 ± 0.15 | 1.22–2.05 |
| LA (cm2) | 782.22 ± 8.92 | 75.01 ± 0.89 | 335.68 ± 107.95 | 125.56–627.33 |
| PA (cm2) | 120.36 ± 0.67 | 18.31 ± 0.47 | 57.30 ± 16.21 | 24.84–100.20 |
| PAI | 6.50 ± 0.16 | 4.1 ± 0.13 | 5.91 ± 1.26 | 3.40–9.07 |
| Total | Average/Plant | ||
|---|---|---|---|
| Raw datas | Reads | 295,559,333 | 1,970,396 |
| Bases (bp) | 88,667,799,900 | 591,118,666 | |
| After filtering datas | Reads | 268,392,401 | 1,789,283 |
| Bases (bp) | 80,374,268,540 | 535,828,456 | |
| Chromosome | Genotyped SNPs in 150 F2 Population | Genotyped SNP from Imputation | Chromosome Size (kb) | SNP Density (SNPs/kb) | Genetic Bins |
|---|---|---|---|---|---|
| A01 | 66,254 | 66,175 | 29,596 | 2.24 | 60 |
| A02 | 65,193 | 65,128 | 31,443 | 2.07 | 51 |
| A03 | 79,985 | 79,882 | 38,154 | 2.09 | 70 |
| A04 | 44,924 | 44,875 | 21,928 | 2.05 | 40 |
| A05 | 62,950 | 62,883 | 28,493 | 2.21 | 63 |
| A06 | 69,416 | 69,307 | 29,168 | 2.38 | 54 |
| A07 | 64,161 | 64,059 | 28,929 | 2.21 | 58 |
| A08 | 52,645 | 52,584 | 22,982 | 2.29 | 45 |
| A09 | 93,194 | 93,082 | 45,157 | 2.06 | 87 |
| A10 | 38,925 | 38,891 | 20,726 | 1.88 | 37 |
| Total | 637,647 | 636,866 | 296,576 | 2.15 | 565 |
| Trait | QTL Name | Chr | Peak Position | LOD | 2-LOD | Bin | Phenotypic Variation R2 (%) | Additive Effect | Dominance Effect |
|---|---|---|---|---|---|---|---|---|---|
| LWT | LWT1 | A02 | 13.7 | 5.79 | 13.0–16.7 | bin_65 | 7.5 | 6.17092 | 0.842102 |
| LWT2 | A03 | 21.8 | 4.42 | 20.5–22.5 | bin_121 | 5.6 | −5.44931 | 6.31162 | |
| LWT3 | A04 | 40.3 | 7.89 | 38.3–42.3 | bin_215 | 10.5 | 7.9973 | 0.337838 | |
| LWT4 | A06 | 98.6 | 12.73 | 98.3–100.3 | bin_330 | 18.4 | 10.5262 | 0.69486 | |
| LWT5 | A08 | 27.9 | 4.65 | 24.9–27.9 | bin_421 | 5.7 | 5.95979 | −0.752932 | |
| LWT6 | A10 | 19.6 | 6.89 | 18.9–20.9 | bin_542 | 9 | 4.59607 | 8.22428 | |
| PWT | PWT1 | A02 | 13.7 | 4.09 | 13.0–17.1 | bin_65 | 5.7 | 3.02863 | 1.6585 |
| PWT2 | A03 | 21.8 | 5.24 | 20.5–22.5 | bin_121 | 7.4 | −4.17852 | 3.39901 | |
| PWT3 | A04 | 39.6 | 5.87 | 36.3–40.3 | bin_214 | 8 | 4.17934 | 0.718021 | |
| PWT4 | A06 | 79.3 | 11.09 | 78.3–79.7 | bin_319 | 17.3 | 5.55048 | 2.59883 | |
| PWT5 | A08 | 26.9 | 5.72 | 24.9–27.9 | bin_421 | 8.2 | 4.26514 | −0.671059 | |
| PWT6 | A10 | 19.6 | 7.17 | 18.9–20.9 | bin_541 | 10.5 | 2.84782 | 5.51818 | |
| PLTI | PLTI1 | A01 | 27.4 | 6.71 | 24.4–27.8 | bin_10 | 12.6 | 0.0685958 | −0.0363513 |
| PLTI2 | A02 | 24.5 | 5.17 | 21.8–26.1 | bin_70 | 9.4 | 0.0403985 | −0.0705622 | |
| PLTI3 | A03 | 21.8 | 6.16 | 20.1–22.5 | bin_121 | 11.4 | 0.0807009 | −0.022673 | |
| PLTI4 | A05 | 71.8 | 5.42 | 70.7–73.4 | bin_270 | 9.9 | −0.0292825 | 0.0991348 | |
| PLTI5 | A05 | 92.7 | 3.56 | 88.3–93.0 | bin_278 | 6.3 | 0.06867 | −0.0152701 | |
| LL | LL1 | A01 | 125.4 | 5.12 | 121.7–126.1 | bin_55 | 4.2 | −1.42783 | 0.132046 |
| LL2 | A04 | 15.5 | 4.06 | 15.1–15.8 | bin_195 | 3.2 | 0.993059 | 0.743498 | |
| LL3 | A05 | 93 | 3.76 | 91.3–97.0 | bin_279 | 3 | 0.990827 | 0.880449 | |
| LL4 | A06 | 45.6 | 8.47 | 45.2–46.6 | bin_305 | 7.2 | 2.53086 | 0.725966 | |
| LL5 | A06 | 99.6 | 5.51 | 98.3–100.3 | bin_330 | 4.1 | 1.9582 | 0.237959 | |
| LL6 | A07 | 19.8 | 6.77 | 18.4–20.4 | bin_363 | 5.7 | 1.12922 | 1.60098 | |
| LL7 | A08 | 20.5 | 6.96 | 19.9–21.2 | bin_415 | 5.8 | 1.61557 | 0.314969 | |
| LL8 | A09 | 13.1 | 6.76 | 8.7–14.1 | bin_445 | 5.5 | 1.65979 | 0.187114 | |
| LL9 | A10 | 18.9 | 12.94 | 16.5–19.6 | bin_540 | 11.6 | 0.881175 | 3.23715 | |
| LW | LW1 | A01 | 37.9 | 6.68 | 36.2–39.9 | bin_19 | 9.4 | 0.908156 | −1.35427 |
| LW2 | A02 | 13 | 7.04 | 10.2–13.7 | bin_64 | 10 | 1.2967 | 0.44839 | |
| LW3 | A04 | 40.3 | 5.24 | 38.3–42.3 | bin_215 | 7.2 | 1.05336 | 0.712885 | |
| LW4 | A06 | 79.3 | 9.48 | 77.7–79.7 | bin_319 | 14 | 1.51683 | 0.740417 | |
| LW5 | A07 | 18.4 | 4.48 | 17.1–19.8 | bin_362 | 6.1 | 0.932614 | 0.686273 | |
| LW6 | A08 | 20.2 | 3.77 | 19.9–20.5 | bin_414 | 5.1 | 0.862757 | 0.600051 | |
| LI | LI1 | A01 | 8.1 | 4.83 | 6.1–8.8 | bin_4 | 7.7 | −0.0992905 | 0.0209628 |
| LI2 | A01 | 110.3 | 6.99 | 109.6–112.0 | bin_46 | 11.5 | −0.120275 | 0.0549563 | |
| LI3 | A02 | 13 | 7.06 | 11.0–17.1 | bin_64 | 11.7 | −0.125431 | −0.0224847 | |
| LI4 | A04 | 40.3 | 4.53 | 36.3–42.3 | bin_215 | 7.2 | −0.0993892 | −0.0127891 | |
| PL | PL1 | A01 | 8.1 | 5.89 | 2.0–8.9 | bin_4 | 10.5 | −1.23337 | 1.07212 |
| PL2 | A01 | 106.3 | 8.13 | 100.4–107.9 | bin_40 | 15.1 | −1.61924 | 0.734733 | |
| PL3 | A06 | 97.3 | 8.48 | 93.2–98.3 | bin_328 | 15.8 | 1.84381 | −0.0259659 | |
| PI | PI1 | A01 | 30.8 | 10.75 | 29.8–31.1 | bin_15 | 18.8 | 0.0961014 | −0.010089 |
| PI2 | A02 | 13 | 7.48 | 11.0–13.7 | bin_64 | 12.4 | 0.0729022 | 0.0110429 | |
| PI3 | A05 | 68.1 | 4.71 | 67.1–68.4 | bin_266 | 7.5 | −0.371918 | −0.457907 | |
| PI4 | A07 | 15.1 | 3.73 | 14.4–16.1 | bin_359 | 5.8 | 0.0496321 | 0.00700846 | |
| PI5 | A09 | 52 | 4.25 | 52.0–53.3 | bin_468 | 6.7 | 0.0541131 | 0.00126054 | |
| LA | LA1 | A02 | 13 | 6.84 | 13.0–16.7 | bin_64 | 7.7 | 39.6736 | 11.7891 |
| LA2 | A04 | 40.3 | 4.37 | 38.3–42.3 | bin_215 | 4.8 | 30.1337 | 9.47975 | |
| LA3 | A05 | 34.9 | 6.37 | 33.2–35.9 | bin_238 | 7.2 | 38.977 | 14.6048 | |
| LA4 | A06 | 46.2 | 14.68 | 45.2–46.6 | bin_305 | 18.9 | 69.4458 | 13.5546 | |
| LA5 | A07 | 19.8 | 5.86 | 18.4–20.4 | bin_363 | 6.6 | 32.2879 | 30.4413 | |
| LA6 | A08 | 26.2 | 5.68 | 24.9–27.9 | bin_421 | 6.3 | 39.7259 | −6.26181 | |
| LA7 | A10 | 19.6 | 10.19 | 18.9–20.9 | bin_541 | 12 | 36.7118 | 57.9985 | |
| PA | PA1 | A02 | 34.9 | 4.22 | 33.9–36.6 | bin_77 | 4.4 | 4.22363 | 2.86465 |
| PA2 | A04 | 39.6 | 5.12 | 36.3–40.3 | bin_214 | 5.3 | 4.89341 | 2.52144 | |
| PA3 | A06 | 45.6 | 18.59 | 44.5–46.2 | bin_305 | 24.6 | 11.2716 | 2.72566 | |
| PA4 | A10 | 8.8 | 18.29 | 6.7–9.4 | bin_535 | 24.1 | 11.1044 | 0.828217 | |
| PAI | PAI1 | A01 | 23.4 | 5.43 | 20.6–27.4 | bin_9 | 9.1 | 0.497833 | −0.225617 |
| PAI2 | A05 | 87.9 | 5.39 | 81.5–88.3 | bin_276 | 8.8 | 0.516711 | 0.148835 | |
| PAI3 | A07 | 0 | 3.55 | 0.0–3.0 | bin_339 | 5.6 | 0.42485 | −0.02513 | |
| PAI4 | A08 | 0 | 3.61 | 0.0–3.7 | bin_397 | 5.7 | 0.408383 | −0.0758493 | |
| PAI5 | A10 | 7.7 | 5.75 | 5.4–8.8 | bin_534 | 9.8 | −0.5165599 | 0.313028 |
| QTL | Bin | Candidate Gene_ID | Other Name |
|---|---|---|---|
| PLTI1 | bin_10 | BraA01g007610.3C | ARF16 |
| LL6, LA5 | bin_363 | BraA07g018740.3C | ARF10 |
| LL4, LA4, PA3 | bin_305 | BraA06g015500.3C | ARF5;IAA2 |
| PWT4, LW4 | bin_319 | BraA06g029140.3C | IAA9 |
| PLTI4 | bin_270 | BraA05g030630.3C | IAA26 |
| LW5 | bin_362 | BraA07g018500.3C | SAUR42 |
| LL4, LA4, PA3, PI4 | bin_305 bin_359 | BraA06g015440.3C BraA07g016220.3C | SAUR53 |
| PWT3, PA2 | bin_214 | BraA04g025760.3C | SAUR45 |
| LWT3, LW3, LA2, LI4 | bin_215 | BraA04g026250.3C | SAUR46 |
| LWT3, LW3, LA2, LI4 | bin_215 | BraA04g026370.3C | RPS5B |
| PL3 | bin_328 | BraA06g037340.3C | RPL12 |
| LWT3, LW3, LA2, LI4 | bin_215 | BraA04g025910.3C | GRF3 |
| PAI4 | bin_397 | BraA08g001700.3C | TCP3 |
| PAI2 | bin_276 | BraA05g035610.3C | WOX5 |
| PWT4, LW4 | bin_319 | BraA06g029220.3C | BAM1 |
| LL7 | bin_415 | BraA08g014070.3C | BAM3 |
| LW5 | bin_362 | BraA07g018220.3C | PGY1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Liu, Z.; Chen, H.; Wu, J.; Cai, X.; Wang, H.; Wang, X.; Liang, J. QTL Mapping of Leaf-Related Traits Using a High-Density Bin Map in Brassica rapa. Horticulturae 2023, 9, 433. https://doi.org/10.3390/horticulturae9040433
Li F, Liu Z, Chen H, Wu J, Cai X, Wang H, Wang X, Liang J. QTL Mapping of Leaf-Related Traits Using a High-Density Bin Map in Brassica rapa. Horticulturae. 2023; 9(4):433. https://doi.org/10.3390/horticulturae9040433
Chicago/Turabian StyleLi, Fengming, Zhiyuan Liu, Haixu Chen, Jian Wu, Xu Cai, Hui Wang, Xiaowu Wang, and Jianli Liang. 2023. "QTL Mapping of Leaf-Related Traits Using a High-Density Bin Map in Brassica rapa" Horticulturae 9, no. 4: 433. https://doi.org/10.3390/horticulturae9040433
APA StyleLi, F., Liu, Z., Chen, H., Wu, J., Cai, X., Wang, H., Wang, X., & Liang, J. (2023). QTL Mapping of Leaf-Related Traits Using a High-Density Bin Map in Brassica rapa. Horticulturae, 9(4), 433. https://doi.org/10.3390/horticulturae9040433








