Identification of PAL Gene in Purple Cabbage and Functional Analysis Related to Anthocyanin Synthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of the PAL Gene Family in Chinese Cabbage
2.2. Physicochemical Analysis and Subcellular Localization Prediction of PAL Gene Family Member Proteins in Chinese Cabbage
2.3. Phylogenetic Analysis of the Cabbage PAL Gene
2.4. Chromosomal Mapping and Synteny Analysis of PAL Gene Family Members in Chinese Cabbage
2.5. Conserved Motifs and Gene Structure Prediction of the PAL Gene Family in Chinese Cabbage
2.6. Analysis of Cis-Acting Elements in the Promoter of the PAL Gene Family in Chinese Cabbage
2.7. Growth and Treatment of Cabbage Seedlings
2.8. Quantitative Real-Time Fluorescent Quantitative PCR and Anthocyanin Content Determination and Data Analysis
3. Results
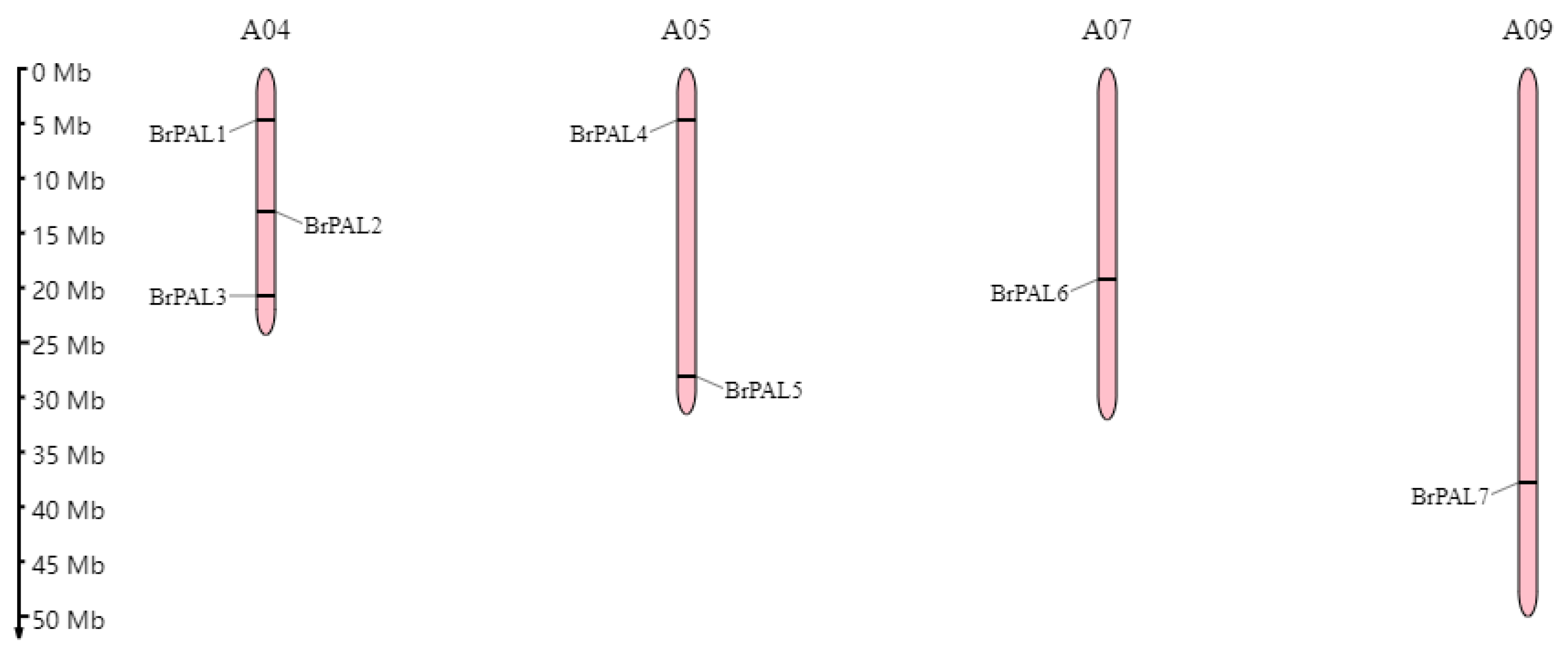
3.1. Analysis of Chromosomal Location and Protein Physicochemical Properties of PAL Gene Family Members in Chinese Cabbage
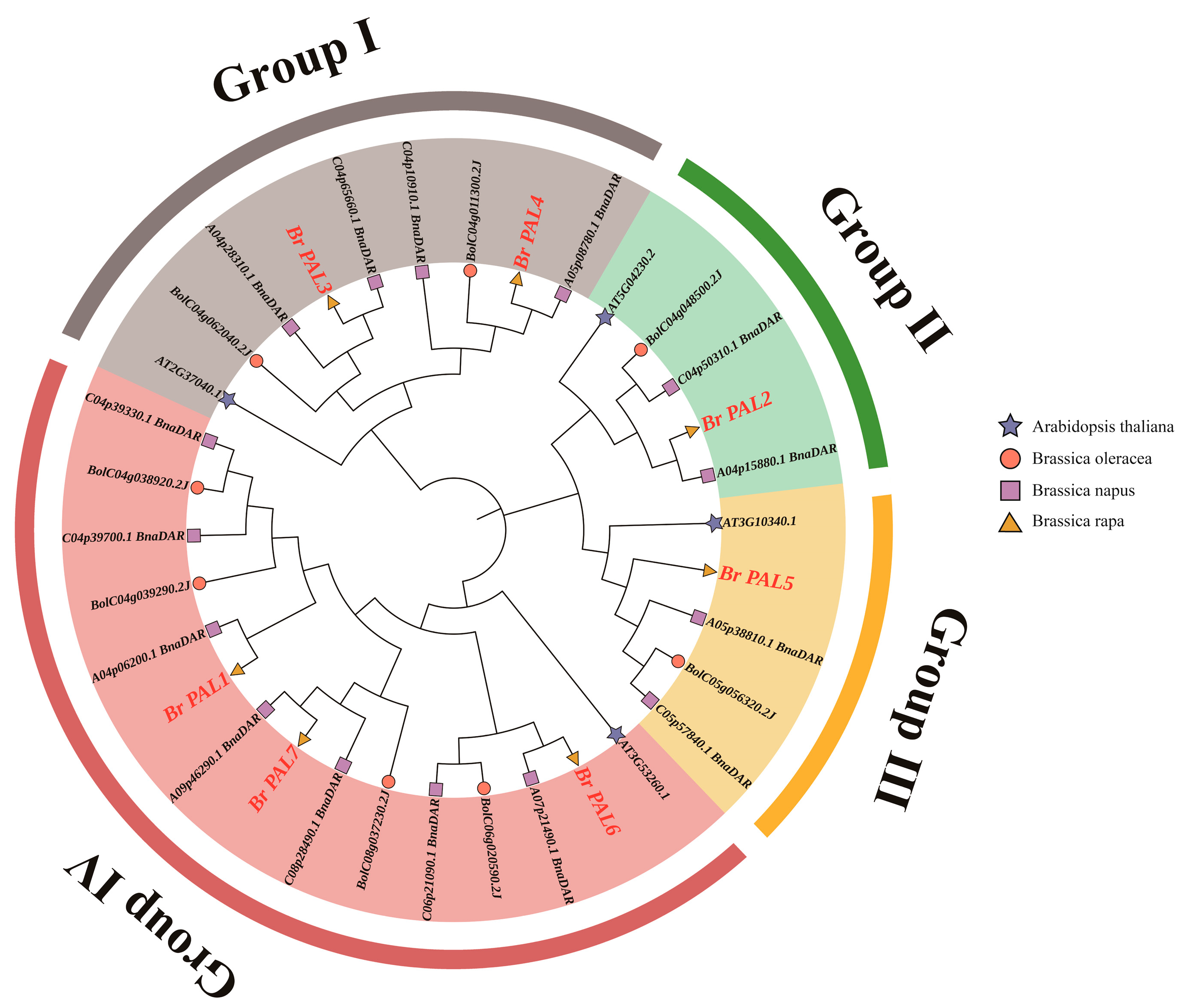
3.2. Phylogenetic Analysis of PAL Gene in Chinese Cabbage
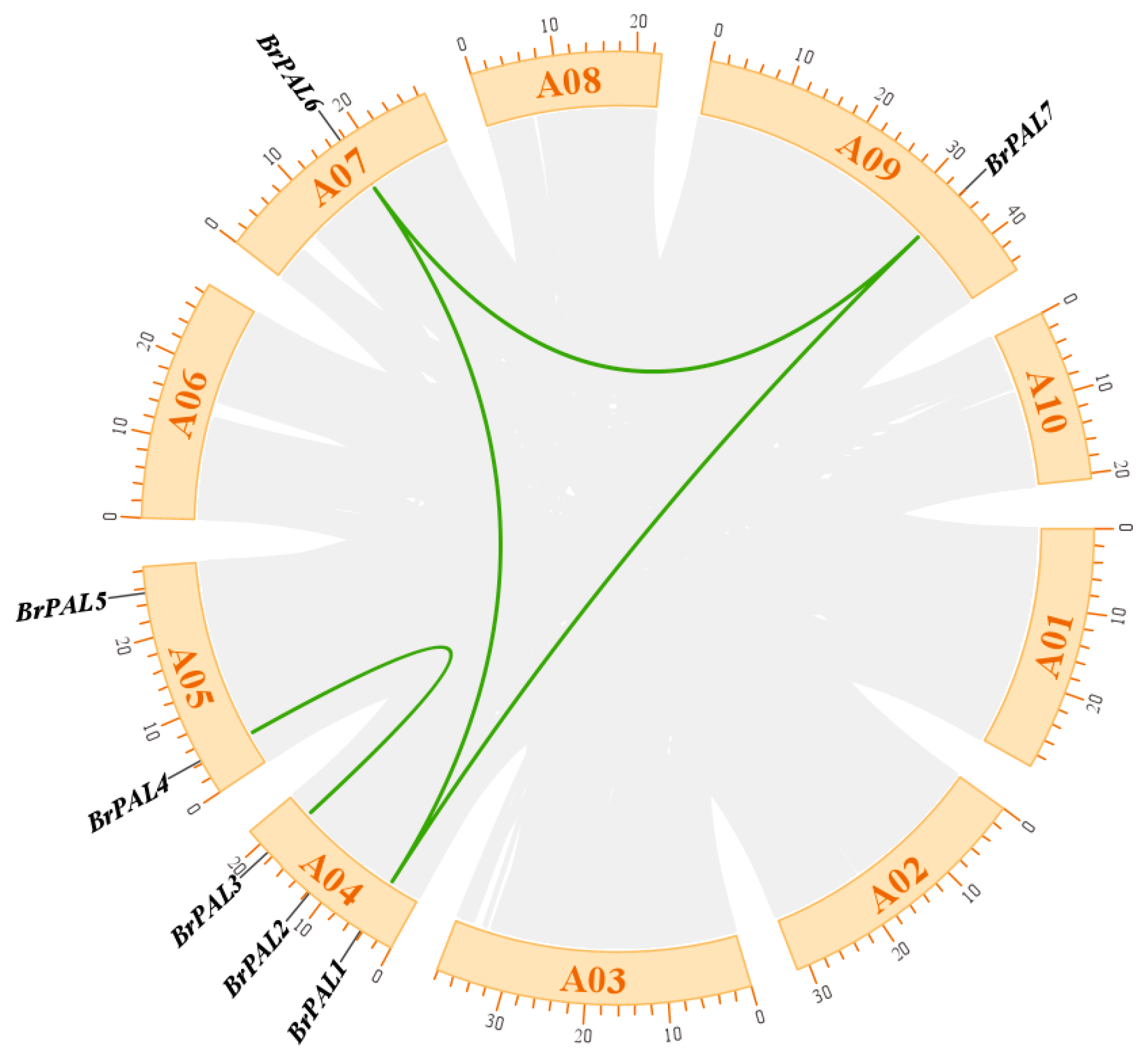
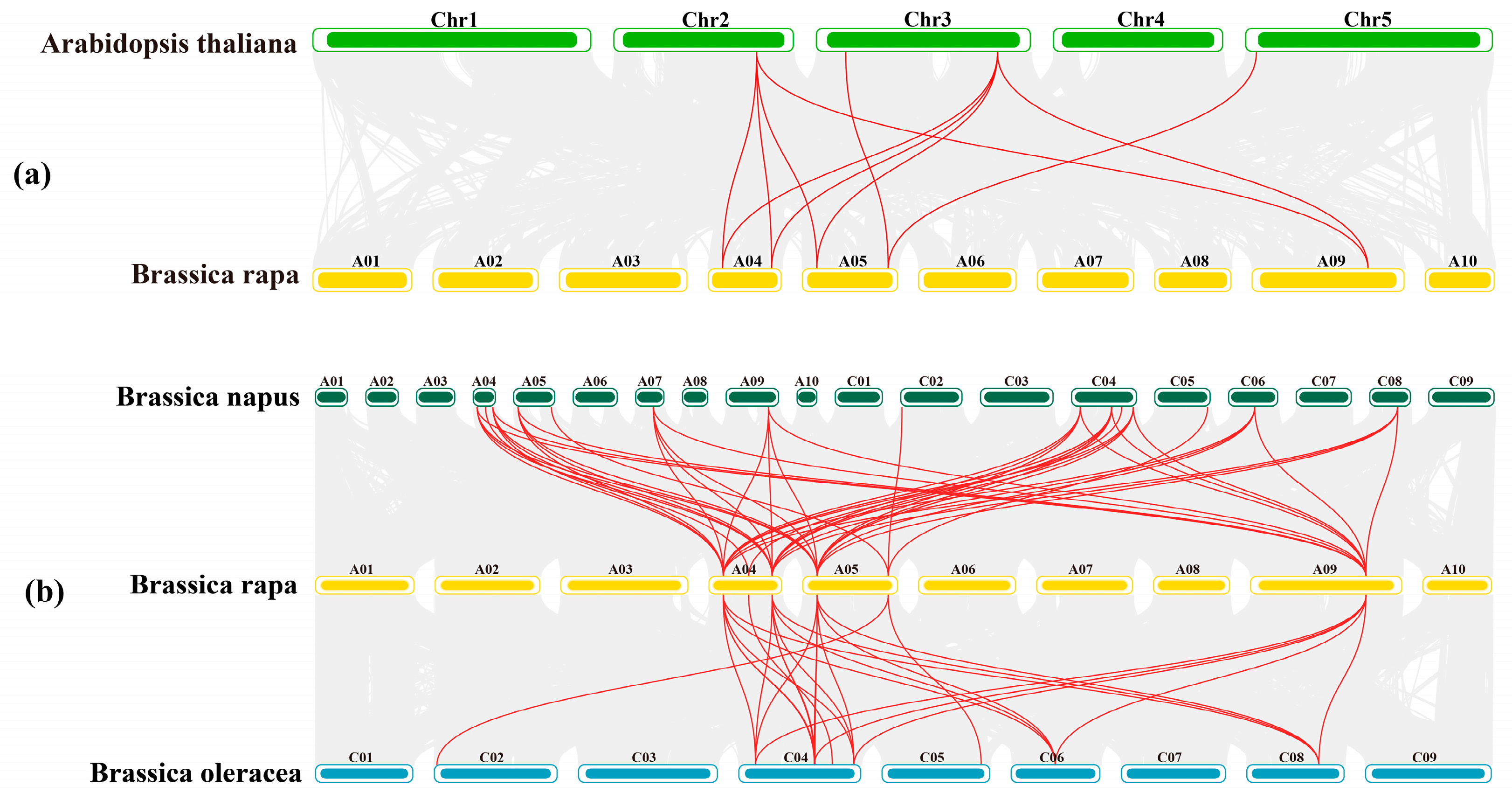
3.3. Collinearity Analysis of PAL Genes in Chinese Cabbage
3.4. Structure and Conserved Motif Analysis of the Cabbage PAL Gene
3.5. Analysis of Cis-Acting Elements in Promoters of the PAL Gene Family in Chinese Cabbage
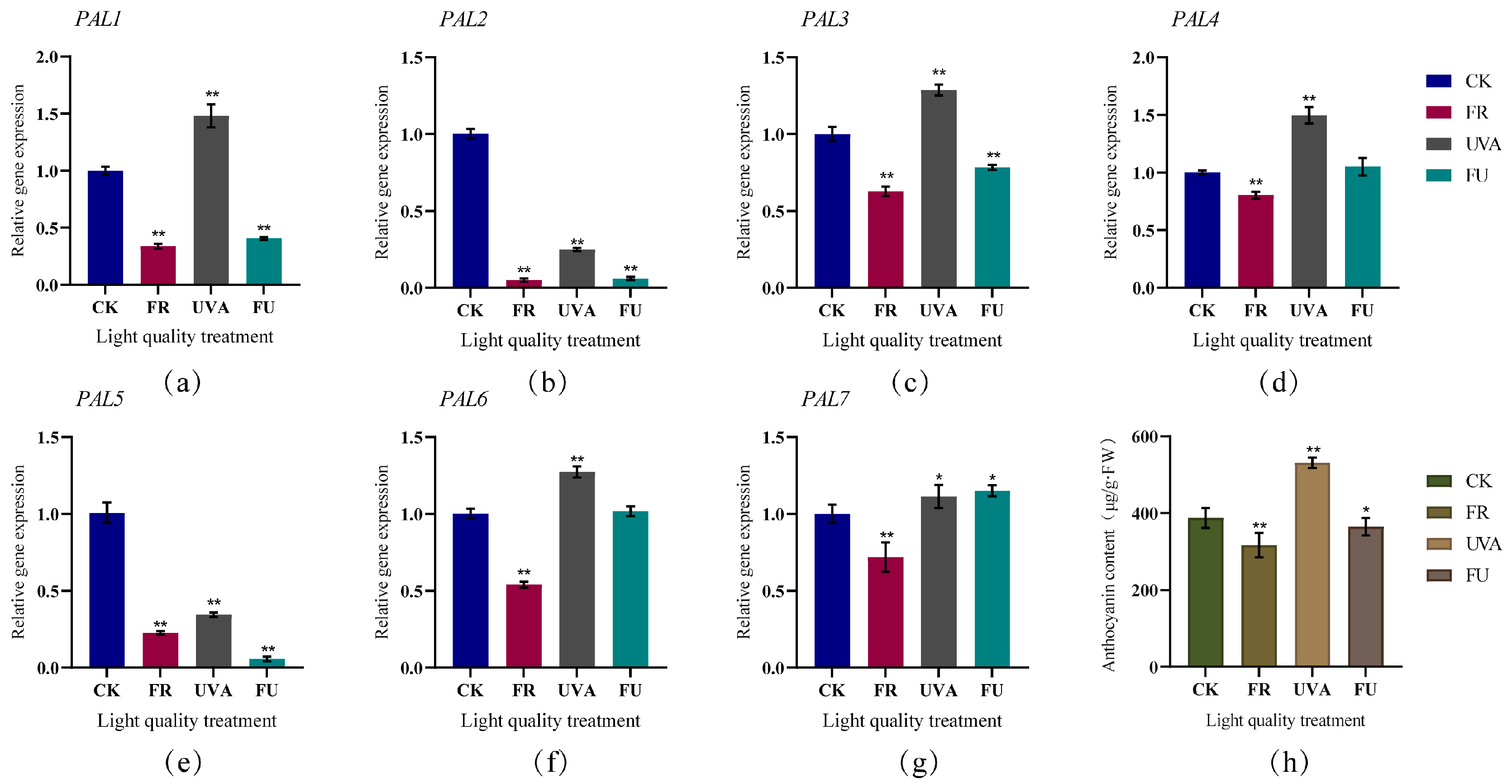
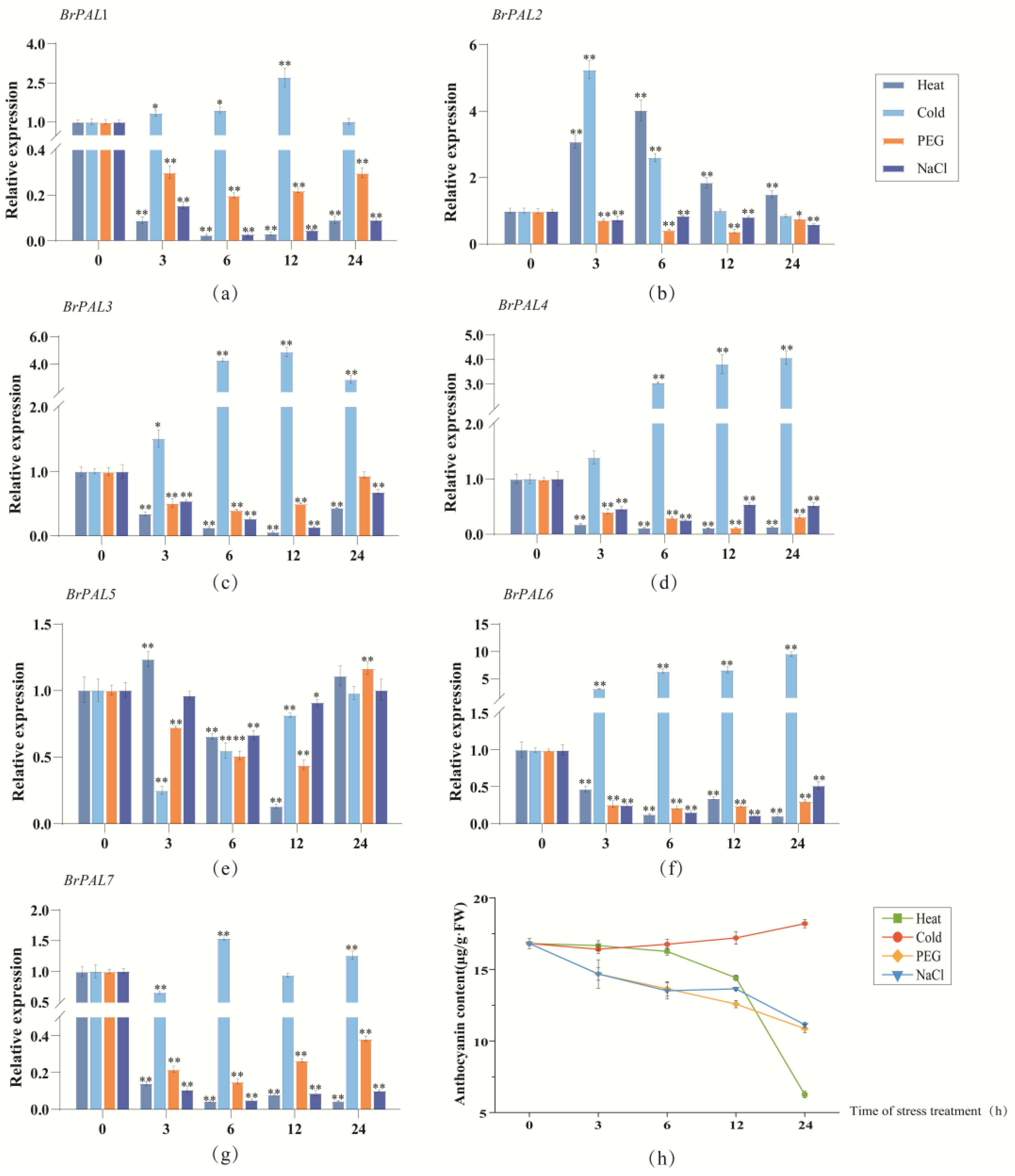
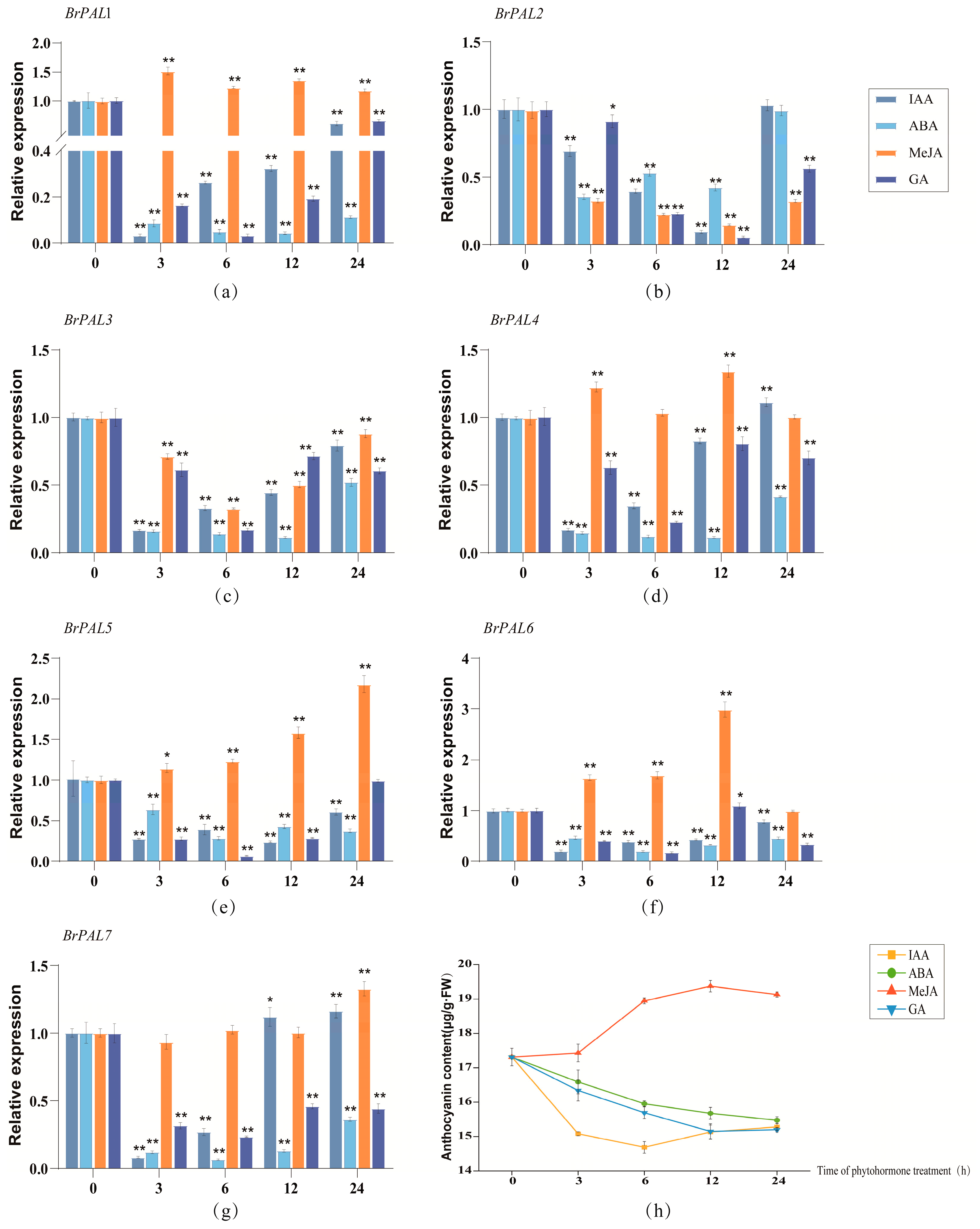
3.6. Expression Patterns of Cabbage PAL Genes under Different Light Quality, Abiotic Stress and Hormone Induction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jones, D.H. Phenylalanine ammonia-lyase: Regulation of its induction, and its role in plant development. Phytochemistry 1984, 23, 1349–1359. [Google Scholar] [CrossRef]
- Ouyang, G.C.; Xue, Y.L. Physiological Significance and Regulation of Phenylpropanoid Metabolism in Plants. Plant Physiol. Commun. 1988, 9–16. [Google Scholar] [CrossRef]
- Koukol, J.; Conn, E.E. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J. Biol. Chem. 1961, 236, 2692–2698. [Google Scholar] [CrossRef]
- De Jong, F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterisation of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Sun, Y.-H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a Systems Approach for Lignin Biosynthesis in Populus trichocarpa: Transcript Abundance and Specificity of the Monolignol Biosynthetic Genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Gui, S.; Wang, S.; Ding, Y. Molecular evolution and functional characterisation of an ancient phenylalanine ammonia-lyase gene (NnPAL1) from Nelumbo nucifera: Novel insight into the evolution of the PAL family in angiosperms. BMC Evol. Biol. 2014, 14, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Q.-M.; Li, L.; Dong, C.-J. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 2012, 236, 1093–1105. [Google Scholar] [CrossRef]
- Joos, H.-J.; Hahlbrock, K. Phenylalanine ammonia-lyase in potato (Solanum tuberosum L.). Genomic complexity, structural comparison of two selected genes and modes of expression. Eur. J. Biochem. 1992, 204, 621–629. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.-J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Mo, F.; Li, L.; Zhang, C.; Yang, C.; Chen, G.; Niu, Y.; Si, J.; Liu, T.; Sun, X.; Wang, S.; et al. Genome-Wide Analysis and Expression Profiling of the Phenylalanine Ammonia-Lyase Gene Family in Solanum tuberosum. Int. J. Mol. Sci. 2022, 23, 6833. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, P.; Gu, M.; Hou, B.; Zhang, C.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. Identification of PAL genes related to anthocyanin synthesis in tea plants and its correlation with anthocyanin content. Hortic. Plant J. 2022, 8, 381–394. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Barros, J.; Dixon, R.A. Plant Phenylalanine/Tyrosine Ammonia-Lyases. Trends Plant Sci. 2020, 25, 66–79. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B. Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [Green Version]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential Expression of Four Arabidopsis PAL Genes; PAL1 and PAL2 Have Functional Specialization in Abiotic Environmental-Triggered Flavonoid Synthesis. J. Plant Physiol. 2008, 165, 1491–1499. [Google Scholar] [CrossRef]
- Zhao, T.; Li, R.; Yao, W.; Wang, Y.; Zhang, C.; Li, Y. Genome-wide identification and characterisation of phenylalanine ammonia-lyase gene family in grapevine. J. Hortic. Sci. Biotechnol. 2021, 96, 456–468. [Google Scholar] [CrossRef]
- Zhan, C.; Li, Y.; Li, H.; Wang, M.; Gong, S.; Ma, D.; Li, Y. Phylogenomic analysis of phenylalanine ammonia-lyase (PAL) multigene family and their differential expression analysis in wheat (Triticum aestivum L.) suggested their roles during different stress responses. Front. Plant Sci. 2022, 13, 982457. [Google Scholar] [CrossRef]
- Zhang, C.; Yao, X.; Ren, H.; Wang, K.; Chang, J. Genome-wide identification and characterization of the phenylalanine ammonia-lyase gene family in pecan (Carya illinoinensis). Sci. Hortic. 2022, 295, 110800. [Google Scholar] [CrossRef]
- Zhao, Y.; Qi, X.; Liu, Z.; Zheng, W.; Guan, J.; Liu, Z.; Ren, J.; Feng, H.; Zhang, Y. Transcriptome and Metabolome Profiling to Explore the Causes of Purple Leaves Formation in Non-Heading Chinese Cabbage (Brassica rapa L. ssp. chinensis Makino var. mutliceps Hort.). Foods 2022, 11, 1787. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Yeo, H.J.; Park, S.-Y.; Kim, J.K.; Park, S.U. Comparative Phytochemical Analyses and Metabolic Profiling of Different Phenotypes of Chinese Cabbage (Brassica Rapa ssp. Pekinensis). Foods 2019, 8, 587. [Google Scholar] [CrossRef] [Green Version]
- Rochfort, S.J.; Imsic, M.; Jones, R.; Trenerry, V.C.; Tomkins, B. Characterization of Flavonol Conjugates in Immature Leaves of Pak Choi [Brassica rapa L. Ssp. chinensis L. (Hanelt.)] by HPLC-DAD and LC-MS/MS. J. Agric. Food Chem. 2006, 54, 4855–4860. [Google Scholar] [CrossRef] [PubMed]
- Favela-Gonzalez, K.M.; Hernandez-Almanza, A.Y.; De la Fuente-Salcido, N.M. The Value of Bioactive Compounds of Cruciferous Vegetables (Brassica) as Antimicrobials and Antioxidants: A Review. J. Food Biochem. 2020, 44, e13414. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, J.; Cai, X.; Li, Y.; Xi, X.; Lin, R.; Liang, J.; Wang, X.; Wu, J. Improved Reference Genome Annotation of Brassica rapa by Pacific Biosciences RNA Sequencing. Front. Plant Sci. 2022, 13, 841618. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013, 41, D348–D352. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2-DeltaDeltaCt Calculation as an Alternate Method of Data Analysis for Quantitative PCR of BCR-ABL P210 Transcripts. Diagn. Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef]
- Ferrer, J.L.; Austin, M.B.; Stewart, C., Jr.; Noel, J.P. Structure and Function of Enzymes Involved in the Biosynthesis of Phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, M.J.; D’Cunha, G.B. A Modern View of Phenylalanine Ammonia Lyase. Biochem. Cell Biol. 2007, 85, 273–282. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, Y.; Mu, K.; Cai, W.; Zhao, Y.; Shen, H.; Wang, X.; Ma, H. Phenylalanine Ammonia Lyase GmPAL1.1 Promotes Seed Vigor under High-Temperature and -Humidity Stress and Enhances Seed Germination under Salt and Drought Stress in Transgenic Arabidopsis. Plants 2022, 11, 3239. [Google Scholar] [CrossRef]
- Rawal, H.C.; Singh, N.K.; Sharma, T.R. Conservation, Divergence, and Genome-Wide Distribution of PAL and POX A Gene Families in Plants. Int. J. Genom. 2013, 2013, 678969. [Google Scholar]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Ji, R.; Havlickova, L.; Wang, L.; Li, Y.; Lee, H.T.; Song, J.; Koh, C.; Yang, J.; Zhang, M.; et al. Genome structural evolution in Brassica crops. Nat. Plants 2021, 7, 757–765. [Google Scholar] [CrossRef]
- Brassica Rapa Genome Sequencing Project Consortium; Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; et al. The Genome of the Mesopolyploid Crop Species Brassica rapa. Nat. Genet 2011, 43, 1035–1039. [Google Scholar]
- Zhou, B.; Li, Y.; Xu, Z.; Yan, H.; Homma, S.; Kawabata, S. Ultraviolet A-specific induction of anthocyanin biosynthesis in the swollen hypocotyls of turnip (Brassica rapa). J. Exp. Bot. 2007, 58, 1771–1781. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, M.-H. Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (Solanum lycopersicum L.). Plant Growth Regul. 2010, 62, 1–8. [Google Scholar] [CrossRef]
- Guo, J.; Wang, M.H. Characterization of the Phenylalanine Ammonia-Lyase Gene (SlPAL5) from Tomato (Solanum lycopersicum L.). Mol. Biol. Rep. 2009, 36, 1579–1585. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Number of Amino Acids (aa) | Molecular Weight (MW/kD) | pI | GRAVY | Subcellular Location |
|---|---|---|---|---|---|---|
| BrPAL1 | BraA04g006770.3.5C.1 | 724 | 78.52 | 6.03 | −0.177 | Cytoplasm |
| BrPAL2 | BraA04g016310.3.5C.1 | 698 | 76.91 | 6.23 | −0.168 | Cytoplasm |
| BrPAL3 | BraA04g027460.3.5C.1 | 722 | 78.30 | 5.9 | −0.163 | Cytoplasm |
| BrPAL4 | BraA05g008230.3.5C.1 | 719 | 78.10 | 5.9 | −0.136 | Cytoplasm |
| BrPAL5 | BraA05g037490.3.5C.1 | 706 | 76.88 | 5.69 | −0.139 | Cytoplasm |
| BrPAL6 | BraA07g021930.3.5C.1 | 587 | 64.30 | 5.93 | −0.164 | Cytoplasm |
| BrPAL7 | BraA09g046240.3.5C.1 | 723 | 78.52 | 5.97 | −0.167 | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, J.; Xu, R.; Chang, K.; Yuan, S.; Huang, C.; Wang, J.; Li, S.; Liu, F.; Zhong, F. Identification of PAL Gene in Purple Cabbage and Functional Analysis Related to Anthocyanin Synthesis. Horticulturae 2023, 9, 469. https://doi.org/10.3390/horticulturae9040469
Tian J, Xu R, Chang K, Yuan S, Huang C, Wang J, Li S, Liu F, Zhong F. Identification of PAL Gene in Purple Cabbage and Functional Analysis Related to Anthocyanin Synthesis. Horticulturae. 2023; 9(4):469. https://doi.org/10.3390/horticulturae9040469
Chicago/Turabian StyleTian, Jun, Ru Xu, Kaizhen Chang, Song Yuan, Chenxin Huang, Jinwei Wang, Shuhao Li, Fazhong Liu, and Fenglin Zhong. 2023. "Identification of PAL Gene in Purple Cabbage and Functional Analysis Related to Anthocyanin Synthesis" Horticulturae 9, no. 4: 469. https://doi.org/10.3390/horticulturae9040469
APA StyleTian, J., Xu, R., Chang, K., Yuan, S., Huang, C., Wang, J., Li, S., Liu, F., & Zhong, F. (2023). Identification of PAL Gene in Purple Cabbage and Functional Analysis Related to Anthocyanin Synthesis. Horticulturae, 9(4), 469. https://doi.org/10.3390/horticulturae9040469





