Transcriptomic Analysis Revealed the Discrepancy between Early-Ripening ‘Geneva Early’ and Late-Ripening ‘Hanfu’ Apple Cultivars during Fruit Development and Ripening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. Determination of Fruit Quality Characteristics
2.3. RNA Extraction and Transcriptome Analysis
2.4. RT-qPCR Analysis
2.5. Statistical Analyses
3. Results
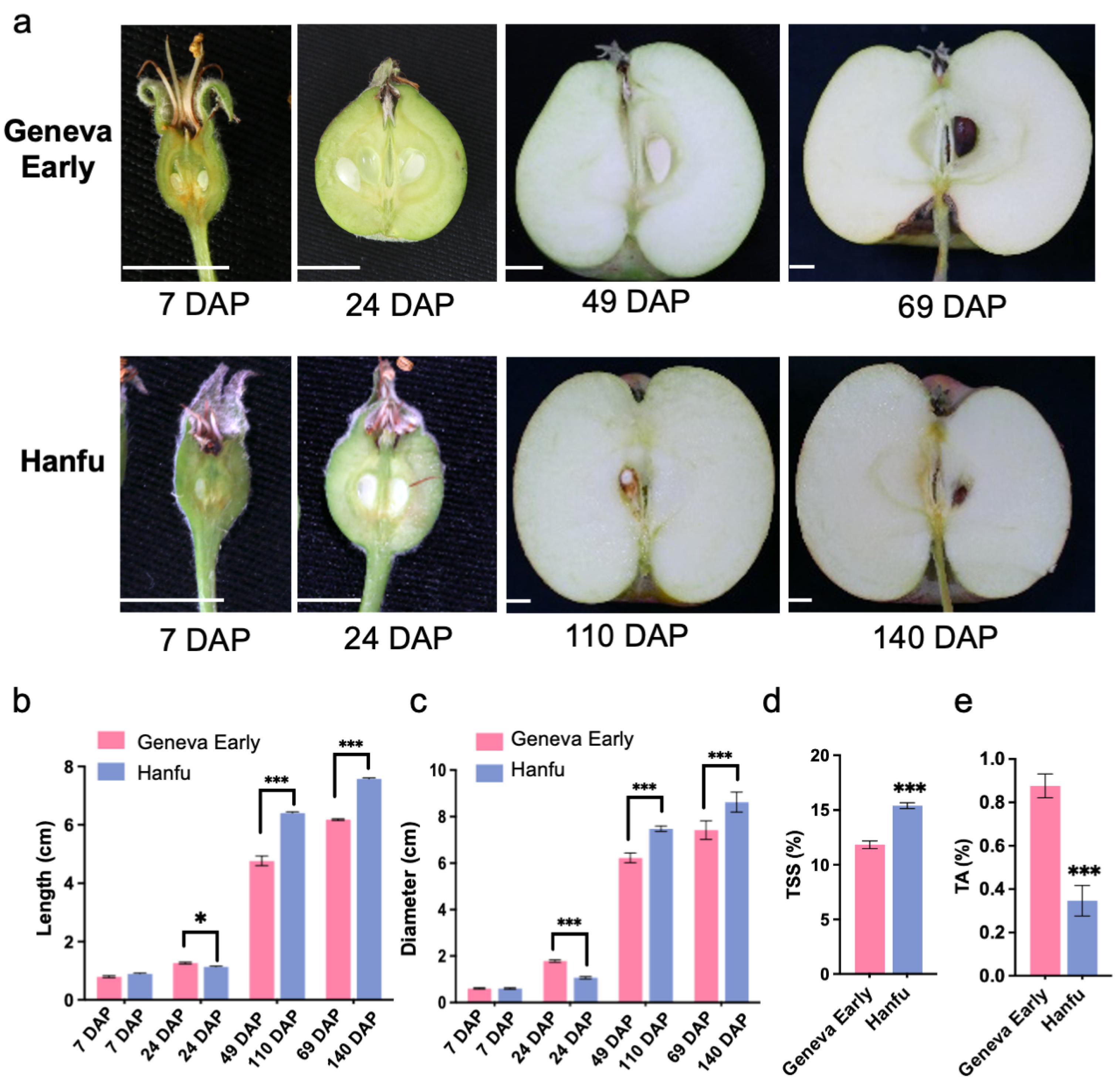
3.1. Apple Fruit Quality Characteristics in ‘Geneva Early’ and ‘Hanfu’
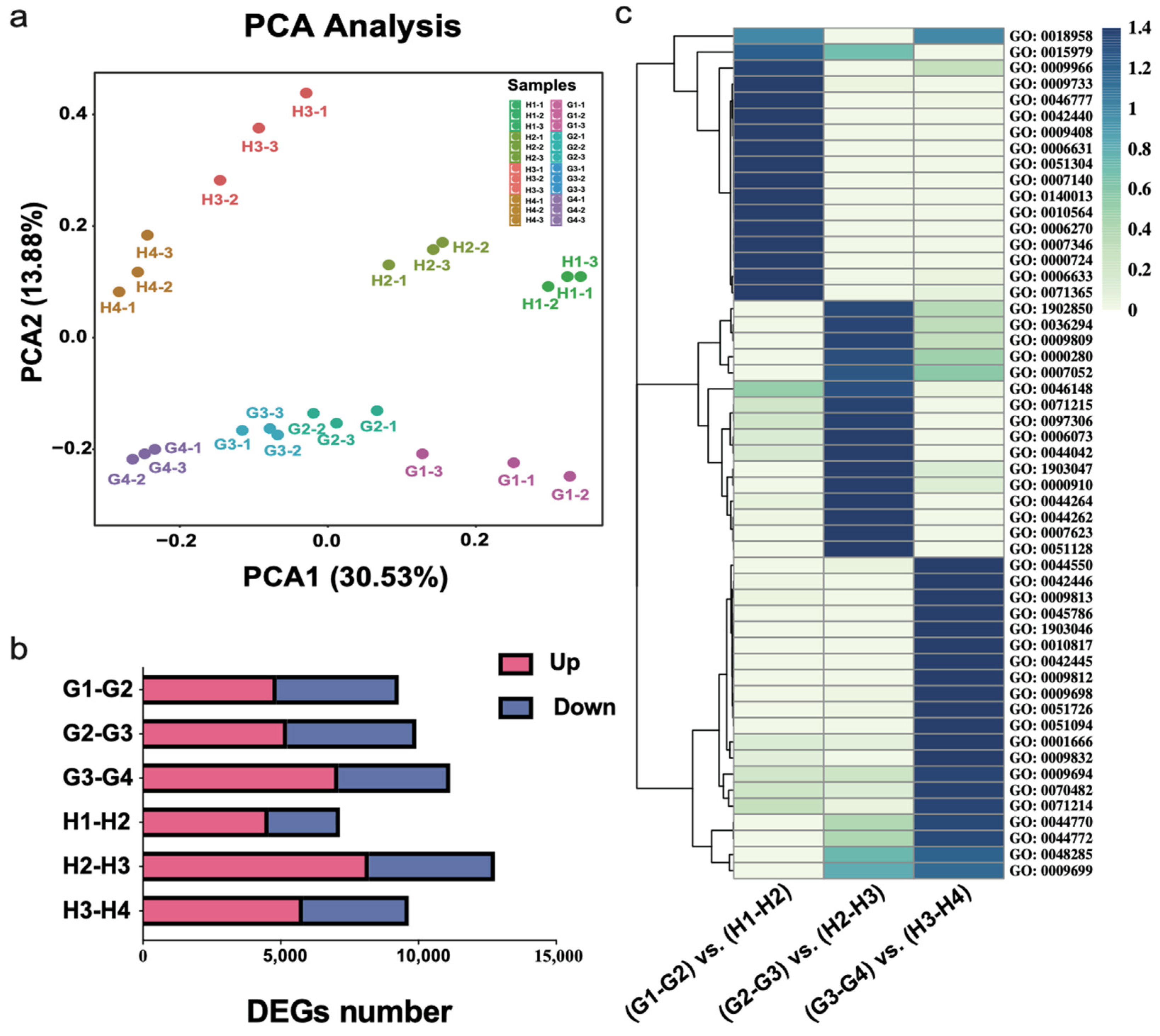
3.2. Transcriptomic Analysis of ‘Geneva Early’ and ‘Hanfu’ during the Four Fruit Developmental Stages
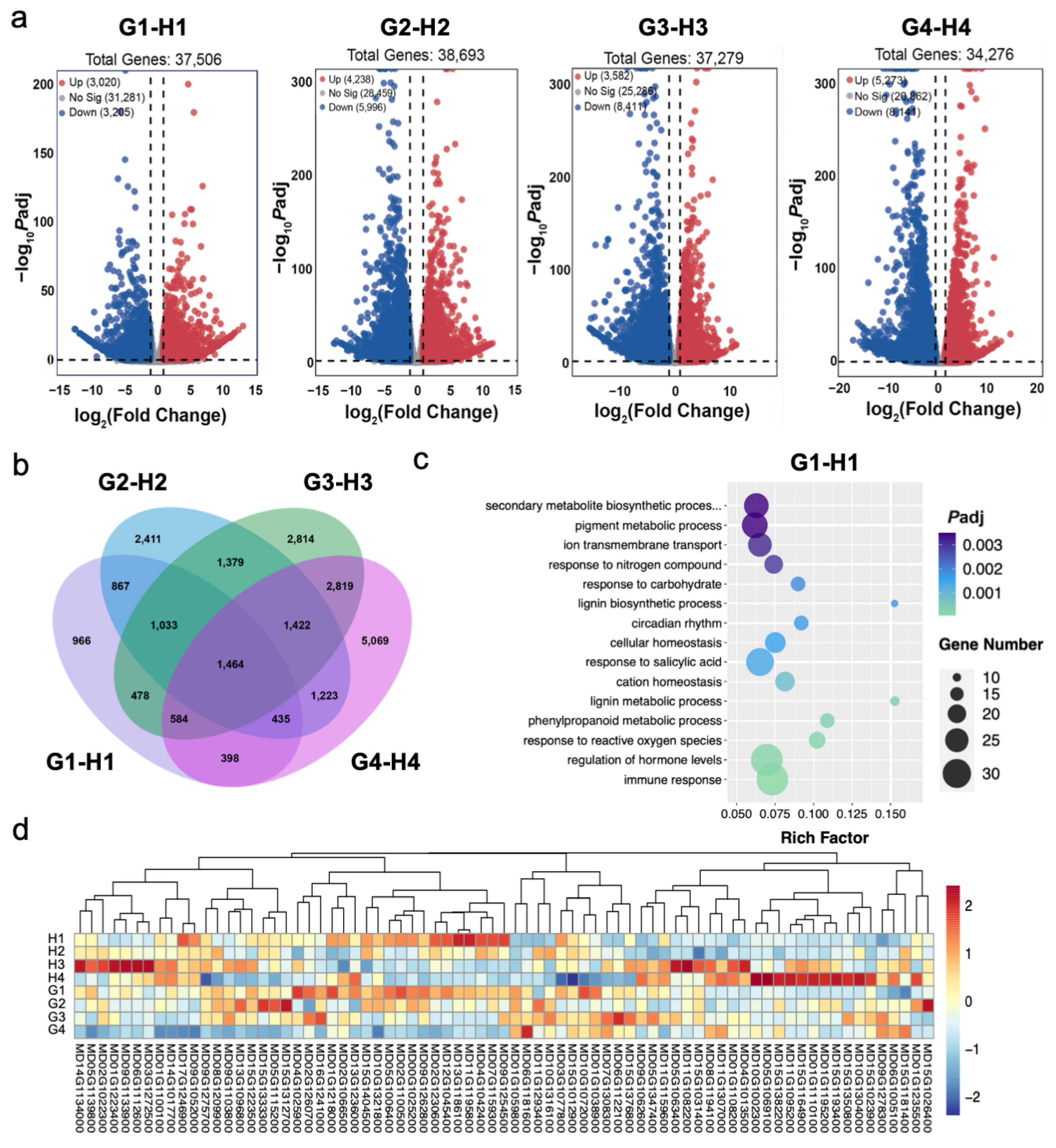
3.3. Expression of DEGs among Different Fruit Development Stages of Apples
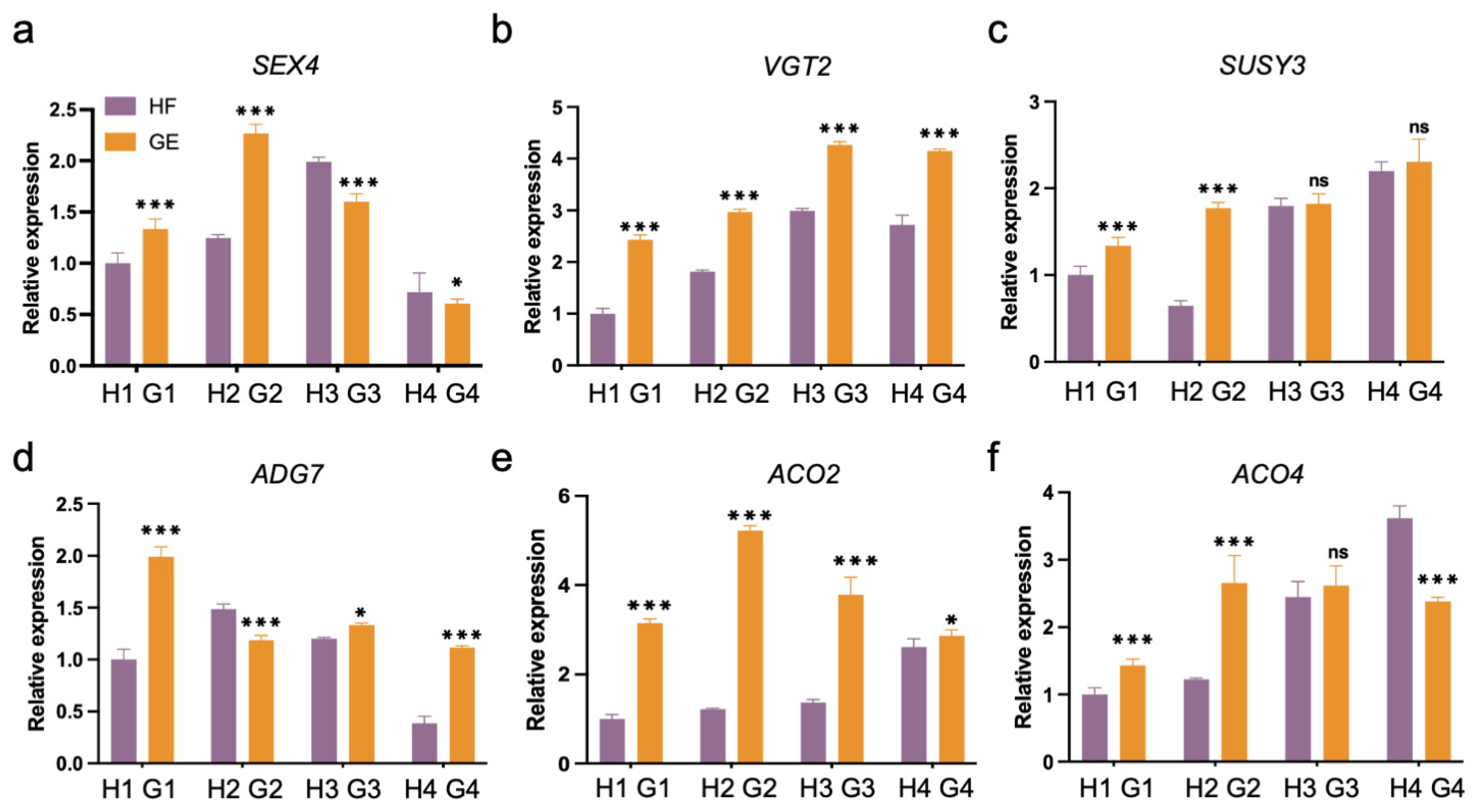
3.4. Expression of Various Functional Groups of Genes during Different Fruit Development Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nkuimi Wandjou, J.G.; Sut, S.; Giuliani, C.; Fico, G.; Papa, F.; Ferraro, S.; Caprioli, G.; Maggi, F.; Dall’Acqua, S. Characterization of nutrients, polyphenols and volatile components of the ancient apple cultivar ‘Mela Rosa Dei Monti Sibillini’ from Marche region, central Italy. Int. J. Food Sci. Nutr. 2019, 70, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Devoghalaere, F.; Doucen, T.; Guitton, B.; Keeling, J.; Payne, W.; Ling, T.J.; Ross, J.J.; Hallett, I.C.; Gunaseelan, K.; Dayatilake, G.A.; et al. A genomics approach to understanding the role of auxin in apple (Malus × domestica) fruit size control. BMC Plant Biol. 2012, 12, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, B.J.; Thodey, K.; Schaffer, R.J.; Alba, R.; Balakrishnan, L.; Bishop, R.; Bowen, J.H.; Crowhurst, R.N.; Gleave, A.P.; Ledger, S.; et al. Global gene expression analysis of apple fruit development from the floral bud to ripe fruit. BMC Plant Biol. 2008, 8, 16. [Google Scholar] [CrossRef] [Green Version]
- Malladi, A.; Hirst, P.M. Increase in fruit size of a spontaneous mutant of ‘Gala’ apple (Malus × domestica Borkh.) is facilitated by altered cell production and enhanced cell size. J. Exp. Bot. 2010, 61, 3003–3013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seymour, G.B.; Østergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef] [Green Version]
- Aghdam, M.S.; Jannatizadeh, A.; Luo, Z.S.; Paliyath, G. Ensuring sufficient intracellular ATP supplying and friendly extracellular ATP signaling attenuates stresses, delays senescence and maintains quality in horticultural crops during postharvest life. Trends Food Sci. Tech. 2018, 76, 67–81. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Li, P.M.; Cheng, L.L. Developmental changes of carbohydrates, organic acids, amino acids, and phenolic compounds in ‘Honeycrisp’ apple flesh. Food Chem. 2010, 123, 1013–1018. [Google Scholar] [CrossRef]
- Li, M.J.; Feng, F.J.; Cheng, L.L. Expression Patterns of Genes Involved in Sugar Metabolism and Accumulation during Apple Fruit Development. PLoS ONE 2012, 7, e33055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, W.L.; Martins, A.O.; Fernie, A.R.; Tohge, T. 2-Oxoglutarate: Linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 2014, 5, 552. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.D.; Yan, J.J.; Li, W.J.; Wang, Q.Y.; Wang, C.X.; Guo, J.X.; Geng, D.L.; Guan, Q.M.; Ma, F.W. Integrative Analyses of Widely Targeted Metabolic Profiling and Transcriptome Data Reveals Molecular Insight into Metabolomic Variations during Apple (Malus domestica) Fruit Development and Ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef]
- Slewinski, T.L. Diverse Functional Roles of Monosaccharide Transporters and their Homologs in Vascular Plants: A Physiological Perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Johnston, J.W.; Gunaseelan, K.; Pidakala, P.; Wang, M.; Schaffer, R.J. Co-ordination of early and late ripening events in apples is regulated through differential sensitivities to ethylene. J. Exp. Bot. 2009, 60, 2689–2699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.-P.; Bouzayen, M. Ethylene Control of Fruit Ripening: Revisiting the Complex Network of Transcriptional Regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.-D.; Cho, Y.; Sheen, J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009, 14, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Wills, R.; McGlasson, W.; Graham, D.; Joyce, D. Physiology and biochemistry. In Postharvest: An Introduction to the Physiology and Handling of Fruit, Vegetables and Ornamentals; CABI: Wallingford, UK, 2007; pp. 28–51. [Google Scholar]
- Gao, J.; Zhang, Y.X.; Li, Z.G.; Liu, M.C. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.-C.; Kuang, J.-F.; Chen, J.-Y.; Liu, X.-C.; Xiao, Y.-Y.; Fu, C.-C.; Wang, J.-N.; Wu, K.-Q.; Lu, W.-J. Banana Transcription Factor MaERF11 Recruits Histone Deacetylase MaHDA1 and Represses the Expression of MaACO1 and Expansins during Fruit Ripening. Plant Physiol. 2016, 171, 1070–1084. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Jiang, Z.; Zhang, L.; Tan, D.; Wei, Y.; Yuan, H.; Li, T.; Wang, A. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 2016, 88, 735–748. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. Molecular and Hormonal Mechanisms Regulating Fleshy Fruit Ripening. Cells 2021, 10, 1136. [Google Scholar] [CrossRef]
- Breitel, D.A.; Chappell-Maor, L.; Meir, S.; Panizel, I.; Puig, C.P.; Hao, Y.; Yifhar, T.; Yasuor, H.; Zouine, M.; Bouzayen, M.; et al. AUXIN RESPONSE FACTOR 2 Intersects Hormonal Signals in the Regulation of Tomato Fruit Ripening. PLoS Genet. 2016, 12, e1005903. [Google Scholar] [CrossRef] [Green Version]
- El-Sharkawy, I.; Sherif, S.M.; Jones, B.; Mila, I.; Kumar, P.P.; Bouzayen, M.; Jayasankar, S. TIR1-like auxin-receptors are involved in the regulation of plum fruit development. J. Exp. Bot. 2014, 65, 5205–5215. [Google Scholar] [CrossRef] [Green Version]
- Yue, P.T.; Lu, Q.; Liu, Z.; Lv, T.X.; Li, X.Y.; Bu, H.D.; Liu, W.T.; Xu, Y.X.; Yuan, H.; Wang, A.D. Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytologist. 2020, 226, 1781–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, K.; Wani, S.H.; Razzaq, A.; Skalicky, M.; Samantara, K.; Gupta, S.; Pandita, D.; Goel, S.; Grewal, S.; Hejnak, V.; et al. Abscisic Acid: Role in Fruit Development and Ripening. Front. Plant Sci. 2022, 13, 817500. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.Z.; Li, C.L.; Han, Y.Y.; Shen, Y.Y. Characterization of the hot pepper (Capsicum frutescens) fruit ripening regulated by ethylene and ABA. BMC Plant Biol. 2018, 18, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, L.; Zhao, F.; Li, R.; Xiao, H. Cross-talk modulation between ABA and ethylene by transcription factor SlZFP2 during fruit development and ripening in tomato. Plant Signal. Behav. 2015, 10, e1107691. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- Messeguer, R.; Ganal, M.W.; Steffens, J.C.; Tanksley, S.D. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol. Biol. 1991, 16, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tieman, D.M.; Jiao, C.; Xu, Y.; Chen, K.; Fei, Z.; Giovannoni, J.J.; Klee, H.J. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. USA 2016, 113, 12580–12585. [Google Scholar] [CrossRef] [Green Version]
- Zou, L.; Liu, W.; Zhang, Z.; Edwards, E.J.; Gathunga, E.K.; Fan, P.; Duan, W.; Li, S.; Liang, Z. Gene body demethylation increases expression and is associated with self-pruning during grape genome duplication. Hortic. Res. 2020, 7, 84. [Google Scholar] [CrossRef]
- Cheng, J.F.; Niu, Q.F.; Zhang, B.; Chen, K.S.; Yang, R.H.; Zhu, J.K.; Zhang, Y.J.; Lang, Z.B. Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 2018, 19, 212. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Liu, R.; Niu, Q.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.; Zhu, J.K.; Lang, Z. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Gao, X.H.; Tong, L.; Liu, M.Z.; Zhou, X.K.; Tahir, M.M.; Xing, L.B.; Ma, J.J.; An, N.; Zhao, C.P.; et al. Multi-omics analyses reveal MdMYB10 hypermethylation being responsible for a bud sport of apple fruit color. Hortic. Res. 2022, 9, uhac179. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.P.; Chen, P.X.; Yan, Y.; Bao, C.N.; Li, X.W.; Wang, L.P.; Shen, X.X.; Li, H.Y.; Liu, X.F.; Niu, C.D.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytologist. 2018, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Way, R.D.; Livermore, K.G.; Aldwinckle, H.S. Geneva Early Apple. Hortscience 1982, 17, 989. [Google Scholar] [CrossRef]
- Baolin, S. Introduction of Zaojie Apple in southern Shandong Province. China Fruit Veg. 2006, 4, 11–12. [Google Scholar]
- Cai, M.; Gao, W.; Chen, J.; Qin, S.; Ma, H.; Lv, D.; Liu, G. Effects of different paper bag treatments on fruit quality development of Hanfu apple. North. Hortic. 2009, 7, 19–21. [Google Scholar]
- Zhao, D.; Liu, G.; Lv, D.; Qin, S.; Ma, H.; Wang, H. Evaluation of the specific characters of Hanfu apple cultivar. J. Fruit Sci. 2009, 26, 6–12. [Google Scholar]
- Li, N.N.; Xu, C.C.; Li-Beisson, Y.H.; Philippar, K. Fatty Acid and Lipid Transport in Plant Cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.J.; Nikovics, K.; To, A.; Lepiniec, L.; Fedosejevs, E.T.; Van Doren, S.R.; Baud, S.; Thelen, J.J. Docking of acetyl-CoA carboxylase to the plastid envelope membrane attenuates fatty acid production in plants. Nat. Commun. 2020, 11, 6191. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Zhang, S.; Liu, J.Y.; Zhao, C.H.; Wu, J.; Cao, Y.P.; Fu, C.X.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.Q.; Lee, C.H.; Zhou, J.L.; McCarthy, R.L.; Ye, Z.H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Shu Huairui, Z.S. History and prospect of Chinese apple industry in 70 years. Deciduous Fruits 2021, 53, 3. [Google Scholar]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833. [Google Scholar] [CrossRef] [Green Version]
- Fait, A.; Hanhineva, K.; Beleggia, R.; Dai, N.; Rogachev, I.; Nikiforova, V.J.; Fernie, A.R.; Aharoni, A. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 2008, 148, 730–750. [Google Scholar] [CrossRef] [Green Version]
- Carrari, F.; Baxter, C.; Usadel, B.; Urbanczyk-Wochniak, E.; Zanor, M.-I.; Nunes-Nesi, A.; Nikiforova, V.; Centero, D.; Ratzka, A.; Pauly, M. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 2006, 142, 1380–1396. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.J.; Pfister, B.; Jenny, C.; Eicke, S.; Zeeman, S.C. Distinct Functions of STARCH SYNTHASE 4 Domains in Starch Granule Formation. Plant Physiol. 2018, 176, 566–581. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Weksler, A.; Friedman, H. Different Preclimacteric Events in Apple Cultivars with Modified Ripening Physiology. Front Plant Sci. 2017, 8, 1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zhu, B.Z.; Fu, D.Q.; Luo, Y.B. RIN transcription factor plays an important role in ethylene biosynthesis of tomato fruit ripening. J. Sci. Food Agric. 2011, 91, 2308–2314. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, J.; Zhu, X.Y.; Song, Y.; Li, Z.P.; Ren, G.D.; Zhou, X.; Kuai, B.K. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.; Yu, S.; Zhu, N.; Chen, Y.R.; Zhou, B.; Pan, Y.; Tzeng, D.; Fabi, J.P.; Argyris, J.; Garcia-Mas, J.; et al. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 2018, 4, 784–791. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, Q.; He, J.; Yang, X.; Cheng, P.; Khan, A.; Shen, W.; Song, Y.; Wang, S.; Ma, F.; Guan, Q. Transcriptomic Analysis Revealed the Discrepancy between Early-Ripening ‘Geneva Early’ and Late-Ripening ‘Hanfu’ Apple Cultivars during Fruit Development and Ripening. Horticulturae 2023, 9, 570. https://doi.org/10.3390/horticulturae9050570
Yue Q, He J, Yang X, Cheng P, Khan A, Shen W, Song Y, Wang S, Ma F, Guan Q. Transcriptomic Analysis Revealed the Discrepancy between Early-Ripening ‘Geneva Early’ and Late-Ripening ‘Hanfu’ Apple Cultivars during Fruit Development and Ripening. Horticulturae. 2023; 9(5):570. https://doi.org/10.3390/horticulturae9050570
Chicago/Turabian StyleYue, Qianyu, Jieqiang He, Xinyue Yang, Pengda Cheng, Abid Khan, Wenyun Shen, Yi Song, Shicong Wang, Fengwang Ma, and Qingmei Guan. 2023. "Transcriptomic Analysis Revealed the Discrepancy between Early-Ripening ‘Geneva Early’ and Late-Ripening ‘Hanfu’ Apple Cultivars during Fruit Development and Ripening" Horticulturae 9, no. 5: 570. https://doi.org/10.3390/horticulturae9050570





