Overexpression of RrGGP2 and RrDHAR Increases Ascorbic Acid Content in Tomato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Total DNA and RNA Extraction, cDNA Synthesis
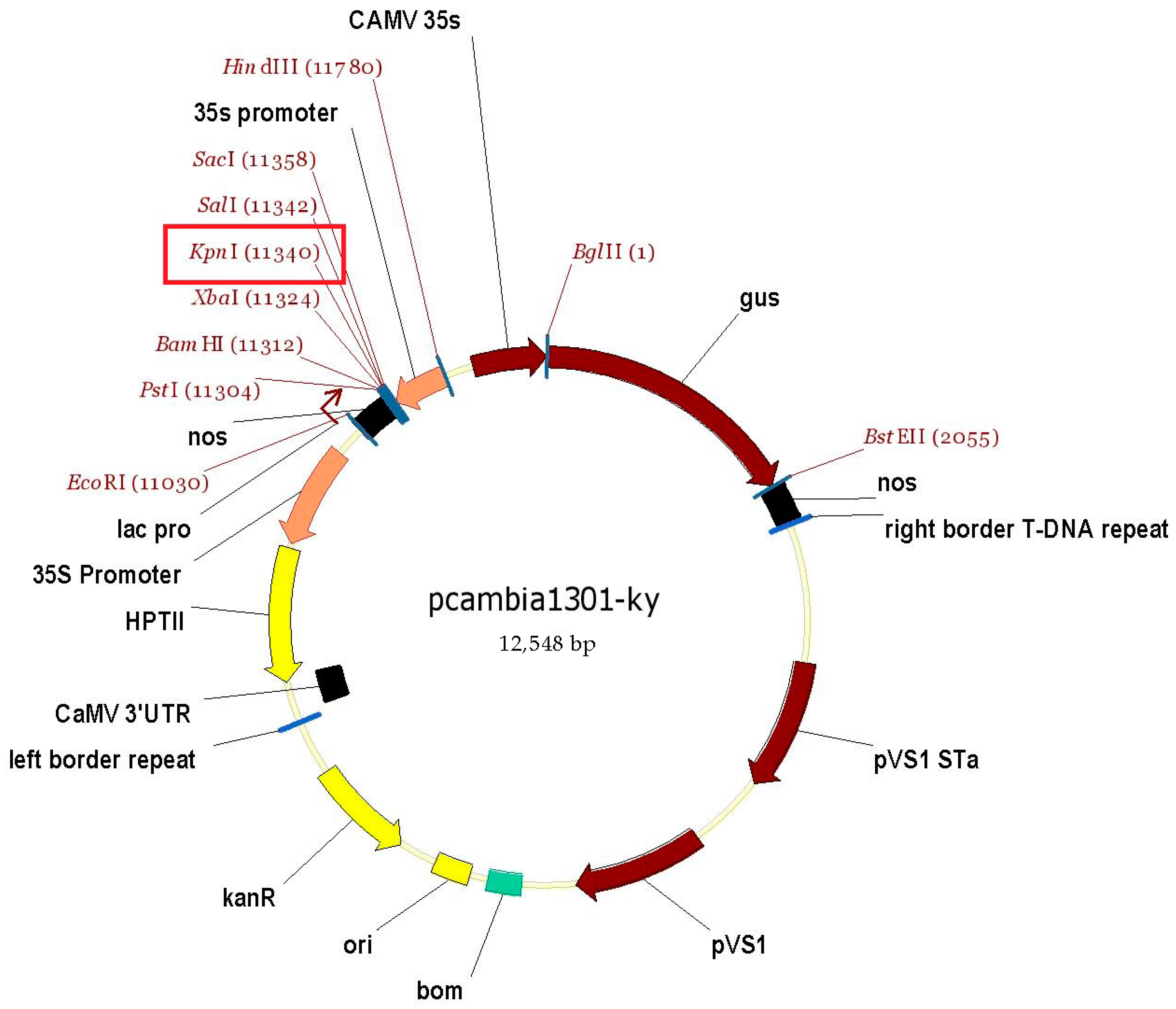
2.3. Cloning of Target Gene and Constructing of Overexpression Vector
2.4. Plant Transformation
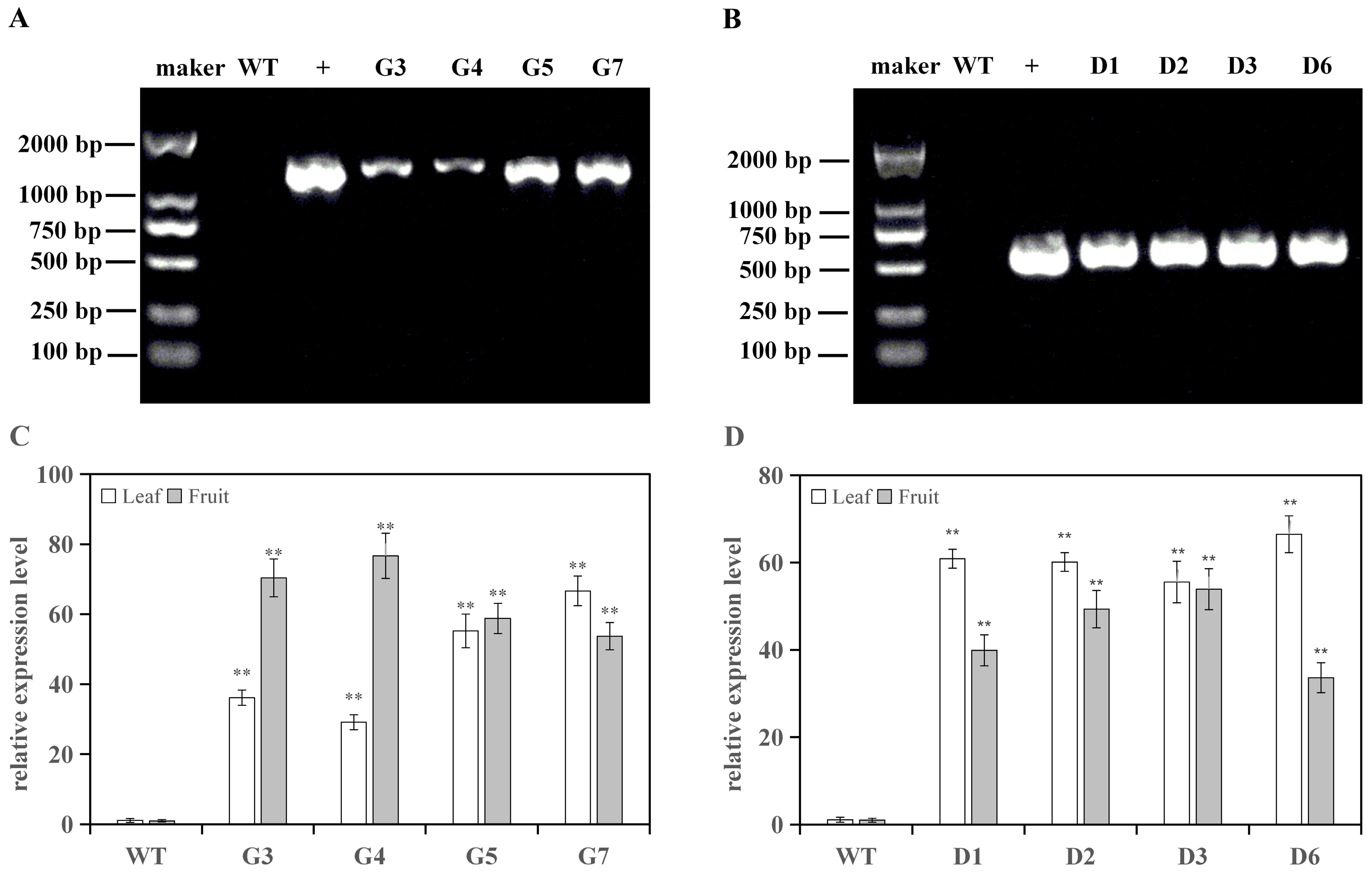
2.5. Transgenic Plants Identification and qRT-PCR Analysis
2.6. Determination of AsA and DHA Content
2.7. Statistical Analysis
3. Results
3.1. Positive Identification of Transgenic Tomato Lines
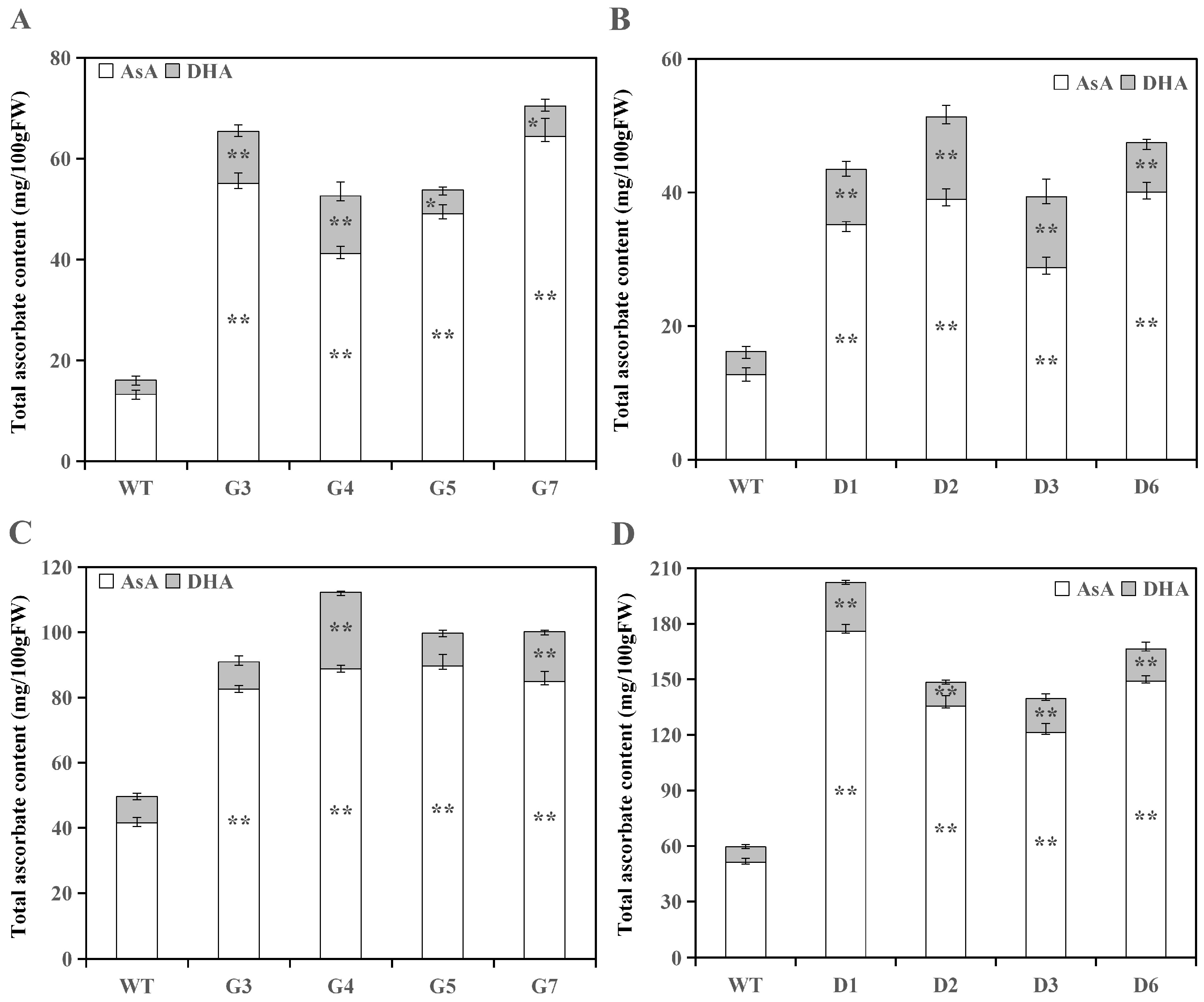
3.2. Analysis of Ascorbate Levels in Transgenic Lines
3.2.1. Analysis of Ascorbate Levels in Transgenic Tomato Fruits
3.2.2. Analysis of Ascorbate Levels in Transgenic Tomato Leaves
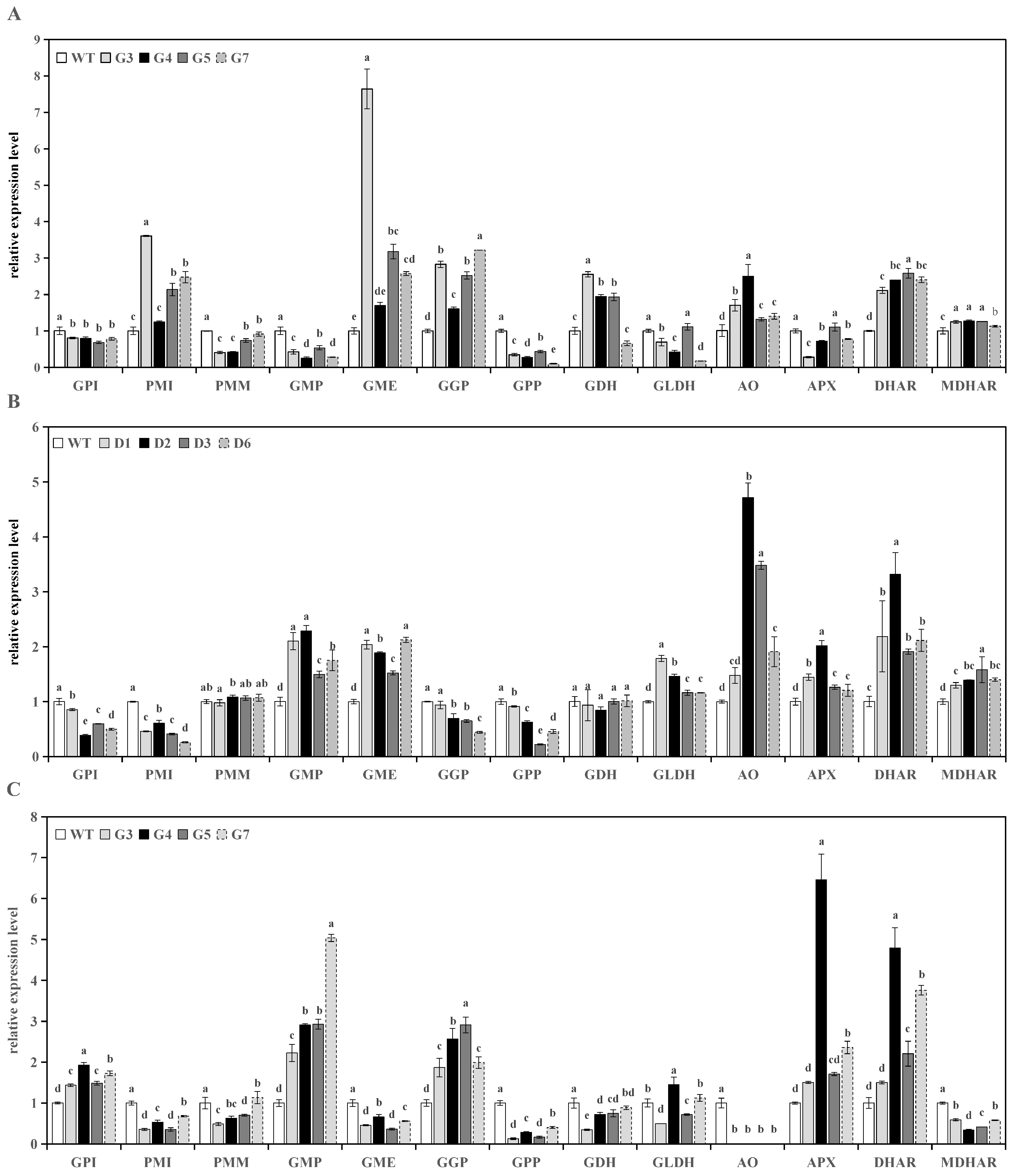
3.3. Analysis of Genes Expression in Transgenic Lines
3.3.1. Analysis of Genes Expression in Transgenic Tomato Fruits
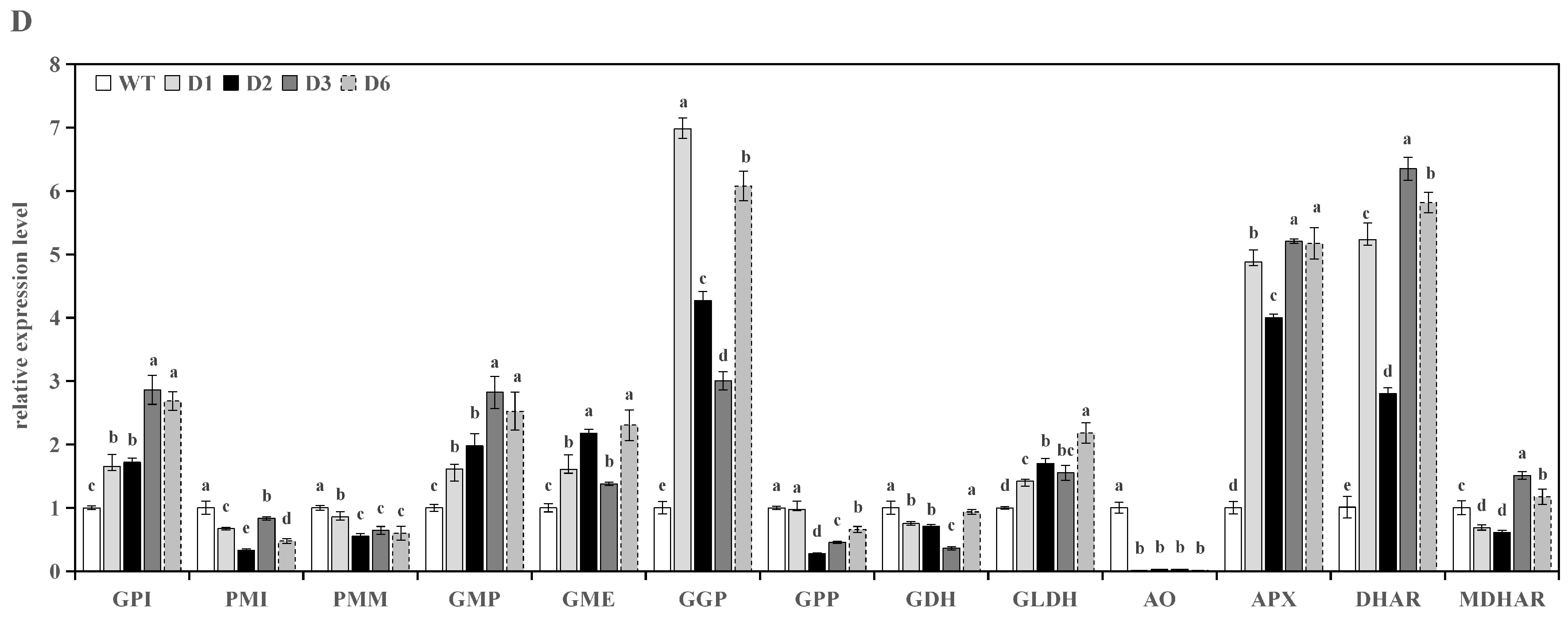
3.3.2. Analysis of Genes Expression in Transgenic Tomato Leaves
3.4. Correlation Analysis between Gene Expression and AsA Content
3.4.1. Correlation Analysis between Gene Expression and AsA Content in Transgenic Tomato Fruits
3.4.2. Correlation Analysis between Gene Expression and AsA Content in Transgenic Tomato Leaves
4. Discussion
4.1. Overexpressing RrGGP2 and RrDHAR in Tomato Indicates That RrGGP2 Is the Key Control Point of AsA Biosynthesis and Metabolism
4.2. Similarities and Differences for the Mechanism of AsA Accumulation in Tomato Fruit and Leaf
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smirnoff, N. Ascorbic acid: Metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 2000, 3, 229–235. [Google Scholar] [CrossRef]
- Broad, R.C.; Bonneau, J.P.; Hellens, R.P.; Johnson, A.A.T. Manipulation of Ascorbate Biosynthetic, Recycling, and Regulatory Pathways for Improved Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 1790. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2012, 64, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Pastori, G.M.; Kiddle, G.; Antoniw, J.; Bernard, S.; Veljovic-Jovanovic, S.; Verrier, P.J.; Noctor, G.; Foyer, C.H. Leaf Vitamin C Contents Modulate Plant Defense Transcripts and Regulate Genes That Control Development through Hormone Signaling. Plant Cell 2003, 15, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Viviani, A.; Fambrini, M.; Giordani, T.; Pugliesi, C. L-Ascorbic acid in plants: From biosynthesis to its role in plant development and stress response. Agrochimica 2021, 65, 151–171. [Google Scholar] [CrossRef]
- Barth, C.; Moeder, W.; Klessig, D.F.; Conklin, P.L. The Timing of Senescence and Response to Pathogens Is Altered in the Ascorbate-Deficient Arabidopsis Mutant Vitamin c-1. Plant Physiol. 2004, 134, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Lachapelle, M.Y.; Drouin, G. Inactivation dates of the human and guinea pig vitamin C genes. Genetica 2011, 139, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.; Shi, Q.; Yu, X. Ascorbic acid contents in transgenic potato plants overexpressing two dehydroascorbate reductase genes. Mol. Biol. Rep. 2011, 38, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Troesch, B.; Hoeft, B.; McBurney, M.; Eggersdorfer, M.; Weber, P. Dietary surveys indicate vitamin intakes below recommendations are common in representative Western countries. Br. J. Nutr. 2012, 108, 692–698. [Google Scholar] [CrossRef]
- Wheeler, G.L.; Jones, M.A.; Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 1998, 393, 5. [Google Scholar] [CrossRef]
- Agius, F.; González-Lamothe, R.; Caballero, J.; Munoz-Blanco, J.; Botella, M.A. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. myo -Inositol Oxygenase Offers a Possible Entry Point into Plant Ascorbate Biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Wolucka, B.A.; Van Montagu, M. GDP-Mannose 3′,5′-Epimerase Forms GDP-L-gulose, a Putative Intermediate for the de Novo Biosynthesis of Vitamin C in Plants. J. Biol. Chem. 2003, 278, 47483–47490. [Google Scholar] [CrossRef]
- Li, M.; Chen, X.; Wang, P.; Ma, F. Ascorbic Acid Accumulation and Expression of Genes Involved in Its Biosynthesis and Recycling in Developing Apple Fruit. J. Am. Soc. Hortic. Sci. 2011, 136, 231–238. [Google Scholar] [CrossRef]
- Bulley, S.M.; Rassam, M.; Hoser, D.; Otto, W.; Schunemann, N.; Wright, M.; MacRae, E.; Gleave, A.; Laing, W. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J. Exp. Bot. 2009, 60, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; An, H.M.; Yang, M. Overexpression of Rosa roxburghii l-galactono-1,4-lactone dehydrogenase in tobacco plant enhances ascorbate accumulation and abiotic stress tolerance. Acta Physiol. Plant. 2013, 35, 1617–1624. [Google Scholar] [CrossRef]
- Liu, F.H.; Wang, L.; Gu, L.; Zhao, W.; Su, H.Y.; Cheng, X.H. Higher transcription levels in ascorbic acid biosynthetic and recycling genes were associated with higher ascorbic acid accumulation in blueberry. Food Chem. 2015, 188, 399–405. [Google Scholar] [CrossRef]
- Mellidou, I.; Kanellis, A.K. Genetic Control of Ascorbic Acid Biosynthesis and Recycling in Horticultural Crops. Front. Chem. 2017, 5, 50. [Google Scholar] [CrossRef]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability: Role of GDP-l-Gal phosphorylase in ascorbate biosynthesis. Plant J. 2007, 52, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Maruta, T.; Yonemitsu, M.; Yabuta, Y.; Tamoi, M.; Ishikawa, T.; Shigeoka, S. Arabidopsis Phosphomannose Isomerase 1, but Not Phosphomannose Isomerase 2, Is Essential for Ascorbic Acid Biosynthesis. J. Biol. Chem. 2008, 43, 28842–28851. [Google Scholar] [CrossRef]
- Qian, W.Q.; Yu, C.M.; Qin, M.; Liu, X.; Zhang, A.M.; Johansen, I.E.; Wang, D.W. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the invoIvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J. 2007, 49, 399–413. [Google Scholar] [CrossRef]
- Conklin, P.L.; Norris, S.R.; Wheeler, G.L.; Williams, E.H.; Smimoff, N.; Last, R.L. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 4198–4203. [Google Scholar] [CrossRef]
- Laing, W.A.; Wright, M.A.; Cooney, J.; Bulley, S.M. The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an l -galactose guanyltransferase, increases leaf ascorbate content. Proc. Natl. Acad. Sci. USA 2007, 104, 9534–9539. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis thaliana VTC4 encodes L-galactose-1-phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 2006, 281, 15662–15670. [Google Scholar] [CrossRef] [PubMed]
- Torabinejad, J.; Donahue, J.L.; Gunesekera, B.N.; Allen-Daniels, M.J.; Gilkpy, G.E. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 2009, 150, 951–961. [Google Scholar] [CrossRef]
- Gatek, S.; Wheeler, G.L.; Smirnoff, N. Antisense suppression of L-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. Plant J. 2002, 30, 541–553. [Google Scholar] [CrossRef]
- Imai, T.; Karita, S.; Shiratori, G.; Hattori, M.; Nunome, T.; Oba, K.; Himi, M. L- galactono-gamma-1actone dehydrogenase from sweet potato: Purification and cDNA sequence analysis. Plant Cell Physiol. 1998, 39, 1350–1358. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawanob, N.; Badawic, G.H.; Kaminakaa, H.; Sanekatad, T.; Inanaga STanaka, K. 0verexpression of monodehydroascorbate reductase in tmnsgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- Yoshida, S.; Tamaoki, M.; Shikano, T.; Nakajima, N.; Ogawa, D.; Ioki, M.; Aono, M.; Kubo, A.; Kamada, H.; Inoue, Y.; et al. Cytosolic dehydroascorbate reducmse is impoaam for ozone tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 304–308. [Google Scholar] [CrossRef]
- Bulley, S.; Wright, M.; Rommens, C.; Yan, H.; Rassam, M.; Lin-Wang, K.; Andre, C.; Brewster, D.; Karunairetnam, S.; Allan, A.C.; et al. Enhancing ascorbate in fruits and tubers through over-expression of the l-galactose pathway gene GDP-l-galactose phosphorylase: Enhanced ascorbate in fruits and tubers. Plant Biotechnol. J. 2012, 10, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, F.; Liang, D.; Li, J.; Wang, Y. Ascorbate Biosynthesis during Early Fruit Development Is the Main Reason for Its Accumulation in Kiwi. PLoS ONE 2010, 5, e14281. [Google Scholar] [CrossRef]
- Mellidou, I.; Keulemans, J.; Kanellis, A.K.; Davey, M.W. Regulation of fruit ascorbic acid concentrations during ripening in high and low vitamin C tomato cultivars. BMC Plant Biol. 2012, 12, 239. [Google Scholar] [CrossRef]
- Gilbert, L.; Alhagdow, M.; Nunes-Nesi, A.; Quemener, B.; Guillon, F.; Bouchet, B.; Faurobert, K.; Gouble, B.; Page, D.; Garcia, V.; et al. GDP-d-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009, 60, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Zhang, Y.; Cai, X.; Gong, P.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 2011, 30, 389–398. [Google Scholar] [CrossRef]
- Koukounaras, A.; Mellidou, I.; Patelou, E.; Kostas, S.; Shukla, V.; Engineer, C.; Papaefthimiou, D.; Amari, F.; Chatzopoulos, D.; Mattoo, A.K.; et al. Over-expression of GGP1 and GPP genes enhances ascorbate content and nutritional quality of tomato. Plant Physiol. Biochem. 2022, 193, 124–138. [Google Scholar] [CrossRef]
- Huang, M.; Xu, Q.; Deng, X.X. L-Ascorbic acid metabolism during fruit development in an ascorbate-rich fruit crop chestnut rose (Rosa roxburghii Tratt). J. Plant Physiol. 2014, 171, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Yang, M.; An, H.M. Expression of GDP-L-galactose pyrophosphatase and its relationship with Ascorbate accumulation in Rosa roxburghii. Acta Hortic. Sin. 2014, 41, 1175–1182. [Google Scholar]
- Yan, Y.; Liu, Y.; Lu, M.; Lu, C.; Ludlow, R.A.; Yang, M.; Huang, W.; Liu, Z.; An, H. Gene expression profiling in Rosa roxburghii fruit and overexpressing RrGGP2 in tobacco and tomato indicates the key control point of AsA biosynthesis. Front. Plant Sci. 2023, 13, 1096493. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lu, M.; Rao, T.; Liu, Z.; Wu, X.; An, H. Comparative analysis of fruit metabolome using widely targeted metabolomics reveals nutritional characteristics of different Rosa roxburghii genotypes. Foods 2022, 11, 850. [Google Scholar] [CrossRef]
- Li, N.; Jiang, L.; Liu, Y.; Zou, S.; Lu, M.; An, H. Metabolomics Combined with Transcriptomics Analysis Revealed the Amino Acids, Phenolic Acids, and Flavonol Derivatives Biosynthesis Network in Developing Rosa roxburghii Fruit. Foods 2022, 11, 1639. [Google Scholar] [CrossRef]
- Wang, L.; Wei, T.; Zheng, L.; Jiang, F.; Ma, W.; Lu, M.; Wu, X.; An, H. Recent Advances on Main Active Ingredients, Pharmacological Activities of Rosa roxbughii and Its Development and Utilization. Foods 2023, 12, 1051. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, H.S.; An, H.M. Chloroplast DNA-based genetic variation of Rosa roxburghii in southwest China: Phylogeography and conservation implications. Hortic. Plant J. 2021, 7, 286–294. [Google Scholar] [CrossRef]
- Gong, L.S.; Lu, M.; An, H.M. Generation of Composite Rosa roxburghii Plants with Transgenic Roots by Agrobacterium-Mediated Transformation. Horticulturae 2022, 8, 1079. [Google Scholar] [CrossRef]
- Lu, M.; An, H.M.; Li, L.L. Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii tratt and leaf ascorbate metabolism genes. PLoS ONE 2016, 11, e0147530. [Google Scholar] [CrossRef]
- Chen, W.F.; Hu, T.X.; Ye, J.; Wang, B.; Liu, G.Z.; Wang, Y.; Yuan, L.; Li, J.M.; Li, F.M.; Ye, Z.B.; et al. CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the d-mannose/l-galactose pathway in tomato. Hortic. Res. 2020, 7, 200. [Google Scholar] [CrossRef]
- Munir, S.; Mumtaz, M.A.; Ahiakpa, J.K.; Liu, G.Z.; Chen, W.F.; Zhou, G.L.; Zheng, W.; Ye, Z.B.; Zhang, Y.Y. Genome-wide analysis of Myo-inositol oxygenase gene family in tomato reveals their involvement in ascorbic acid accumulation. BMC Genom. 2020, 21, 284. [Google Scholar] [CrossRef]
- Takahama, U.; Oniki, T. Regulations of peroxidase-dependent oxidation of phenolics in the apoplast of spinach leaves by ascorbate. Plant Cell Physiol. 1992, 33, 379–387. [Google Scholar]
- Li, G.; Lin, G.; Hu, Y.; Zhang, H.; Bai, J.; An, H.; Lu, M. Sequence polymorphisms of GGP and DHAR genes and their association with vitamin C content in R. roxburghii. Jiangsu Agric. Sci. 2020, 48, 63–69. [Google Scholar]
- Badejo, A.A.; Eltelib, H.A.; Fujikawa, Y.; Esaka, M. Genetic Manipulation for Enhancing Vitamin C Content in Tobacco Expressing Acerola (Malpighia glabra) GDP-L-galactose phosphorylase Gene. Hort. Environ. Biotechnol. 2009, 50, 329–333. [Google Scholar]
- Zhou, Y.; Tao, Q.C.; Wang, Z.N.; Fan, R.; Li, Y.; Sun, X.F.; Tang, K.X. Engineering ascorbic acid biosynthetic pathway in Arabidopsis leaves by single and double gene transformation. Biol. Plant. 2012, 56, 451–457. [Google Scholar] [CrossRef]
- Fenech, M.; Amorim-Silva, V.; del Valle, A.E.; Arnaud, D.; Ruiz-Lopez, N.; Castillo, A.G.; Smirnoff, N.; Botella, M.A. The role of GDP- l -galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol. 2021, 185, 1574–1594. [Google Scholar] [CrossRef]
- Wang, L.Y.; Meng, X.; Yang, D.Y.; Ma, N.N.; Wang, G.D.; Meng, Q.W. Overexpression of tomato GDP-l-galactose phosphorylase gene in tobacco improves tolerance to chilling stress. Plant Cell Rep. 2014, 33, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Laing, W.A.; Martinez-Sanchez, M.; Wright, M.A.; Bulley, S.M.; Brewster, D.; Dare, A.P.; Rassam, M.; Wang, D.; Storey, R.; Macknight, R.C. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 2015, 27, 772–786. [Google Scholar] [CrossRef]
- Broad, R.C.; Bonneau, J.P.; Beasley, J.T.; Roden, S.; Philips, J.G.; Baumann, U.; Hellens, R.P.; Johnson, A.A.T. Genome-wide identification and characterization of the GDP-L-galactose phosphorylase gene family in bread wheat. BMC Plant Biol. 2019, 19, 515. [Google Scholar] [CrossRef]
- Li, X.; Ye, J.; Munir, S.; Yang, T.; Chen, W.; Liu, G.; Zheng, W.; Zhang, Y. Biosynthetic Gene Pyramiding Leads to Ascorbate Accumulation with Enhanced Oxidative Stress Tolerance in Tomato. Int. J. Mol. Sci. 2019, 20, 1558. [Google Scholar] [CrossRef]
- Yang, D.Y.; Zhuang, K.Y.; Ma, N.N. Overexpression of SlGGP-LIKE gene enhanced the resistance of tomato to salt stress. Protoplasma 2023, 260, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Y.; Liu, R.R.; Zhang, C.Q.; Tang, K.X.; Sun, M.F.; Yan, G.H.; Liu, Q.Q. Manipulation of the Rice L-Galactose Pathway: Evaluation of the Effects of Transgene Overexpression on Ascorbate Accumulation and Abiotic Stress Tolerance. PLoS ONE 2015, 10, e0125870. [Google Scholar] [CrossRef]
- Ali, B.; Pantha, S.; Acharya, R.; Ueda, Y.; Wu, L.B.; Ashrafuzzaman, M.; Ishizaki, T.; Wissuwa, M.; Bulley, S.; Frei, M. Enhanced ascorbate level improves multi-stress tolerance in a widely grown indica rice variety without compromising its agronomic characteristics. J. Plant Physiol. 2019, 240, 152998. [Google Scholar] [CrossRef]
- Liu, X.; Xie, X.; Zhong, C.; Li, D. Comparative Transcriptome Analysis Revealed the Key Genes Regulating Ascorbic Acid Synthesis in Actinidia. Int. J. Mol. Sci. 2021, 22, 12894. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef]
- Imai, T.; Ban, Y.; Yamamoto, T.; Moriguchi, T. Ectopic overexpression of peach GDP-d-mannose pyrophosphorylase and GDP-d-mannose-3′,5′-epimerase in transgenic tobacco. Plant Cell Tissue Organ Cult. 2012, 111, 1–13. [Google Scholar] [CrossRef]
- Zhang, C.J.; Ouyang, B.; Yang, C.X.; Zhang, X.H.; Liu, H.; Zhang, Y.Y.; Zhang, J.H.; Li, H.X.; Ye, Z.B. Reducing AsA Leads to Leaf Lesion and Defence Response in Knock-Down of the AsA Biosynthetic Enzyme GDP-D-Mannose Pyrophosphorylase Gene in Tomato Plant. PLoS ONE 2013, 8, e61987. [Google Scholar] [CrossRef] [PubMed]
- Haroldsen, V.M.; Chi-Ham, C.L.; Kulkarni, S.; Lorence, A.; Bennett, A.B. Constitutively expressed DHAR and MDHAR influence fruit, but not foliar ascorbate levels in tomato. Plant Physiol. Biochem. 2011, 49, 1244–1249. [Google Scholar] [CrossRef]
- Fenech, M.; Amaya, I.; Valpuesta, V.; Botella, M.A. Vitamin C Content in Fruits: Biosynthesis and Regulation. Front. Plant Sci. 2019, 9, 2006. [Google Scholar] [CrossRef] [PubMed]
- Tyapkina, D.U.; Kochieva, E.Z.; Slugina, M.A. Identification and Analysis of VTC2 Homologs Encoding the Key Enzyme of L-Ascorbic Acid Biosynthesis in Tomato Species (Solanum Section of Lycopersicon). Dokl. Biochem. Biophys. 2018, 483, 374–378. [Google Scholar] [CrossRef]
- Muñoz, P.; Castillejo, C.; Gómez, J.A.; Miranda, L.; Lesemann, S.; Olbricht, K.; Petit, A.; Chartier, P.; Haugeneder, A.; Trinkl, J.; et al. QTL analysis for ascorbic acid content in strawberry fruit reveals a complex genetic architecture and association with GDP-L-galactose phosphorylase. Hortic. Res. 2023, 10, uhad006. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Hao, Z.; Huang, C. Molecular evolution of GDP-L-galactose phosphorylase, a key regulatory gene in plant ascorbate biosynthesis. AoB Plants 2020, 12, plaa055. [Google Scholar] [CrossRef]
- Li, J.; Liang, N.; Li, M.; Ma, F. Light and abiotic stresses regulate the expression of GDP-l-galactose phosphorylase and levels of ascorbic acid in two kiwifruit genotypes via light-responsive and stress-inducible cis-elements in their promoters. Planta 2013, 238, 535–547. [Google Scholar] [CrossRef]
- Zhang, S.X. Cloning, Analysis and Functional Verification of GGP Gene Promoter in Rosa roxburghii Tratt. Master’s Thesis, Guizhou University, Guizhou, China, 2018. [Google Scholar]
- Maruta, T. How does light facilitate vitamin C biosynthesis in leaves? Biosci. Biotechnol. Biochem. 2022, 86, 1173–1182. [Google Scholar] [CrossRef]
- Deslous, P.; Bournonville, C.; Decros, G.; Okabe, Y.; Mauxion, J.-P.; Jorly, J.; Gadin, S.; Brès, C.; Mori, K.; Ferrand, C.; et al. Overproduction of ascorbic acid impairs pollen fertility in tomato. J. Exp. Bot. 2021, 72, 3091–3107. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Li, C.; Yu, X. Enhanced ascorbic acid accumulation through overexpression of dehydroascorbate reductase confers tolerance to methyl viologen and salt stresses in tomato. Czech J. Genet. Plant Breed. 2012, 48, 74–86. [Google Scholar] [CrossRef]
- Huang, C.; He, W.; Guo, J.; Chang, X.; Su, P.; Zhang, L. Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J. Exp. Bot. 2005, 56, 3041–3049. [Google Scholar] [CrossRef]
- Suekawa, M.; Fujikawa, Y.; Inoue, A.; Kondo, T.; Uchida, E.; Koizumi, T.; Esaka, M. High levels of expression of multiple enzymes in the Smirnoff-Wheeler pathway are important for high accumulation of ascorbic acid in acerola fruits. Biosci. Biotechnol. Biochem. 2019, 83, 1713–1716. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free. Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Li, H.X.; Shu, W.B.; Zhang, C.J.; Zhang, W.; Ye, Z.B. Suppressed Expression of Ascorbate Oxidase Gene Promotes Ascorbic Acid Accumulation in Tomato Fruit. Plant Mol. Biol. Report. 2011, 29, 638–645. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Shu, W.; Zhang, C.; Ye, Z. RNA interference of a mitochondrial APX gene improves vitamin C accumulation in tomato fruit. Sci. Hortic. 2011, 129, 220–226. [Google Scholar] [CrossRef]
- Lee, Y.P.; Baek, K.H.; Lee, H.S.; Kwak, S.S.; Bang, J.W.; Kwon, S.Y. Tobacco seeds simultaneously over-expressing Cu/Zn-superoxide dismutase and ascorbate peroxidase display enhanced seed longevity and germination rates under stress conditions. J. Exp. Bot. 2010, 61, 2499–2506. [Google Scholar] [CrossRef]
- Wang, Y.; Wisniewski, M.; Meilan, R.; Cui, M.; Fuchigami, L. Transgenic tomato (Lycopersicon esculentum) overexpressing cAPX exhibits enhanced tolerance to UV-B and heat stress. J. Appl. Hortic. 2006, 8, 87–90. [Google Scholar] [CrossRef]
- Murgia, I.; Tarantino, D.; Vannini, C.; Bracale, M.; Carravieri, S.; Soave, C. Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J. 2004, 38, 940–953. [Google Scholar] [CrossRef]
- Xu, W.; Shi, W.; Ueda, A.; Takabe, T. Mechanisms of salt tolerance in transgenic Arabidopsis thaliana carrying a peroxisomal ascorbate peroxidase gene from barley. Pedosphere 2008, 18, 486–495. [Google Scholar] [CrossRef]
- Davey, M.W.; Montagu, M.V.; Inze, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.J.J.; Strain, J.J.; Favell, D.; Fletcher, J. Plant L-ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agr. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Potters, G.; Horemans, N.; Caubergs, R.J.; Asard, H. Ascorbate and dehydroascorbate influence cell cycle progression in a tobacco cell suspension. Plant Physiol. 2000, 124, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, J.J. Completing a pathway to plant vitamin C synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 9109–9110. [Google Scholar] [CrossRef] [PubMed]
- Harb, J.; Khraiwesh, B.; Streif, J.; Reski, R.; Frank, W. Characterization of blueberry monodehydroascorbate reductase gene and changes in levels of ascorbic acid and the antioxidative capacity of water soluble antioxidants upon storage of fruits under various conditions. Sci. Hortic. 2010, 125, 390–395. [Google Scholar] [CrossRef]
- Melino, V.J.; Soole, K.L.; Ford, C.M. Ascorbate metabolism and the developmental demand for tartaric and oxalic acids in ripening grape berries. BMC Plant Biol. 2009, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xiao, Y.; Chen, W.; Tang, K.; Zhang, L. Increased Vitamin C Content Accompanied by an Enhanced Recycling Pathway Confers Oxidative Stress Tolerance inArabidopsis. J. Integr. Plant Biol. 2010, 52, 400–409. [Google Scholar] [CrossRef]
| Experiment Purpose | Gene Abbreviation | Gene ID | Primer Sequences (5′-3′) |
|---|---|---|---|
| Homologous recombination clone | RrGGP2 | evm.model.Contig360.207 | F:GCGGGTCGACGGTACCATGCTGAAGATCAAGAGGGTGCGGGTCGACGTACCATGGGT R:TAGACATATGGGTACCTTACTGCAGAACGAGGCATTCTAGATATATGGGGTATTC |
| RrDHAR | evm.model.Contig319.146 | F:GCGGGTCGACGGTACCATGGCTCTTGAGGTTGCTGCGCGGTCGACGTACCATGGCTGC R:TAGACATATGGGTACCCTATTTGGGGTTGACTTTCGGTAGACATATGGGTACCCTATTTGGGGTTGACTTTCGG | |
| PCR identification | RrGGP2 | evm.model.Contig360.207 | F:ATGCTGAAGATCAAGAGGGTTCCCAC R:TTACTGCAGAACGAGGCATTCCTGT |
| RrDHAR | evm.model.Contig319.146 | F:ATGGCTCTTGAGGTTGCTGC R:CTATTTGGGGTTGACTTTCGGCT | |
| qRT-PCR | RrGGP2 | evm.model.Contig360.207 | F:AAGCTCCTGGCTGAGGTCTCT R:CCATCATCGCCACCACAAGCAAT |
| RrDHAR | evm.model.Contig319.146 | F:ACAAGCCCCAATGGTTTACAGA R:CCTCAGAATCCCAGCAAGCAC | |
| SlGPI | Solyc04g076090 | F:TGCTCTTCAAAAGCGTGTCC R:CGGCAATAAGTGCTCTGTCA | |
| SlPMI | Solyc02g086090 | F:TACATTGTGGTGGAACGAGGA R:ACCCCATTTGGCAAGAACAG | |
| SlPMM | Solyc05g048760 | F:TTTACCCTCCATTACATTGCTGA R:CTTCTTGACTACAGTTTCTCCCA | |
| SlGMP | Solyc03g096730 | F:AAACCTGAAATCGTGATGTGAGA R:TGAAGAAGAGGAGAACTGGAAAC | |
| SlGME | Solyc01g097340 | F:AATCCGACTTCCGTGAGCC R:CTGAGTTGCGACCACGGAC | |
| SlGGP | Solyc06g073320 | F:GAAATCTGGTCTGTTCCTCTGTGA R:TTCACACACCAACTCCACATTACA | |
| SlGPP | Solyc04g014800 | F:AGCCGCTACAAACCCTCATCT R:TGTCCGCTTTCCATCTCCTAT | |
| SlGDH | Solyc01g106450 | F:CTTCTTACTGAGGCTGGTGGTC R:AACCTCTTTAACAGACTTCATCCC | |
| SlGLDH | Solyc10g079470 | F:ATTGAGGTTCCCAAGGACATAG R:ATGTTATTAGATAGGATGCGGTTT | |
| SlAO | Solyc04g054690 | F:AGGATGGCTCAGAGTGTT R:ATCAGGTAAGGCGTATGG | |
| SlAPX | Solyc06g005150 | F:TGGAGCCCATTAGGGAGCA R:GCCAGGGTGAAAGGGAACAT | |
| SlDHAR | Solyc05g054760 | F:CCTACCTTCGTCTCATTTCCG R:TGAACAAACATTCTGCCCATT | |
| SlMDHAR | Solyc09g009390 | F:GGTGATGTTGCCACTTTTCCTTT R:CGACAGACTTCCCTTGCTCACT | |
| Actin | Solyc11g005330 | F:GTCCTCTTCCAGCCATCCA R:ACCACTGAGCACAATGTTACCG |
| Correlation Coefficient | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GPI | PMI | PMM | GMP | GME | GGP | GPP | GDH | GLDH | AO | APX | DHAR | MDHAR | ||
| RrGGP2 | AsA | −0.802 | 0.739 | −0.328 | −0.854 | 0.521 | 0.951 * | −0.938 * | 0.171 | −0.574 | 0.270 | −0.416 | 0.848 | 0.603 |
| DHA | −0.270 | 0.228 | 0.030 | −0.708 | 0.361 | 0.097 | −0.55 | 0.617 | −0.500 | 0.953 * | −0.699 | 0.445 | 0.697 | |
| RrDHAR | AsA | −0.789 | −0.826 | 0.485 | 0.878 | 0.965 ** | −0.664 | −0.433 | −0.397 | 0.555 | 0.481 | 0.634 | 0.83 | 0.677 |
| DHA | −0.806 | −0.526 | 0.636 | 0.762 | 0.540 | −0.447 | −0.600 | −0.637 | 0.444 | 0.910 * | 0.819 | 0.874 | 0.819 | |
| Correlation Coefficient | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GPI | PMI | PMM | GMP | GME | GGP | GPP | GDH | GLDH | AO | APX | DHAR | MDHAR | ||
| RrGGP2 | AsA | 0.844 | −0.869 | −0.435 | 0.684 | −0.882 * | 0.885 * | −0.943 ** | −0.521 | 0.002 | −0.990 ** | 0.463 | 0.626 | −0.952 ** |
| DHA | 0.856 | −0.091 | −0.039 | 0.452 | −0.007 | 0.455 | −0.239 | 0.097 | 0.856 | −0.423 | 0.965 ** | 0.962 ** | −0.623 | |
| RrDHAR | AsA | 0.457 | −0.658 | −0.501 | 0.468 | 0.663 | 0.956 * | −0.214 | −0.275 | 0.629 | −0.906 * | 0.872 | 0.718 | −0.265 |
| DHA | 0.379 | −0.180 | −0.106 | 0.326 | 0.203 | 0.816 | 0.147 | −0.366 | 0.274 | −0.697 | 0.761 | 0.765 | −0.066 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Rao, T.; Ludlow, R.A.; Yan, Y.; Lu, M.; An, H. Overexpression of RrGGP2 and RrDHAR Increases Ascorbic Acid Content in Tomato. Horticulturae 2023, 9, 587. https://doi.org/10.3390/horticulturae9050587
Liu Z, Rao T, Ludlow RA, Yan Y, Lu M, An H. Overexpression of RrGGP2 and RrDHAR Increases Ascorbic Acid Content in Tomato. Horticulturae. 2023; 9(5):587. https://doi.org/10.3390/horticulturae9050587
Chicago/Turabian StyleLiu, Zeyang, Tianzhi Rao, Richard A. Ludlow, Yali Yan, Min Lu, and Huaming An. 2023. "Overexpression of RrGGP2 and RrDHAR Increases Ascorbic Acid Content in Tomato" Horticulturae 9, no. 5: 587. https://doi.org/10.3390/horticulturae9050587
APA StyleLiu, Z., Rao, T., Ludlow, R. A., Yan, Y., Lu, M., & An, H. (2023). Overexpression of RrGGP2 and RrDHAR Increases Ascorbic Acid Content in Tomato. Horticulturae, 9(5), 587. https://doi.org/10.3390/horticulturae9050587




