Abstract
The Trihelix is a plant-specific transcription factor family and has critical roles in plant growth and development and stress resistance. There is less information about Trihelix transcription factor genes and their potential functions in strawberries (Fragaria vesca). In the present study, we performed a detailed bioinformatics analysis of the Trihelix family in strawberries including physicochemical properties, chromosomal location, exon–intron distribution, domain arrangement, and subcellular localization. Thirty Trihelix family members were identified and divided into five subfamilies. The expression of FvTrihelix genes in different tissues/organs, i.e., root, stolon, leaf, flower, and fruit, was measured in strawberries after infection with Colletotrichum. gloeosporioides and foliar applications of salicylic acid (SA) and jasmonic acid (JA). Most of the genes showed differential expression responses following C. gloeosporioides infection and hormone treatments (SA and JA), suggesting critical roles in disease resistance and hormonal signaling pathways. As anticipated, the ectopic expression of FvTrihelix6 in Arabidopsis thaliana increased resistance against Colletotrichum. higginsianum infection. FvTrihelix6 protein was localized in the nucleus. We surmise that FvTrihelix6 enhances resistance against pathogens through the SA and JA signaling pathways. This study provides novel insights into the strawberry Trihelix transcription factor genes and new candidates for disease-resistance breeding of strawberries.
1. Introduction
Transcription factors (TFs) control the expression of genes by adhering to certain DNA sequences and form complex structures in the promoter regions of target genes. The Trihelix gene family was one of the earliest transcription factor families discovered in plants. Trihelix TFs possess a highly conserved triple helix structure. The Trihelix gene family is also known as the GT factor family because this unique domain can bind to light-responsive GT elements. GT protein domains have highly conserved amino acid sequences.
Pea (Pisum sativum) was identified as having the first Trihelix transcription factor [1]. Most recently, research has led to the characterization of the Trihelix transcription factor family in other species. Trihelix family members have been reported in Arabidopsis thaliana (30 family members), Oryza sativa (41), Chrysanthemum morifolium (20), Solanum lycopersicum (36), Glycine max (Linn.) Merr. (71), Osmanthus fragrans (56), Sorghum bicolor (40), Chenopodium quinoa (47), Populus trichocarpa (56), and Ananas comosus (L.) Merr (23) [2,3,4,5,6,7,8,9,10,11]. In most species, such as A. thaliana, rice, wheat, and tree peony, the Trihelix family has been divided into five subfamilies: GT-1, GT-2, GTγ, SH4, and SIP1 [6,12,13]. In some species, more than five Trihelix subfamilies have been reported. In tomato and soybean, for example, Trihelix members have been divided into six subfamilies [2,7], and seven subfamilies have been identified in sorghum. The identification of additional subfamilies suggests that these genes may perform unique functions [8]. In the “Beni Hoppe” strawberry, FaGT-2 plays an important role in coping with salt, drought, cold, and other abiotic stress [14].
Studies of the Trihelix family of transcription factors first concentrated on the control of genes that respond to light. In addition, Trihelix genes have been involved in plant growth and development, such as early embryonic development and stomatal development [15,16], and the regulation of cell wall formation [17]. Trihelix TFs have been found to be essential for mitigating biotic and abiotic stressors. The OsGTγ-2 gene in rice is an important positive regulator against salt stress [18]. ShCIGT enhances cold and drought resistance in cultivated tomatoes [19], while GhGT26 improves tolerance to salt stress in transgenic A. thaliana plants [20].
Trihelix TFs have been implicated in the response to pathogen stress [5]. PTI is the immune system of plants in the process of being attacked by pathogens, which are triggered by microbial patterns through local pattern recognition receptors (PRRs) on the cell surface [21]. ASR3 of the SH4 subfamily has been shown to have a negative regulatory effect on pattern-triggered immunity (PTI) in A. thaliana. AlTF1 of the SIP1 subfamily interacts with ASR3 to co-regulate the plant immune response against pathogens. ASR3 is a transcriptional repressor located downstream of MRK4 that can finetune the transcription of immunity genes [22]. GTL1 of the GT-2 subfamily also plays a vital part in plant immunity. GTL1 coordinates related genes involved in salicylic acid (SA) metabolism, transport, and response, and is an important part of the MPK4 pathway that positively regulates bacterial-triggered immunity and SA homeostasis [23]. In maize, knockdown of the ZmGT-3b gene has resulted in significant up-regulation of many defense-related genes, increased cell wall content, and increased resistance to Fusarium graminearum [24]. In Populus trichocarpa, all members of the Trihelix gene family were shown to be strongly expressed in response to treatments such as SA, methyl jasmonate (MeJA), and pathogen infection, and inhibition of PtrGT10 resulted in increased reactive oxygen species’ scavenging capacity and reduced cell death [4]. The expression of SCaM-4 by pathogens in soybean and A. thaliana is significantly influenced by the interaction between the GT-1 cis-element and GT-1-like transcription factor [25]. Further, after infection with the blast fungus (Magnaporthe grisea), the expression of rml1 in rice was significantly up-regulated [26]. Despite extensive characterization of the Trihelix TFs in plants, to our knowledge, the Trihelix family has not been characterized in fruit trees.
The strawberry (Fragaria × ananassa) harboring (2 n = 8 x = 56) and genome size of the strawberry genus, ∼240 Mb, is a perennial herb that contains nutrients such as minerals, vitamins, fatty acids, and dietary fiber. The phenolic substances contained in strawberries can prevent cancer and cardiovascular and other diseases [27,28]. Strawberries are one of the most widely grown fruit crops in the world. However, the production and cultivation of strawberries are often exposed to various stresses such as salt, drought, and both fungal and bacterial pathogens. Anthracnose is one of the most devastating diseases caused by several species of fungi in the genus Colletotrichum that hinders strawberry production [29]. During growth and development, it will affect the root, fruit, leaf, stolon, and other strawberry organs [30]. So far, little is known about the strawberry TTF genes, especially concerning their responses to common stresses.
Here, TTF genes were identified in the strawberry and detailed bioinformatic analyses were performed. The identified genes were divided into subfamilies based on phylogenetic analysis. Expression analysis of genes against C. gloeosporioides infection and treatments of SA and JA were calculated. Based on the results, FvTrihelix6 was selected and functionally characterized. Ectopic expression of FvTrihelix6 in A. thaliana enhanced resistance against C. higginsianum via modulating the SA and JA signaling pathways. This study provides the groundwork for the functional characterization of strawberry TTF genes.
2. Materials and Methods
2.1. Plant Materials and Seedlings Treatment
The plant materials F. vesca ssp. accession Hawaii 4 (National Clonal Germplasm Repository accession #PI551572) and A. thaliana (Col-0) were collected from the School of Horticulture, Fujian Agricultural and Forestry University. Incubators were used to grow strawberries at 85% humidity and 23–25 °C temperature. For 4 weeks, A. thaliana was grown under controlled conditions with a dark cycle at 20–25 °C, 12 h light, and 70% RH. Three technical replicates of each independent experiment were performed, with six plants per replicate. Two strains of the pathogen, C. gloeosporioides and C. higginsianum, were prepared as spore suspensions at a concentration of 1 × 106/mL. Approximately 2 mL of C. gloeosporioides spore suspension was sprayed on strawberries, and samples were collected at 0, 3, 6, 12, and 24 h post inoculation (hpi). RNA samples were placed in a −80 °C refrigerator for cryopreservation. C. higginsianum was cultured and inoculated according to previous methods [31,32]. C. higginsianum spore suspension was sprayed on wild-type and transgenic A. thaliana to assess the biological activity of FvTrihelix6 at a concentration of 1 × 106/mL against anthracnose. Samples were collected at 0, 6, 12, 24, and 48 hpi and stored at −80 °C. For exogenous hormone treatment, 1.5 mL of either 50 mM methyl jasmonate (MeJA) or 5 mM salicylic acid (SA) solution was sprayed on strawberry leaves, and distilled water was used as a control. RNA extraction was performed at 0, 3, 6, 12, and 24 hpt and stored at −80 °C. RNA from roots, stolons, leaves, flowers, and fruits of strawberries was sampled and extracted. The obtained samples were immediately frozen in liquid nitrogen and kept at −80 °C. For each experiment, three biological and technical replicates were used.
2.2. Identification of Trihelix Genes in the F. vesca Genome
The amino acid sequences of the Trihelix genes from O. sativa and A. thaliana were retrieved from PlantTFDB (http://planttfdb.gao-lab.org, accessed on 20 April 2023). From the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html, accessed on 20 April 2023), the genomic sequence and gene annotation files of F. vesca V4.0.a2 were downloaded, while the presence of an intact Myb/SANT-like protein domain in all putative Trihelix genes was checked through TBtools and the conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 20 April 2023). After eliminating redundant genes, a total of 30 FvTrihelix genes were selected for further study. The physicochemical properties, instability index, amino acid count, molecular mass, and isoelectric point of the FvTrihelix genes were obtained using the ProtParam tool in the ExPASy database (https://web.expasy.org/protparam/, accessed on 20 April 2023), while the Plant-mPLoc Server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 20 April 2023) was used for the prediction of the subcellular position of FvTrihelix proteins.
2.3. Phylogenetic Analysis, Gene Structure, Motif Analysis, and Multiple Sequence Alignment
The phylogenetic tree among TTF genes of A. thaliana, F. vesca, and O. sativa was generated by MEGA7.0 using the maximum likelihood (ML) method with 1000 bootstrap values. The conserved motifs of TTF members were identified with a limit of 10 motifs through MEME (http://meme-suite.org/tools/meme, accessed on 20 April 2023), whereas the gene structure map of TTF genes was developed through TBtools [33].
2.4. Chromosomal Distribution and Promoter Analysis
The genome annotation files (https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/, accessed on 20 April 2023) of F. vesca were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/, accessed on 20 April 2023). TBtools software was used to show the chromosomal locations of the genes. Putative promoter sequences (2kb upstream of transcription start sites) of all TTF genes were obtained from the strawberry genome database. PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 20 April 2023) was used for cis-element analysis.
2.5. RNA Extraction and PCR Analysis
Reverse transcription and RNA extraction were carried out using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) and Prime ScriptTM RT Kit (Takara, Dalian, China), respectively. Quantitative Real-Time PCR was conducted using TB Green® Premix Ex TaqTM II kit (Takara, Dalian, China). Each reaction mixture’s total volume was 10 μL and consisted of 5 μL TB Green® Premix Ex TaqTM II, 0.4 μL ROX Reference Dye, 0.8 μL of each primer, 2 μL sterile water, and 1 μL cDNA. The reaction conditions were as follows: 95 °C for 30 s, 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 15 s, for a total of 38 cycles. The strawberry FvActin gene (GenBank accession number: AB116565) was used as an internal reference gene and each reaction was carried out with three biological and technical replicates. Primer sequences information is shown in Table S1.
2.6. Plasmid Construction and Transformation
RNA was extracted from the strawberries and reverse transcribed into cDNA using the PrimerScriptTM II cDNA synthesis kit (Takara Bio Inc., Dalian, China). The product was ligated with pMD18-T (Takara Bio Inc., Dalian, China). The samples were sequenced and analyzed by FuZhou ShangYa BioInc. (FuZhou, 155 China). FvTrihelix6 was transformed into the target vector pCMBIA1300-HA, and the constructed pCMBIA1300-HA-FvTrihelix6 vector was transformed into Agrobacterium tumefaciens GV3101. The floral dip method was used to transform FvTrihelix6-GV3101 into A. thaliana [34]. The inflorescences of all the Arabidopsis plants were dipped for a few seconds into the 5% sucrose solution containing 0.05% (v/v). Silwet L-77 and resuspended Agrobacterium cells carrying 35S::construct FvTrihelix6. The transformed A. thaliana plants were grown on a solid MS medium supplemented with 50 mg/L hygromycin and, at maturity, seeds were collected and grown for the next generations [35].
2.7. Subcellular Localization of FvTrihelix6
The open reading frame (ORF) region of FvTrihelix6 was amplified using specific primers FvTrihelix6-GFP-F and FvTrihelix6-GFP-R and restriction sites (BamH I and Kpn I). The resultant product was inserted into the pGFPc vector. The sequence-verified fusion vector was transformed into onion epidermal cells with A. tumefaciens GV3101. The empty vector was also transformed as a control. After two days of dark culture, the GFP (Green Fluorescent Protein) fluorescence response in the onion epidermis was observed using laser scanning confocal microscopy (OLYMPUS IX83-FV3000) [36].
2.8. Cloning of FvTrihelix6 Promoter, GUS Protein Staining, and GUS Activity Assay
The bacterial β-glucuronidase (GUS) gene is often introduced into plants as a reporter gene fused to a promoter because of its advantages over other reporter genes [37]. The promoter fragments of FvTrihelix6 were amplified using specific primers P-FvTrihelix6-GUS-F and P-FvTrihelix6-GUS-R containing EcoR I and Pst I restriction sites, and the product was incorporated into the GUS reporter plasmid to create the pFvTrihelix6::GUS vector. The pCaMV35S::GUS and pC0380::GUS vectors were used as positive and negative controls, respectively. The fusion vectors pFvTrihelix6::GUS, pCaMV35S::GUS, and pC0380::GUS were transformed into A. tumefaciens GV3101, resuspended, and then vacuum-infiltrated into tobacco (Nicotiana tabacum) leaves. Before infiltration, tobacco leaves were kept in freshly prepared X-Gluc staining solution for two days at 25 °C in low light. The leaves were removed after sufficient staining, submerged in 90% ethanol, and placed in boiling water until the green color faded. This was repeated 3–5 times using 8–10 tobacco leaves each time [36,38]. Agrobacterium suspension containing the pFvTrihelix6::GUS fusion vector was injected. The Agrobacterium suspension contained the positive control pCaMV35S::GUS, and SA into the back of the well-grown tobacco leaves, which were then cultured in darkness at 25 °C for two days. The leaf samples were collected and ground into powder, and the GUS enzyme activity was measured using the microplate reader (TECAN M200 PRO).
2.9. Statistical Analyses
At least three independent replicates were used to generate averages and standard deviations in all trials. The statistical analysis was performed with the help of SPSS 21.0. The differences in mean expression levels were assessed with the Student’s t-test. Significant differences were denoted by the symbols * p < 0.05 and ** p < 0.01, respectively.
3. Results
3.1. Identification of Strawberry TTF Members
Thirty FvTrihelix family members were identified from the strawberry genome. The genes were named based on their chromosomal locations, i.e., FvTrihelix1 to FvTrihelix30. Physicochemical properties, instability index, isoelectric point (pI), molecular weight (MW), amino acid sequences, and other related information is shown in Table 1 and Data S1. The FvTrihelix genes’ coding sequences ranged in length from 684 to 2871 bp, the pI values ranged from 4.68 to 9.78, the MW values ranged from 25.75 to 99.00 kDa, and the instability index varied from 35.38 to 72.34. Twenty-nine FvTrihelix genes were supposed to reside in the nucleus. However, there were exceptions. For example, FvTrihelix21 was expected to be in the chloroplast, whereas FvTrihelix25 and FvTrihelix30 were predicted to be in the chloroplast and nucleus and FvTrihelix14 was predicted to be localized in the chloroplast and cytoplasm.

Table 1.
Specifics of the Trihelix gene family found in Fragaria vesca.
3.2. The Distribution of FvTrihelix Genes across Chromosomes and Evolutionary Relationships
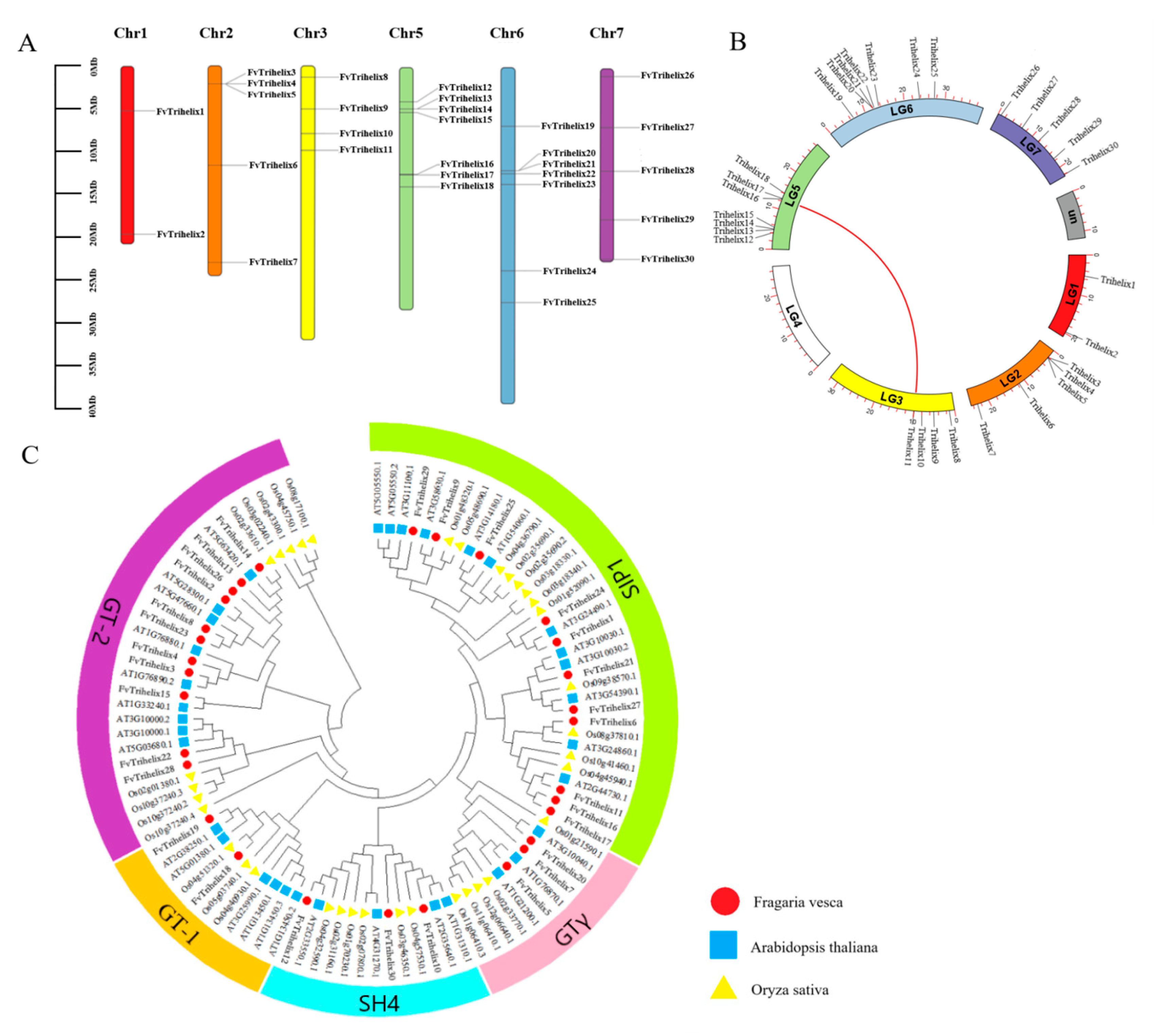
The 30 FvTrihelix genes were found to be unevenly distributed across 6 chromosomes of the strawberry (Fragaria vesca), with no genes on chromosome 4. The highest FvTrihelix genes were found on chromosomes 5 and 6, and the lowest were found on chromosome 1. Clusters of FvTrihelix genes were found on chromosomes 2, 5, and 6, indicating possible origins via duplication events (Figure 1A, Table 1). Studying gene replication events can provide further insights into the evolution of species. Four tandem (FvTrihelix3/FvTrihelix4/FvTrihelix5, FvTrihelix13/FvTrihelix14, FvTrihelix16/FvTrihelix17, FvTrihelix20/FvTrihelix21) and only one pair of segmental duplication (FvTrihelix11 and FvTrihelix16) was identified among FvTrihelix genes (Figure 1B). To explore the evolutionary relationships among different species, the phylogenetic tree was constructed among Trihelix members of A. thaliana, O. sativa, and F. vesca. The phylogenetic tree was constructed using MEGA7 with the maximum likelihood method (ML) with default parameters. All genes were divided into five subfamilies (GT-1, GT-2, SIP1, SH4, and GTγ) based on their phylogenetic relationships (Figure 1C).
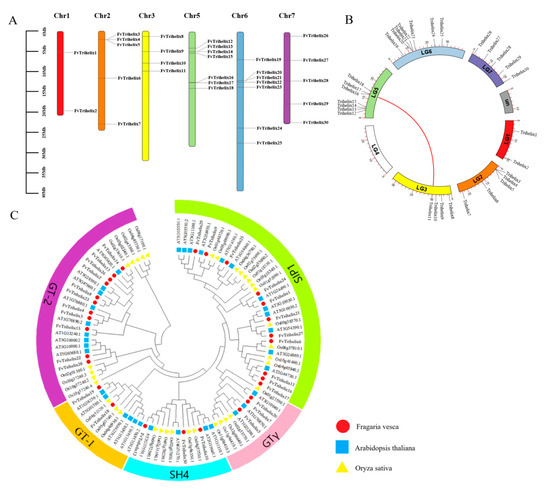
Figure 1.
Phylogenetic linkages and analysis of collinearity. (A) Distribution of FvTrihelix genes across chromosomes. A vertical scale denotes chromosome size (Mb). (B) Tandem and segmental duplications in the FvTrihelix family. The arc length represents the length of each chromosome and the red line indicates the duplicated Trihelix gene pair. (C) Phylogenetic tree among TTF genes of O. sativa, A. thaliana, and F. vesca. The red circle, blue square, and yellow triangle denotes O. sativa, A. thaliana, and F. vesca genes, respectively. Different subfamilies are represented with different colors.
3.3. Phylogenetic and Gene Structure Analysis
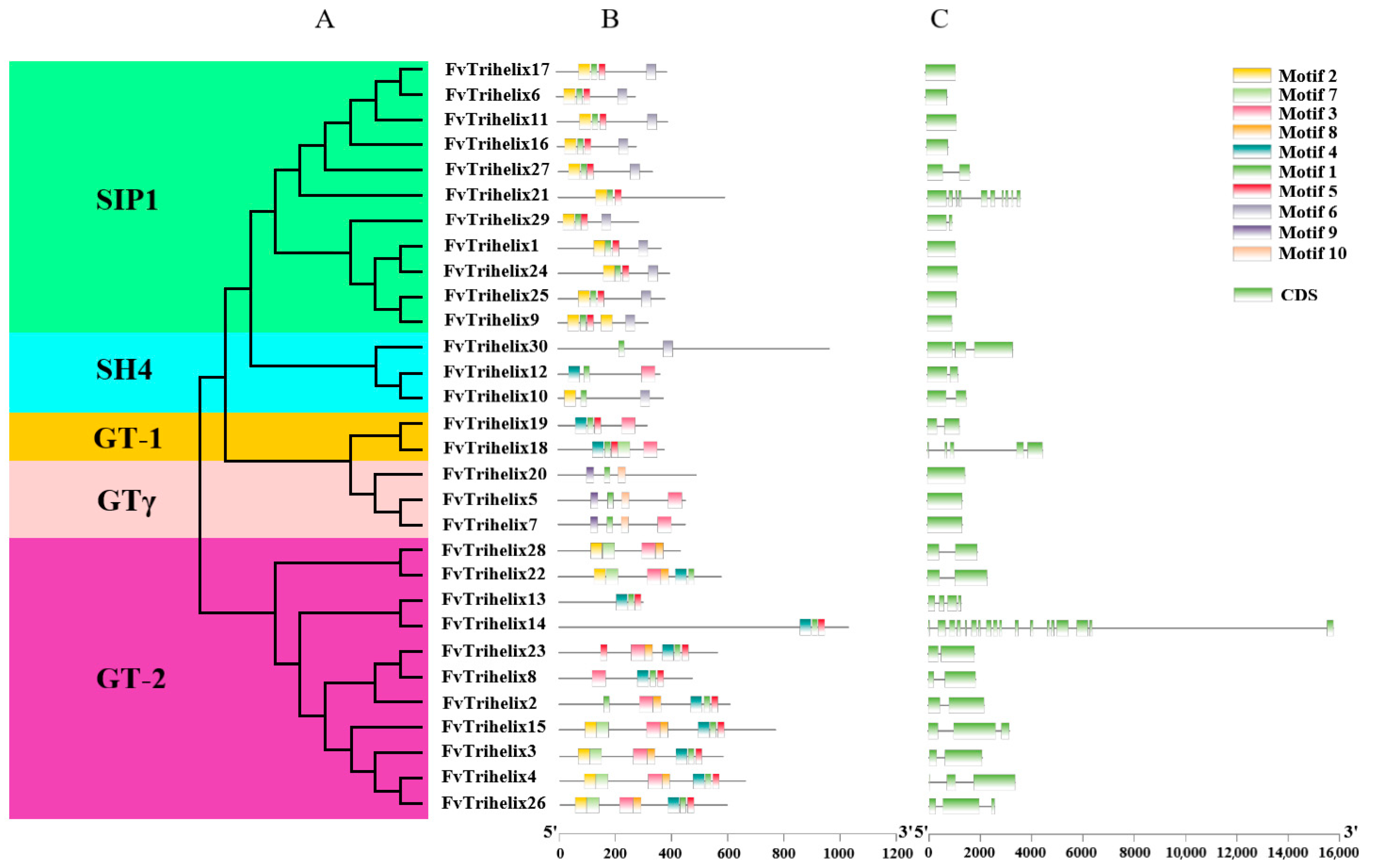
Thirty FvTrihelix genes were unevenly distributed among the five subfamilies. For example, there were 2, 11, 11, 3, and 3 genes in GT-1, GT-2, SIP1, SH4, and GTγ, respectively. The MEME suite was used to identify the conserved motifs in the FvTrihelix proteins (Figure 2B). The conserved motifs within each subfamily were largely similar. Except for FvTrihelix21, all other members of the SIP1 subfamily contained similar motifs (motifs 1, 2, 5, and 6). Similarly, the three FvTrihelix members in the SH4 subfamily contained motif 1, though other motifs were not shared among all three members. There were 2 members of the GT-1 subfamily that contained similar motifs (1, 3, 4, and 5). All members of the GT-2 subfamily contained motif 3 except FvTrihelix13 and FvTrihelix14. Moreover, with the exception of FvTrihelix28, all GT-2 family members contained motif 1 and motif 4. This indicates that different motifs may be associated with unique functions within each subfamily. Nineteen FvTrihelix genes contained more than one intron (Figure 2C).
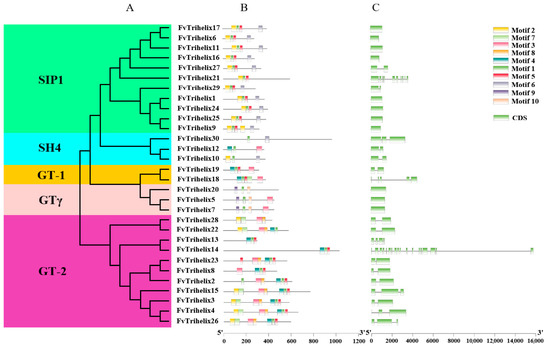
Figure 2.
Phylogenetic, conserved motif, and gene structure analysis of FvTrihelix family members. (A) Phylogenetic tree of FvTrihelix genes. Green indicates subfamily SIP1, blue indicates subfamily SH4, orange indicates subfamily GT-1, pink indicates subfamily GTγ, and purple indicates subfamily GT-2. (B) Distributions of motifs in FvTrihelix genes. (C) Exon–intron distributions and coding domains of FvTrihelix genes.
3.4. Cis-Acting Elements Analysis
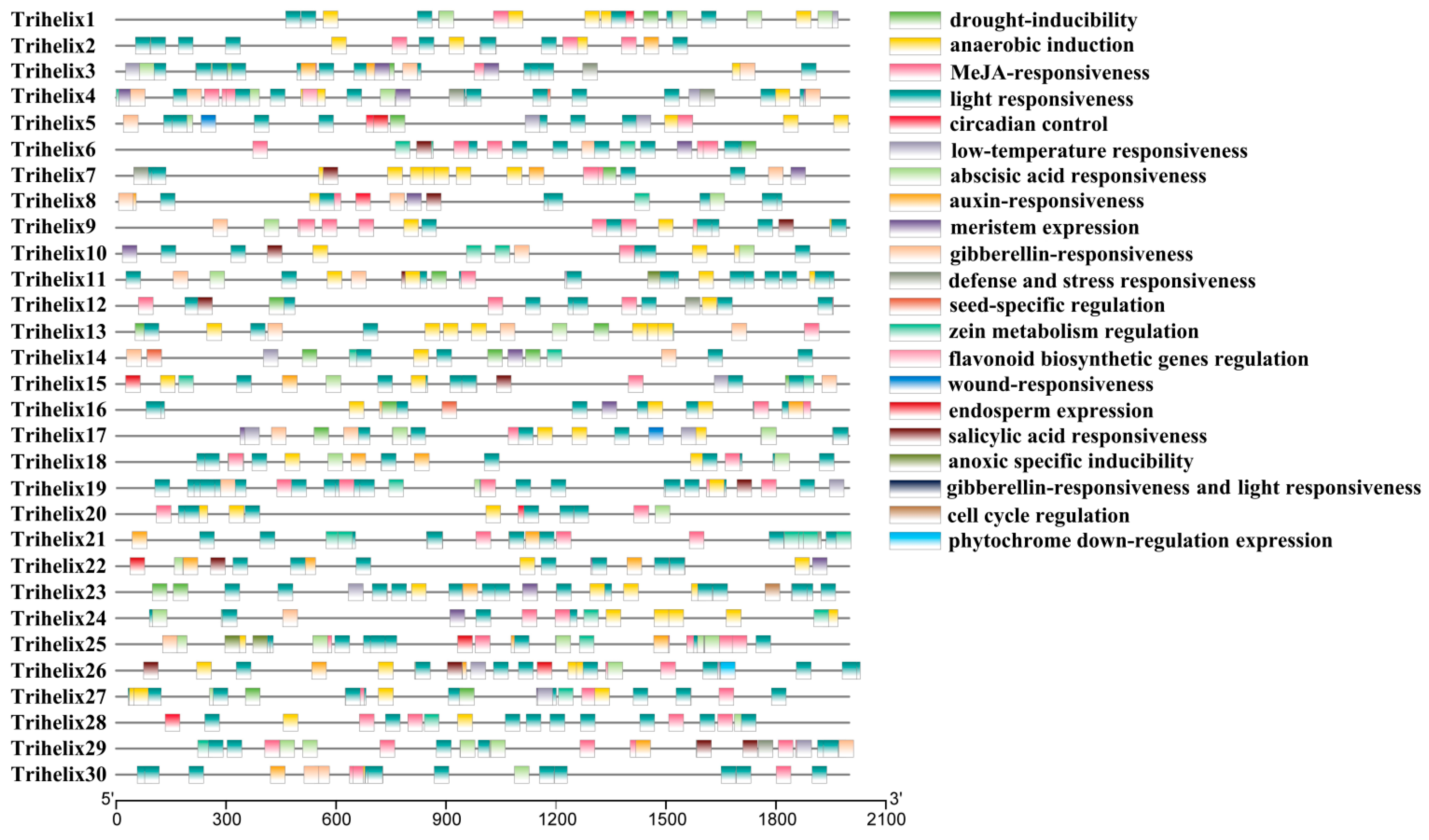
PlantCARE was used to find potential cis-acting elements in the promoter regions of TTF genes (Figure 3). Most of the genes have stress and hormone-responsive cis-elements. For example, elements responsive to methyl jasmonate, salicylic acid, abscisic acid, drought, anaerobic induction, defense, and low temperature stress were detected (Table S2). The FvTrihelix promoter regions contained G-box elements, CGTCA motifs, and TGACG motifs associated with methyl jasmonate and salicylic acid induction. They also contained anaerobic-inducible elements (ARE elements), elements associated with drought stress (MBS elements), and abscisic acid-inducible elements (ABRE elements). Taken together, these results indicate a strong association between the FvTrihelix family and biotic and abiotic stresses. Moreover, TTF members also contained circadian control, auxin responsiveness, meristem expression, seed-specific regulation, flavonoid biosynthesis, and endosperm expression responsive elements, suggesting that the Trihelix genes may be crucial for plant growth and development.
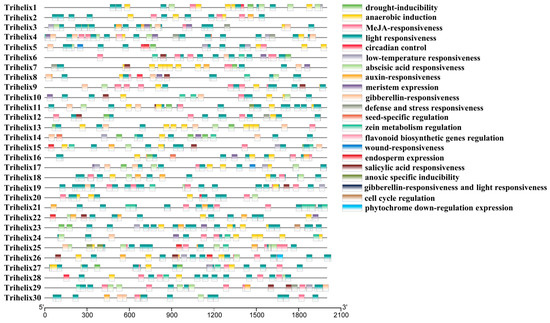
Figure 3.
Identification of putative cis-acting regulatory elements in the promoter regions of 30 FvTrihelix genes using PlantCARE.
3.5. Expression Analysis of TTF Genes in Various Parts of the Strawberry
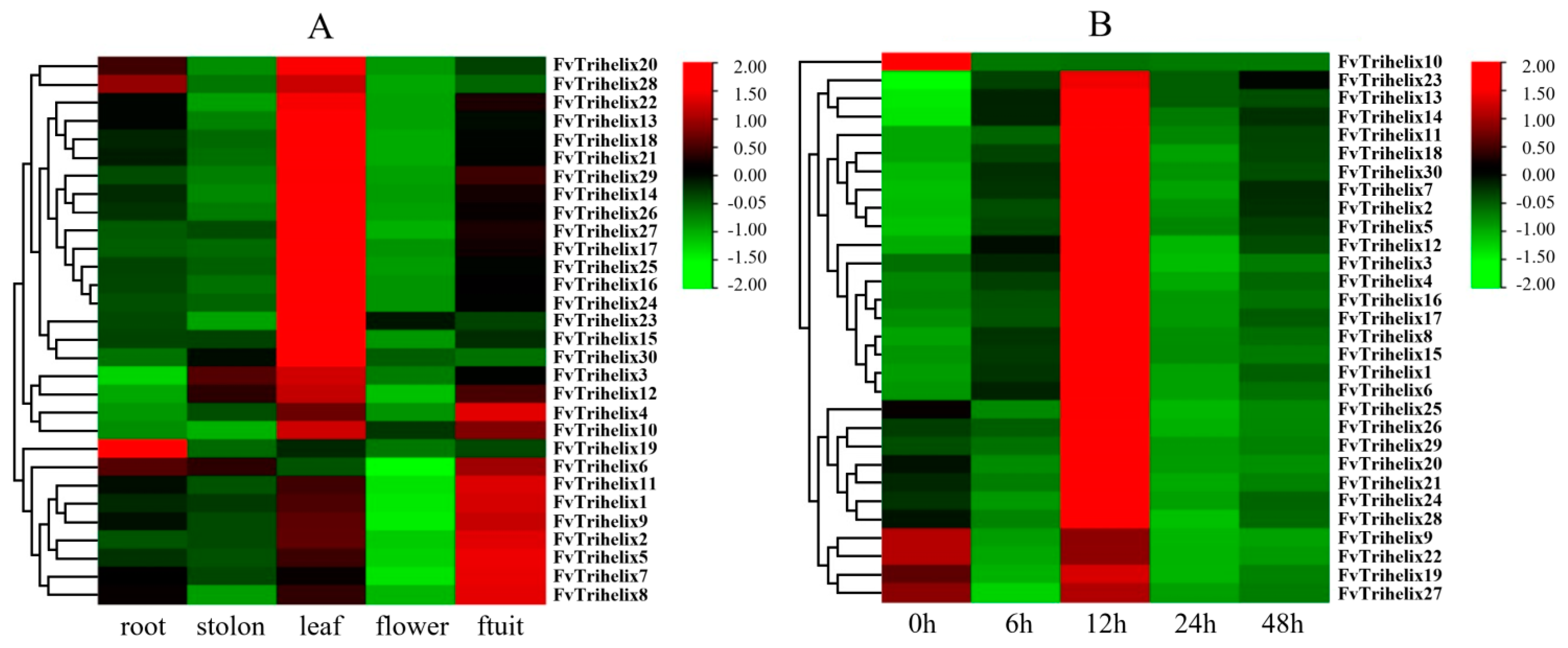
qRT-PCR was employed to analyze the expression of all 30 genes in 5 tissues/organs (leaf, stolon, root, flower, and fruit) of the strawberry. Most of the genes showed a different expression in different plant parts (Figure 4A). FvTrihelix1, 2, 4, 5, 6, 7, 8, 9, 10, and 11 showed high expression in fruit. Only Trihelix19 and FvTrihelix28 were expressed in roots. FvTrihelix4, 10, and 12 were expressed in both leaves and fruits. FvTrihelix3, 10, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, and 30 were all highly expressed in leaves. FvTrihelix11 and FvTrihelix16 (segmentally duplicated) were highly expressed in fruits and leaves, respectively, indicating that functional divergence occurred after duplication.
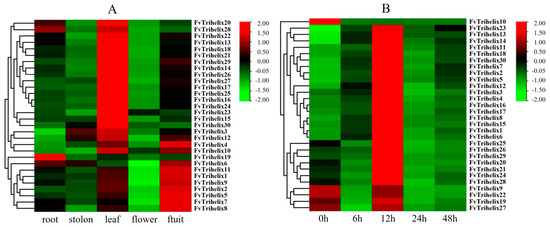
Figure 4.
(A) Expression of Trihelix genes in different tissues of Fragaria vesca (root, stolon, leaf, flower, and fruit). (B) Expression of strawberry Trihelix genes in strawberry leaves following C. gloeosporioides infections. Red color represents up−regulation and green color represents down−regulation.
3.6. Evaluation of the FvTrihelix Genes’ Expression following C. gloeosporioides Inoculation
According to the results of qRT-PCR, we observed four trends in the expression response of FvTrihelix genes in strawberry leaves after inoculation with C. gloeosporioides (Figure 4B). The most common trend (trend I) was observed in 18 genes (FvTrihelix1, 2, 3, 4, 5, 6, 7, 8, 11, 12, 13, 14, 15, 16, 17, 18, 23, and 30). These genes showed up-regulation and displayed peak expression at 12 hpi, followed by subsequent down-regulation and then modest up-regulation at 48 hpi. There were 7 FvTrihelix genes (FvTrihelix20, 21, 24, 25, 26, 28, and 29) that were first down-regulated, then up-regulated to peak expression at 12 hpi, and then gradually down-regulated thereafter (trend II). Interestingly, all genes (trend I and trend II) had the highest expression at 12 hpi; however, the peak expression at 12 hpi was significantly less in trend II genes than trend I genes. This shows that genes associated with trend I are particularly crucial for preventing C. gloeosporioides infection. Four genes (FvTrihelix9, 19, 22, and 27) showed two consecutive periods of up-regulation followed by down-regulation (trend III). These genes showed only slight up-regulation at 12 hpi, suggesting that trend III genes do not play more roles in the plant response to pathogens. FvTrihelix10 gene was down-regulated over time after inoculation with C. gloeosporioides (trend IV).
3.7. Expression Profiling of Selected Strawberry TTF Genes in Strawberry Leaves following SA and MeJA Treatments
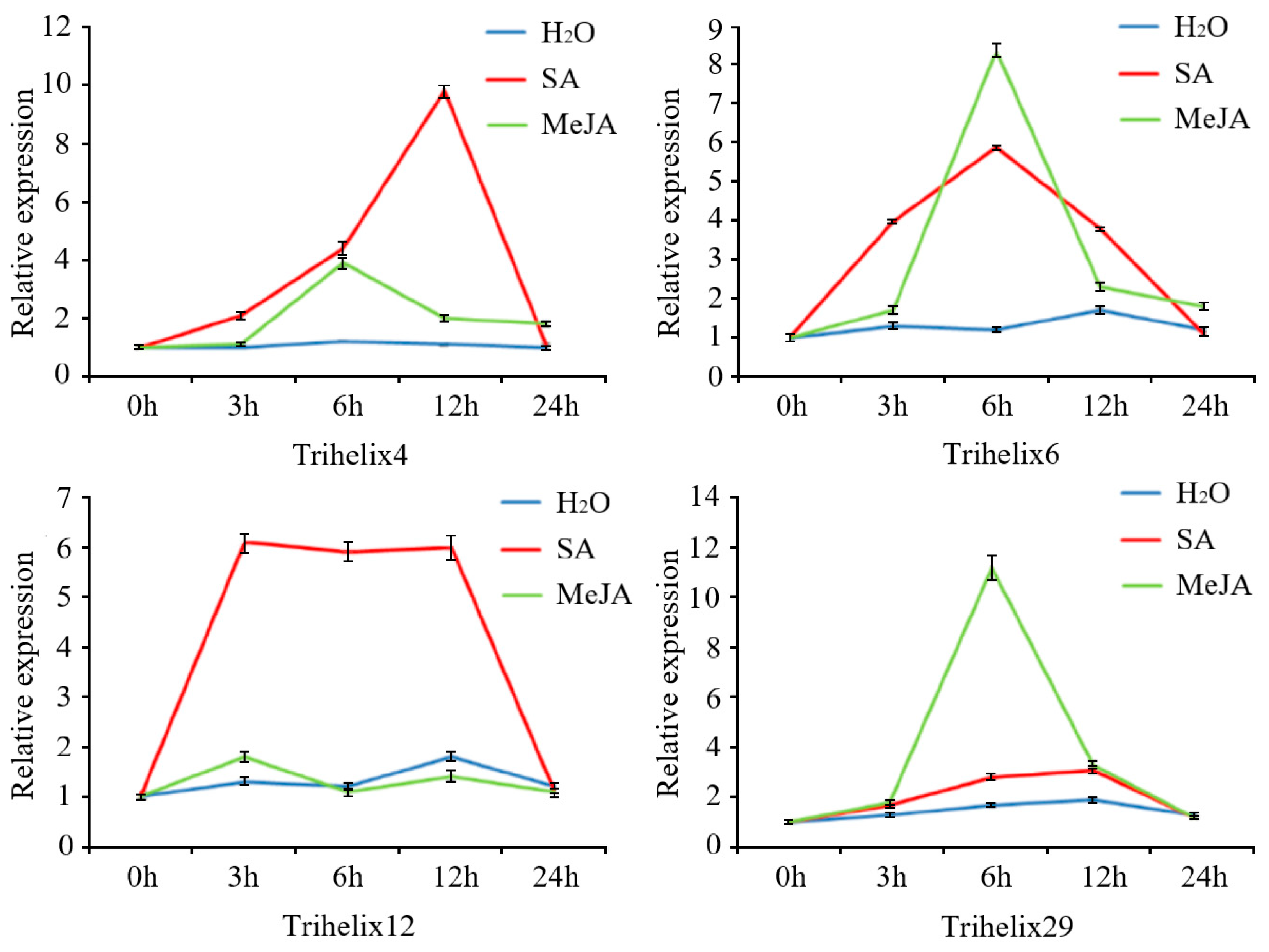
Four genes were examined by qRT-PCR after SA and MeJA treatments (Figure 5). These genes were selected based on the presence of MeJA- and salicylic-acid-responsive cis-acting elements in their promoter regions’ high expression levels against C. gloeosporioides infection. All genes showed strong expression responses to SA or MeJA treatments. FvTrihelix4 showed a strong response to SA treatment and peak expression at 12 hpt (9.78-fold) followed by down-regulation at 24 hpt. The response of FvTrihelix4 to MeJA was less with peak expression at 6 hpt (4.15-fold) followed by down-regulation. FvTrihelix6 expression was induced by C. gloeosporioides infection as well as by SA and MeJA treatments, indicating its involvement in pathogen resistance. FvTrihelix12 showed a strong response to SA treatment with high expression at 3, 6, and 12 hpt (6.18-fold, 6.02-fold, and 6.13-fold, respectively), but no significant response to JA treatment. FvTrihelix29 responded strongly to JA, showing up-regulation with peak expression at 6 hpt (11.31-fold) and subsequent down-regulation. The response of FvTrihelix29 to SA was not high, with a slight up-regulation followed by down-regulation.
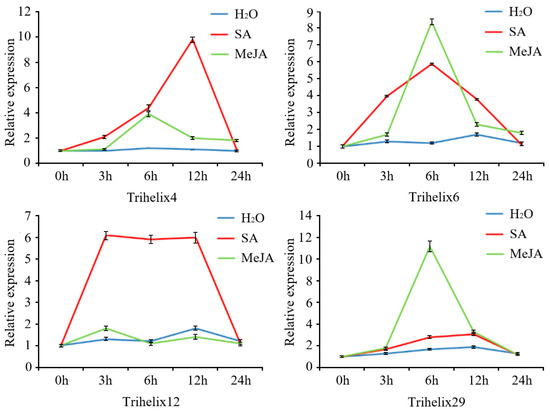
Figure 5.
Four FvTrihelix genes’ expression patterns in response to SA, MeJA, and water treatments. For exogenous hormone treatment, methyl jasmonate or salicylic acid solution was sprayed on strawberry leaves and distilled water was used as a control. Standard deviations (SD) from three biological replicates are shown by error bars.
3.8. FvTrihelix6 Ectopic Expression in A. thaliana Enhanced Resistance to C. higginsianum
Based on FvTrihelix6 expression against C. gloeosporioides infection, and SA and MeJA treatments, FvTrihelix6 (840 bp, Acc. NO. OQ319481) was cloned in A. thaliana for further investigation (Figure S1 in Supplementary Material). FvTrihelix6 belongs to the SIP1 subfamily and is located on chromosome 2. It has a molecular mass of 32.40 kDa and pI of 5.91 and is predicted to be localized in the nucleus. Three transgenic plants (T3-2, T3-10, and T3-23) showing the highest expression levels of FvTrihelix6 were selected for further studies.
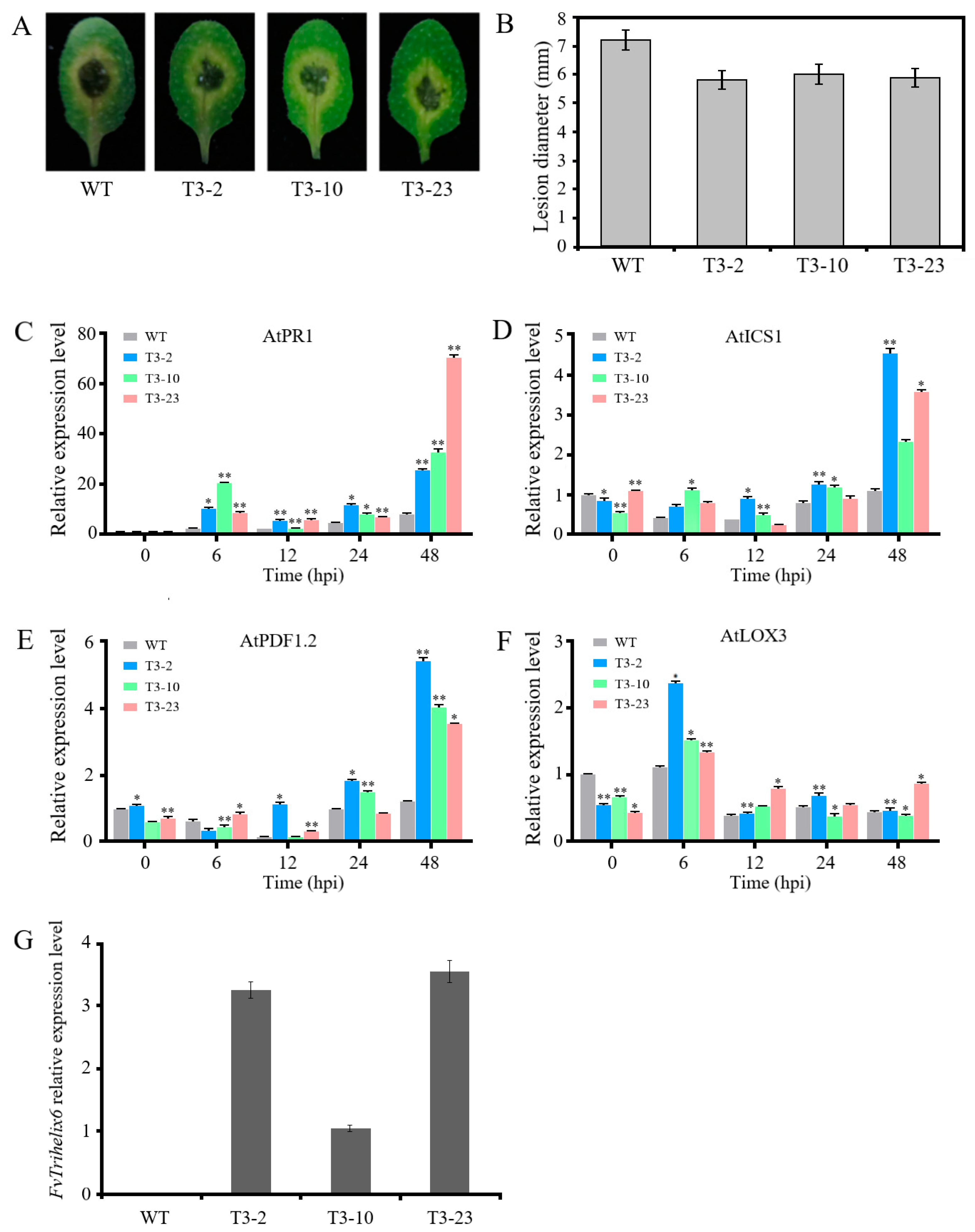
Among the three transgenic lines, the expression level of FvTrihelix6 was highest in T3-2 and lowest in T3-10, and no FvTrihelix6 expression was shown in WT (Figure 6G). These transgenic lines and control plants (WT) were then inoculated with C. higginsianum. All leaves displayed disease symptoms, with brown necrotic spots appearing in the center of the leaves. However, the lesions developed in transgenic plants were smaller than the lesions developed in WT plants, and transgenic plants showed less severe disease signs than WT plants (Figure 6A,B). We also investigated the expression patterns of genes involved in JA (AtPDF1.2 and AtLOX3) and SA (AtPR1 and AtICS1) signaling for exploring the molecular basis of the C. higginsianum resistance mechanism. AtPR1 was up-regulated at 6 hpi and 48 hpi. AtPR1 displayed 8.30-fold more expression at 6 hpi expression in the T3-10 line compared with WT, while AtPR1 expression in T3-23 was up-regulated 8.70-fold at 48 hpi (Figure 6C). AtICS1 was expressed 4.13-fold higher in T3-2 than the WT control at 48 hpi (Figure 6D). AtPDF1.2 was up-regulated 4.30-fold in T3-2 at 48 hpi compared to the WT control (Figure 6E). The expression of AtLOX3 in T3-23 was almost 2-fold higher at 48 hpi compared to the WT control (Figure 6F).
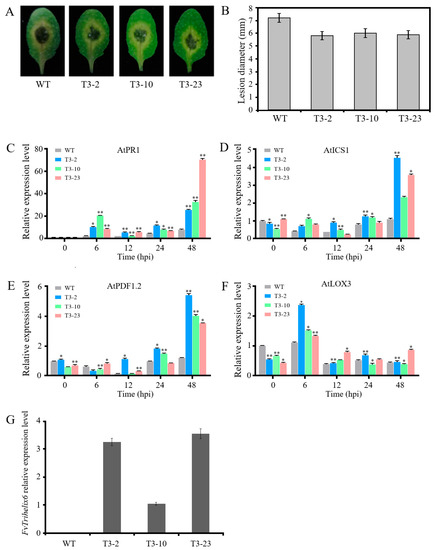
Figure 6.
Expression comparison of genes in transgenic A. thaliana and wild type lines following C. higginsianum infection. (A) Disease symptoms in wild type and transgenic A. thaliana lines after two days of C. higginsianum inoculation. (B) Disease lesion diameter in wild type and transgenic A. thaliana leaves after two days of inoculation. (C) Expression of AtPR1 via qRT-PCR. (D) qRT-PCR analysis of AtICS1. (E) qRT-PC analysis of AtPDF1.2. (F) qRT-PCR analysis of AtLOX3. Asterisks represent significant differences between wild type and transgenic A. thaliana (* p < 0.05, ** p < 0.01, Student’s t-test). (G) Expression of FvTrihelix6 in transgenic A. thaliana and wild type lines following C. higginsianum infection.
3.9. Subcellular Localization of FvTrihelix6
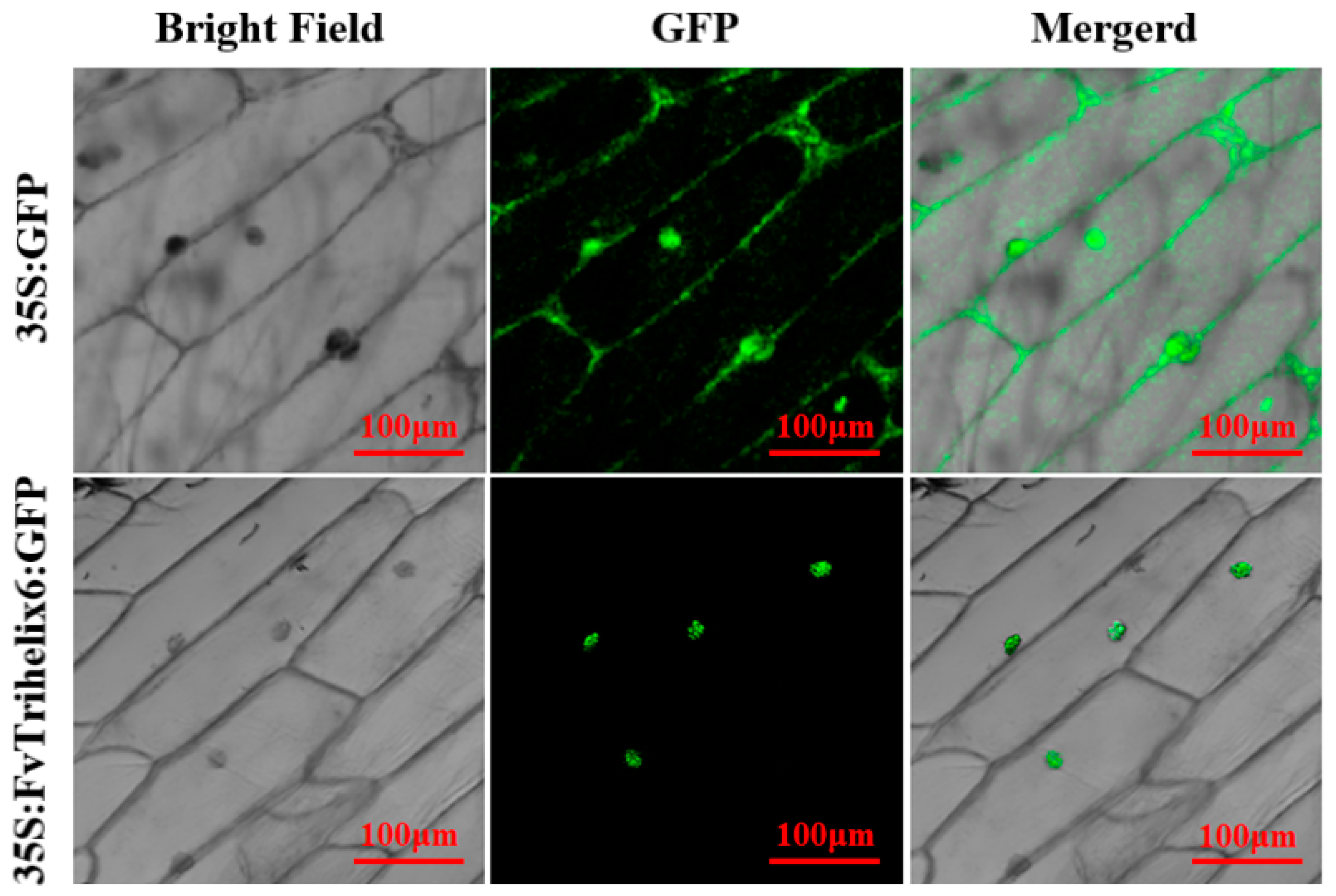
To identify the subcellular location of FvTrihelix6, a 35S:FvTrihelix6-GFP fusion protein was constructed and transiently expressed in onion epidermis (Figure 7). We observed 35S:FvTrihelix6-GFP in onion epidermal cells using laser scanning confocal microscopy. As shown, the control 35S:GFP was detected as green fluorescence in the nucleus and in the cell membrane. In addition, 35S:FvTrihelix6-GFP fluorescence was detected in the nucleus, indicating that FvTrihelix6 does not localize in the cell membrane and may localize in the nucleus.
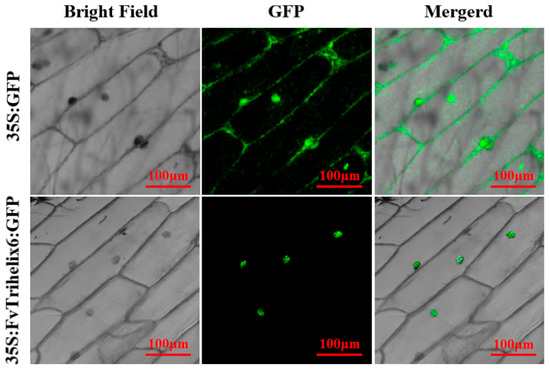
Figure 7.
Subcellular localization of 35S:GFP control and 35S:FvTrihelix6-GFP in onion epidermis.
3.10. Response of the FvTrihelix6 Promoter to SA and C. gloeosporioides Using a GUS Reporter
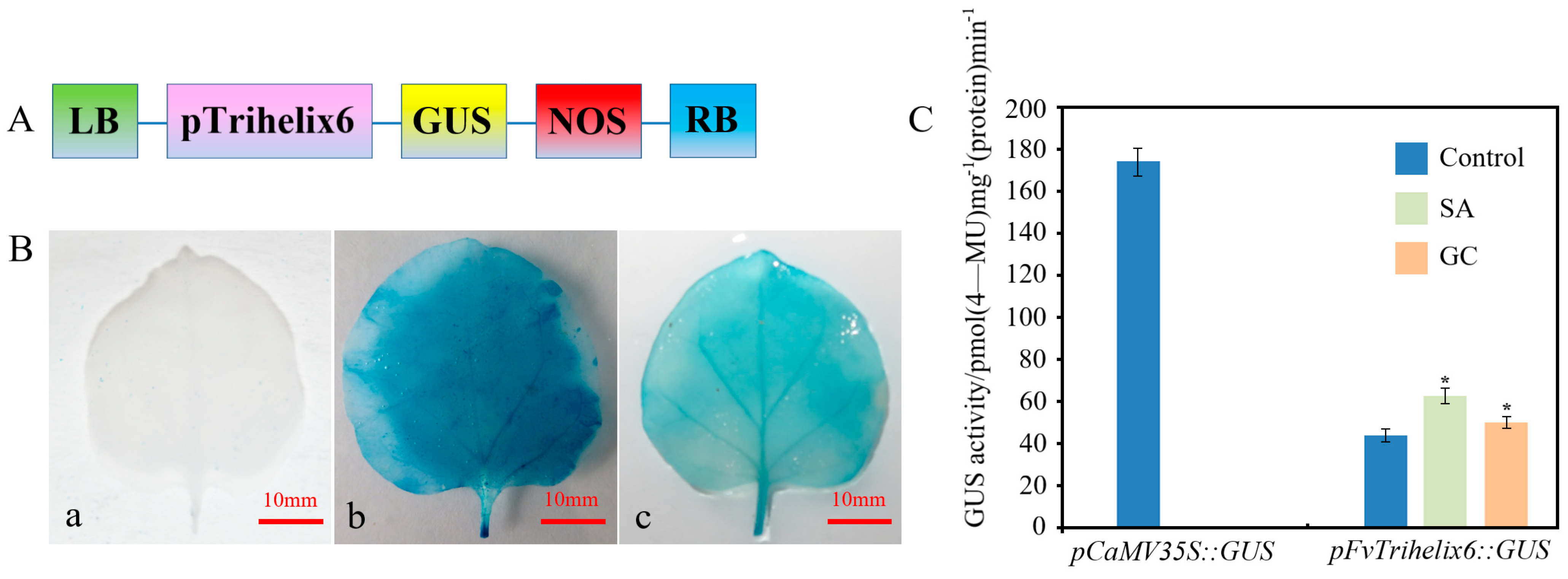
PlantCARE software was used to identify putative cis-acting elements in the promoter region of FvTrihelix6 (1099 bp, Acc. NO. OQ357817, Figure S2). There were light-, stress-, drought-, salicylic-acid-, and MeJA-responsive cis-elements (Figure S2 and Table S2). This indicates that the FvTrihelix6 promoter may play important roles in plant growth, development, and resistance to biotic and abiotic stresses. We fused the FvTrihelix6 promoter to a GUS reporter to generate the pFvTrihelix6::GUS vector (Figure 8A). pFvTrihelix6::GUS tobacco leaves showed blue staining but it was lighter than the positive control leaves. The negative control leaves were white (Figure 8B). This indicated that the pFvTrihelix6 gene promoter can drive GUS expression and FvTrihelix6 gene expression. The pFvTrihelix6::GUS fusion vector was transferred into tobacco leaves by injection. Three treatments were performed: a control group, an SA treatment group, and a C. gloeosporioides inoculation group. The pCaMV35S::GUS vector was also transformed as a positive control and GUS activity was measured after two days of dark culture. GUS activity driven by pFvTrihelix6 was 1.43-fold that of the control after inoculation with C. gloeosporioides solution. The GUS activity of pFvTrihelix6 was 1.24-fold compared to the control after treatment with SA. The GUS activity in the pFvTrihelix6 leaves was significantly lower than that of the positive control. These results indicated that the FvTrihelix6 promoter could respond to both C. gloeosporioides and SA by positively regulating gene expression (Figure 8C).
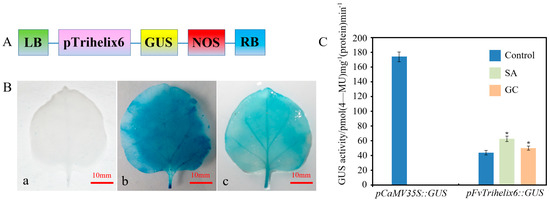
Figure 8.
Detection of GUS enzyme activity in tobacco leaves transformed with the pFvTrihelix6::GUS vector, a GUS reporter under the control of the FvTrihelix6 promoter. (A) Structure of the promoter−FvTrihelix6::GUS construct. LB is left border, RB is right border, NOS is nos−terminator and GUS is β−glucuronidase. (B) (a) staining indicating GUS activity after pC0380::GUS was transformed into tobacco leaves (b) staining indicating GUS activity after pCaMV35S::GUS was transformed into tobacco leaves (c) staining indicating GUS activity after pFvTrihelix6::GUS was transformed into tobacco leaves (C) Transient expression of GUS enzyme activity in tobacco. Control represents control group, SA represents SA treatment group, and GC represents C. gloeosporioides inoculation.
4. Discussion
Strawberry TTF members contained a large number of light-responsive elements, low-temperature-responsive elements, stress and defense elements, drought-inducibility elements, and hormone (MeJA, SA, GA, ABA, and IAA) response-related elements in their promoter regions (Figure 3 and Table S2). In Arabidopsis, AtGT-1 was found to contain many light-responsive elements [39]. In maize, SA- and MeJA-responsive cis-acting elements were also found in the ZmGT-3b gene, which is associated with resistance to Fusarium graminearum [24]. These findings suggest that the TTF genes have potential roles in stress tolerance and plant growth and development.
Some FvTrihelix genes showed differential expression patterns following MeJA and SA treatments. For example, the expression of FvTrihelix4, 6, and 29 showed up-regulation, while FvTrihelix12 expression was down-regulated. In C. quinoa, Trihelix genes were strongly up-regulated in response to SA [10]. OsTrihelix20 in rice showed significant up-regulation after treatment with MeJA [40]. Our results are in line with previous studies and support the idea that TTF genes play roles in disease resistance via regulating the SA and JA signaling pathways. In addition, we further investigated the changes in gene expression of FvTrihelix family members after C. gloeosporioides infection. Most of the genes showed up-regulation after C. gloeosporioides infection (Figure 4). They showed significantly high up-regulation following C. gloeosporioides inoculation, and SA and MeJA treatments. Based on these results, FvTrihelix6 was selected for functional analysis and further research. FvTrihelix6 is a member of the SIP1 subfamily and there is information about the roles of SIP1 genes in disease-resistance mechanisms. In the promoter region of FvTrihelix6, we identified a W-box element (TTGACT), which has been shown to play an important role in plant resistance to pathogenic infection [41,42]. The Trihelix disease-resistance gene rml1 also contains W-box elements [26]. The W-box cis-elements in the promoter of the FvTrihelix6 promoter may play potential roles in the C. higginsianum resistance mechanisms. The presence of JA- and SA-responsive cis-elements also supports the involvement of FvTrihelix6 in biotic stress tolerance. This result is consistent with the results of Zhang et al. [24]. The localization of FvTrihelix6 in the nucleus implies that FvTrihelix6 may defend against pathogens by regulating the transcription of disease-resistance target genes [9].
The ectopic expression of FvTrihelix6 improved resistance to C. higginsianum infection. Previously, the Trihelix family member rml1 has been shown to be up-regulated in response to Magnaporthe grisea infection [26]. In maize, ZmGT-3b improved disease resistance to Fusarium graminearum [24]. We also found that ectopic expression of FvTrihelix6 increased disease resistance in transgenic A. thaliana lines (Figure 6A,B). SA- and JA-mediated signaling pathways play an important role in plant disease resistance. SA-mediated signaling pathways respond in the defense against hemibiotrophic fungal pathogens, which is associated with resistance to C. higginsianum in A. thaliana [43]. JA-mediated signaling pathways respond in the defense against necrotrophic fungal pathogens. Plant resistance to pathogenic bacteria is usually achieved via a complex defense network that is mediated through salicylic acid (SA) and jasmonic acid (JA) [6,44]. In Arabidopsis, ICS is the key enzyme for SA synthesis. Infection by pathogens may result in increased levels of AtICS1 transcripts in plants [45,46]. The PR1 gene has been recognized as a marker gene for the SA signal pathway. AtPDF1.2 is commonly used to detect a JA response [47,48]. To support our results, expression profiling of genes having presumed roles in the JA and SA signaling pathways was performed.
The expression of AtPR1 was up-regulated at 6 hpi and 48 hpi (Figure 6C). The expression of AtICS1 gene in the three transgenic lines was higher than that of WT A. thaliana at almost every time point (Figure 6D). AtPDF1.2 reached its peak at 48 hpi (Figure 6E). AtLOX3 showed higher expression patterns in three transgenic lines than WT (Figure 6F). These findings may indicate that the SA signaling pathway-related gene AtPR1 is activated and up-regulated first, followed by the JA signaling pathway-related gene AtPDF1.2. This pattern is consistent with the hemibiotrophic mode of infection found in previous studies [49]. These findings imply that ectopic expression of FvTrihelix6 in A. thaliana increased resistance against C. higginsianum infection via activating the SA and JA signaling pathways. This study provides information about the potential role of the FvTrihelix6 gene in the disease-resistance mechanism. It also provides a basis for the functional characterization of strawberry TTF genes.
5. Conclusions
The 30 Trihelix genes in F. vesca were divided into 5 subfamilies. Most Trihelix genes exhibited differential expression in different organs of strawberries and were up-regulated after infection with C. gloeosporioides. Ten genes werere induced after hormone (SA and JA) treatments. The ectopic expression of FvTrihelix6 in A. thaliana increased resistance against C. higginsianum infection. Further, pFvTrihelix6-GUS activity increased following C. gloeosporioides and SA treatments. The FvTrihelix6 protein was localized in the nucleus, and a pFvTrihelix6-GUS reporter indicated that the promoter of FvTrihelix6 could drive transcription of downstream genes. This study provides strawberries’ candidate genes for disease-resistance breeding and a basis for studying the disease resistance of the Trihelix transcription factor family.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae9060633/s1, Data S1: Amino acid sequences information of FvTrihelix genes; Table S1: Primers sequences information; Figure S1: The nucleotide sequence and amino acid sequence of FvTrihelix6; Figure S2: The promoter sequence and cis-element prediction of FvTrihelix6; Table S2: Putative cis-acting elements of FvTrihelix genes.
Author Contributions
Investigation, J.F., F.J., H.S., T.H., Y.L. and G.J.; Resources, Q.C. and Z.W.; Writing—original draft preparation, J.F.; Writing—review and editing, J.F., B.A., S.A.M.B. and Z.W. All authors have read and agreed to the published version of this manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32072526).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable for this article.
Acknowledgments
C. gloeosporioides and C. higginsianum strain Ch-1 were provided by Han Yongchao and Zheng Lu, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Green, P.J.; Kay, S.A.; Chua, N.H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987, 6, 2543–2549. [Google Scholar] [CrossRef]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Wu, D.; Fan, Q.; Tian, C.; Chen, S.; Guan, Z.; Xin, J.; Zhao, K.; Chen, F. Transcriptome-Wide Identification and Expression Profiling Analysis of Chrysanthemum Trihelix Transcription Factors. Int. J. Mol. Sci. 2016, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Q.; Wang, H.; Zhang, H.; Xu, X.; Li, C.; Yang, C. Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Sci. Rep. 2016, 6, 36274. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Wang, J.; Liu, H.; Zheng, H.; Zhen, Z.; Zhao, H.; et al. Genome-Wide Characterization and Identification of Trihelix Transcription Factor and Expression Profiling in Response to Abiotic Stresses in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 251. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Y.; Li, W.; Lin, Y.; Wang, C.; Xu, R.; Zhang, L. Genome-wide characterization and expression analysis of soybean trihelix gene family. PeerJ 2020, 8, e8753. [Google Scholar] [CrossRef]
- Li, K.; Duan, L.; Zhang, Y.; Shi, M.; Chen, S.; Yang, M.; Ding, Y.; Peng, Y.; Dong, Y.; Yang, H.; et al. Genome-wide identification and expression profile analysis of trihelix transcription factor family genes in response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genom. 2021, 22, 738. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Bin, J.; Ding, H.; Pan, D.; Tian, Q.; Yang, X.; Wang, L.; Yue, Y. Insights into the trihelix transcription factor responses to salt and other stresses in Osmanthus fragrans. BMC Genom. 2022, 23, 334. [Google Scholar] [CrossRef]
- Li, K.; Fan, Y.; Zhou, G.; Liu, X.; Chen, S.; Chang, X.; Wu, W.; Duan, L.; Yao, M.; Wang, R.; et al. Genome-wide identification, phylogenetic analysis, and expression profiles of trihelix transcription factor family genes in quinoa (Chenopodium quinoa Willd.) under abiotic stress conditions. BMC Genom. 2022, 23, 499. [Google Scholar] [CrossRef]
- Wang, J.; Ouyang, Y.; Wei, Y.; Kou, J.; Zhang, X.; Zhang, H. Identification and Characterization of Trihelix Transcription Factors and Expression Changes during Flower Development in Pineapple. Horticulturae 2022, 8, 894. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Pan, Q.; Chen, S.; Feng, C.; Hai, J.; Li, H. Comparison of Trihelix transcription factors between wheat and Brachypodium distachyon at genome-wide. BMC Genom. 2019, 20, 142. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Hu, J.; Behera, J.R.; Kilaru, A.; Yuan, Y.; Zhai, Y.; Xu, Y.; Xie, L.; Zhang, Y.; Zhang, Q.; et al. A Tree Peony Trihelix Transcription Factor PrASIL1 Represses Seed Oil Accumulation. Front. Plant Sci. 2021, 12, 796181. [Google Scholar] [CrossRef]
- Feng, C.; Song, X.; Tang, H. Molecular cloning and expression analysis of GT-2-like genes in strawberry. 3 Biotech 2019, 9, 105. [Google Scholar] [CrossRef]
- Tzafrir, I.; Pena-Muralla, R.; Dickerman, A.; Berg, M.; Rogers, R.; Hutchens, S.; Sweeney, T.C.; McElver, J.; Aux, G.; Patton, D.; et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004, 135, 1206–1220. [Google Scholar] [CrossRef]
- Weng, H.; Yoo, C.Y.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. Poplar GTL1 is a Ca2+/calmodulin-binding transcription factor that functions in plant water use efficiency and drought tolerance. PLoS ONE 2012, 7, e32925. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.H.; Chen, F.; Torres-Jerez, I.; Tang, Y.; Wang, M.; Du, Q.; Cheng, X.; Wen, J.; Dixon, R. Transcriptome analysis of secondary cell wall development in Medicago truncatula. BMC Genom. 2016, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Negm, M.; Wu, D.; Li, J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Song, L.; Song, J.; Ouyang, B.; Guo, L.; Shang, L.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. ShCIGT, a Trihelix family gene, mediates cold and drought tolerance by interacting with SnRK1 in tomato. Plant Sci. 2018, 270, 140–149. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Z.; Dong, Y.; Xie, Z. Trihelix Transcriptional Factor GhGT26 of Cotton Enhances Salinity Tolerance in Arabidopsis. Plants 2022, 11, 2694. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, M.; Zhang, Y.; Huang, M.; Wei, L.; Lin, Y.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D.; et al. Coordinated regulation of plant defense and autoimmunity by paired trihelix transcription factors ASR3/AITF1 in Arabidopsis. New Phytol. 2023, 237, 914–929. [Google Scholar] [CrossRef] [PubMed]
- Völz, R.; Kim, S.K.; Mi, J.; Mariappan, K.G.; Guo, X.; Bigeard, J.; Alejandro, S.; Pflieger, D.; Rayapuram, N.; Al-Babili, S.; et al. The Trihelix transcription factor GT2-like 1 (GTL1) promotes salicylic acid metabolism, and regulates bacterial-triggered immunity. PLoS Genet. 2018, 14, e1007708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhong, T.E.L.; Xu, M.; Dai, W.; Sun, S.; Ye, J. GT Factor ZmGT-3b Is Associated With Regulation of Photosynthesis and Defense Response to Fusarium graminearum Infection in Maize Seedling. Front. Plant Sci. 2021, 12, 724133. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hong, G.; Han, B. Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 2004, 324, 105–115. [Google Scholar] [CrossRef]
- Giampieri, F.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Alvarez-Suarez, J.M.; Afrin, S.; Bompadre, S.; Quiles, J.L.; Mezzetti, B.; Battino, M. Strawberry as a health promoter: An evidence based review. Food Funct. 2015, 6, 1386–1398. [Google Scholar] [CrossRef]
- Miller, K.; Feucht, W.; Schmid, M. Bioactive Compounds of Strawberry and Blueberry and Their Potential Health Effects Based on Human Intervention Studies: A Brief Overview. Nutrients 2019, 11, 1510. [Google Scholar] [CrossRef]
- Morkeliūnė, A.; Rasiukevičiūtė, N.; Valiuškaitė, A. Conditions in a Temperate Climate for Colletotrichum acutatum, Strawberry Pathogen Distribution and Susceptibility of Different Cultivars to Anthracnose. Agriculture 2021, 11, 80. [Google Scholar] [CrossRef]
- Chen, X.Y.; Dai, D.J.; Zhao, S.F.; Shen, Y.; Wang, H.D.; Zhang, C.Q. Genetic Diversity of Colletotrichum spp. Causing Strawberry Anthracnose in Zhejiang, China. Plant Dis. 2020, 104, 1351–1357. [Google Scholar] [CrossRef]
- Casado-Díaz, A.; Encinas-Villarejo, S.; Santos, B.d.l.; Schilirò, E.; Yubero-Serrano, E.-M.; Amil-Ruíz, F.; Pocovi, M.I.; Pliego-Alfaro, F.; Dorado, G.; Rey, M.; et al. Analysis of strawberry genes differentially expressed in response to Colletotrichum infection. Physiol. Plant. 2006, 128, 633–650. [Google Scholar] [CrossRef]
- Wen, Z.; Bai, J.; Wang, L.; Yao, L.; Ahmad, B.; Hanif, M.; Chen, Q. Over expression of a Chitinase 2 gene from Chinese Wild Strawberry improves resistance to anthracnose disease in transgenic Arabidopsis thaliana. Plant Biotechnol. Rep. 2020, 14, 725–736. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Jiang, W.; Yang, B.; Weeks, D.P. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 2014, 9, e99225. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Shen, Y.; Ahmad, B.; Yao, L.; He, T.; Fan, J.; Liu, Y.; Chen, Q.; Wen, Z. Genome-wide identification and expression analysis of dirigent gene family in strawberry (Fragaria vesca) and functional characterization of FvDIR13. Sci. Hortic. 2022, 297, 110913. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y.; Nakajima, K.; Arai, Y.J.P. An improved assay for ß-glucuronidase (GUS) in transformed cells: Methanol almost suppresses a putative endogenous GUS activity. Plant Sci. 1990, 70, 133–140. [Google Scholar] [CrossRef]
- Wang, L.; Huang, M.; Yao, L.; Bai, J.; Chen, Q.; Wen, F. Cloning and Expression Analysis of FnMYB24 Transcription Factor Gene and Promoter from Fragaria nilgerrensis Schltdl (Chinese). Acta Bot. Boreali-Occident. Sinica 2020, 40, 1646–1654. [Google Scholar]
- Le Gourrierec, J.; Li, Y.F.; Zhou, D.X. Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA-TBP-TATA complex. Plant J. 1999, 18, 663–668. [Google Scholar] [CrossRef]
- Ji, J.H.; Zhou, Y.J.; Wu, H.H.; Yang, L.M. Genome-wide analysis and functional prediction of the Trihelix transcription factor family in rice. Yi Chuan Hered. 2015, 37, 1228–1241. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Schmelzer, E.; Hahlbrock, K.; Somssich, I.E. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Reinstädler, A.; Lipka, V.; Lippok, B.; Somssich, I.E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 2002, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shi, L.; Yang, S.; Lin, Y.; Weng, Y.; Li, X.; Hussain, A.; Noman, A.; He, S. Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection. Int. J. Mol. Sci. 2017, 18, 1661. [Google Scholar] [CrossRef]
- Amil-Ruiz, F.; Garrido-Gala, J.; Gadea, J.; Blanco-Portales, R.; Muñoz-Mérida, A.; Trelles, O.; de Los Santos, B.; Arroyo, F.T.; Aguado-Puig, A.; Romero, F.; et al. Partial Activation of SA- and JA-Defensive Pathways in Strawberry upon Colletotrichum acutatum Interaction. Front. Plant Sci. 2016, 7, 1036. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Strawn, M.A.; Marr, S.K.; Inoue, K.; Inada, N.; Zubieta, C.; Wildermuth, M.C. Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 2007, 282, 5919–5933. [Google Scholar] [CrossRef]
- Uknes, S.; Mauch-Mani, B.; Moyer, M.; Potter, S.; Williams, S.; Dincher, S.; Chandler, D.; Slusarenko, A.; Ward, E.; Ryals, J. Acquired resistance in Arabidopsis. Plant Cell 1992, 4, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14 (Suppl. S1), S153–S164. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Maeda, S.; Sugano, S.; Ohtake, M.; Hayashi, N.; Ichikawa, T.; Kondou, Y.; Kuroda, H.; Horii, Y.; Matsui, M.; et al. Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2011, 9, 466–485. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).