At the Intersection of Natural Structural Coloration and Bioengineering
Abstract
:1. Introduction
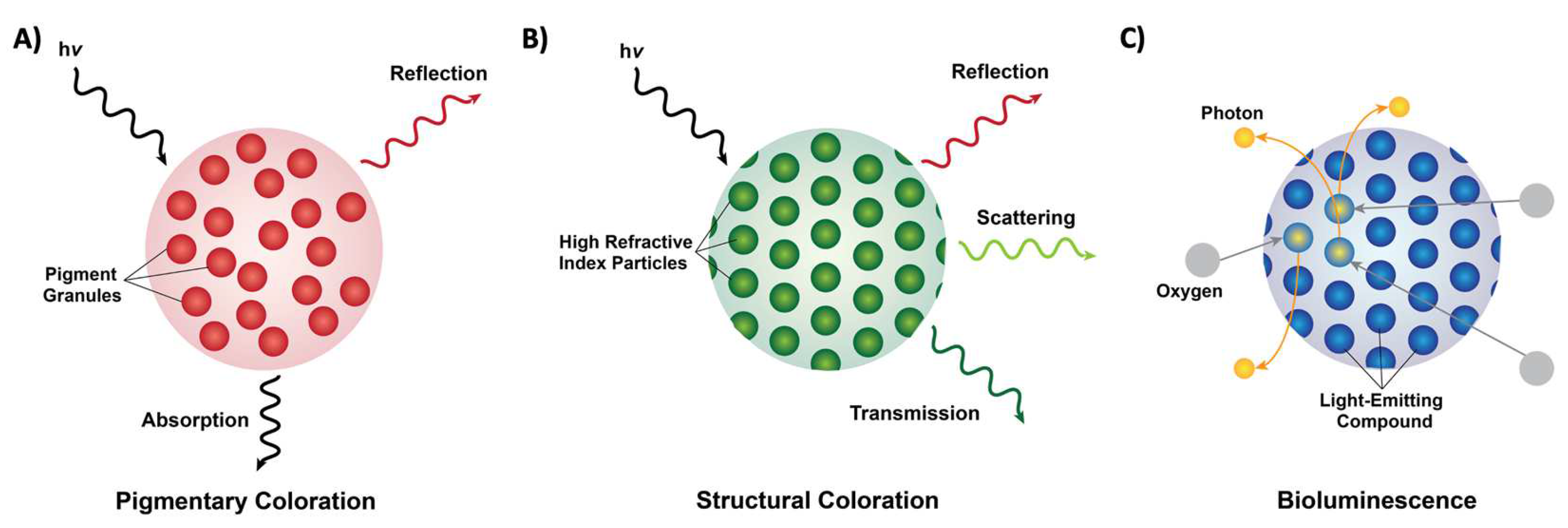
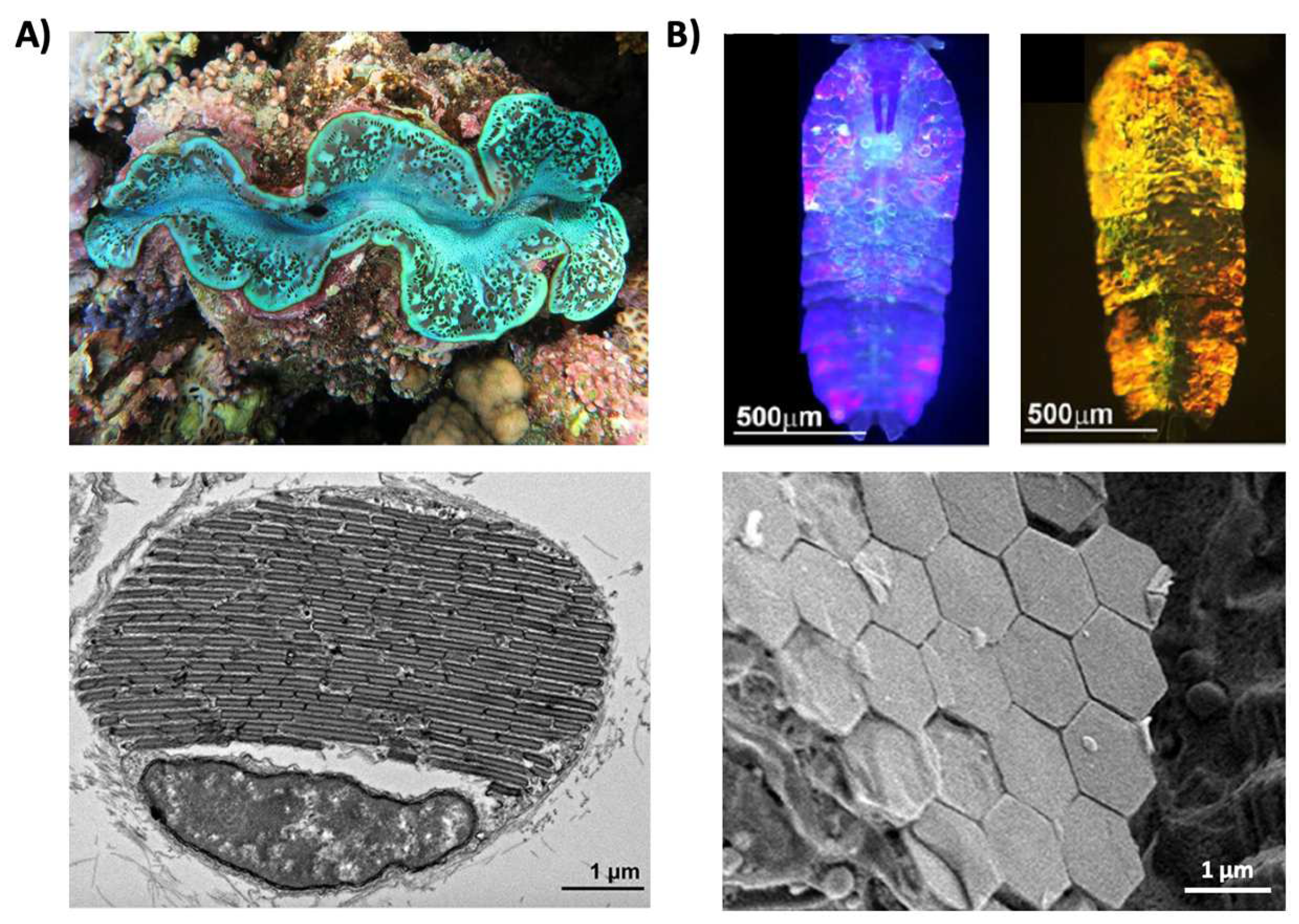
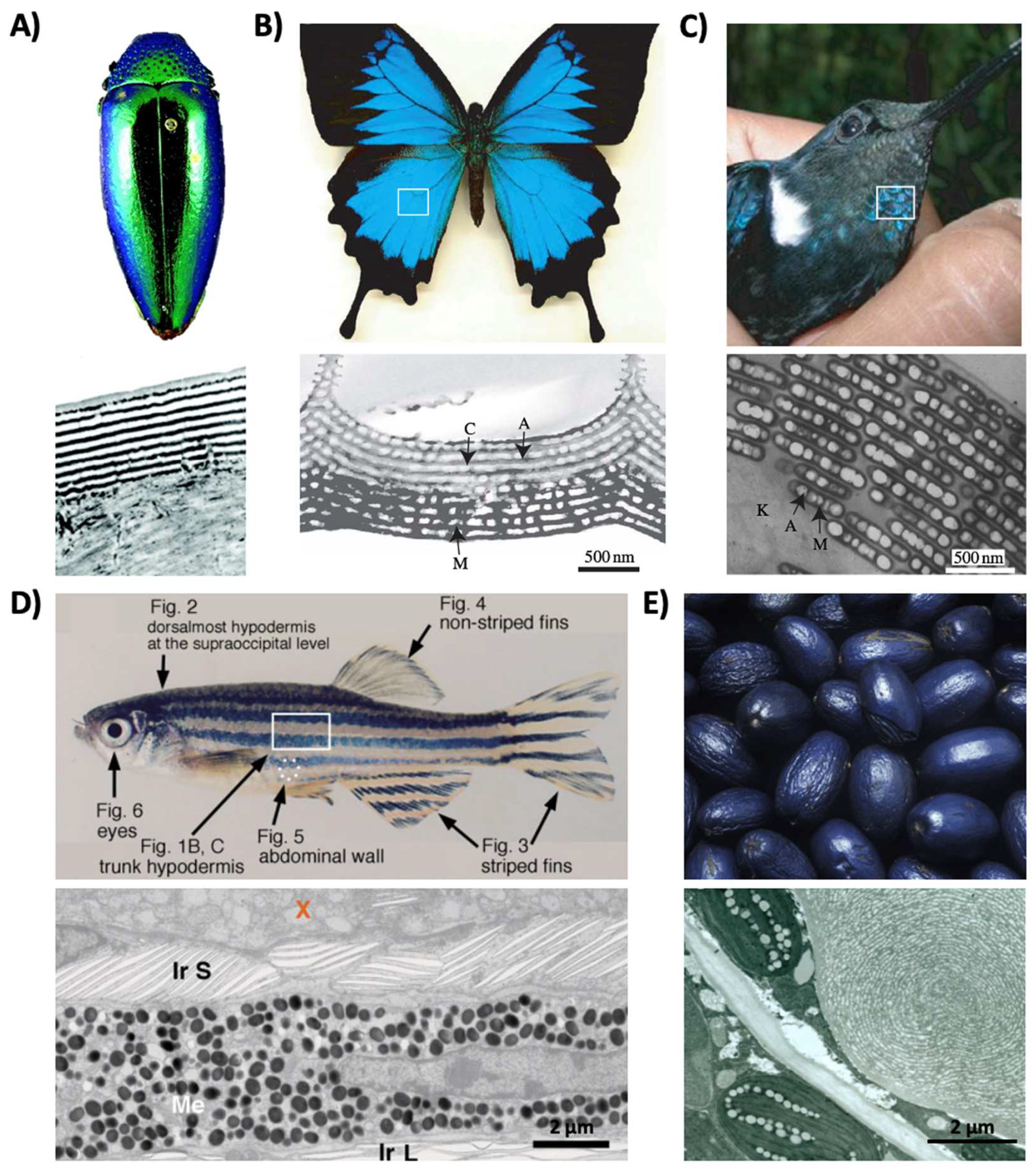
2. Pigmentary and Structural Coloration in Nature
3. Cephalopods as Model Organisms for Dynamic Structural Coloration
4. Techniques for the Structural and Optical Characterization of Proteins
5. Future Directions for the Study of Optically-Active Proteins
6. Conclusions
Funding
Conflicts of Interest
References
- Barrows, F.P.; Bartl, M.H. Photonic structures in biology: A possible blueprint for nanotechnology. Nanomater. Nanotechnol. 2014, 4, 1. [Google Scholar] [CrossRef]
- Jacques, S.L. Optical properties of biological tissues: A review. Phys. Med. Biol. 2013, 58, 37. [Google Scholar] [CrossRef] [PubMed]
- Vukusic, P.; Sambles, J.R. Photonic structures in biology. Nature 2003, 424, 852–855. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Bhushan, B.; Tong, J. Structural coloration in nature. RSC Adv. 2013, 3, 14862–14889. [Google Scholar] [CrossRef]
- Parker, A.R. 515 million years of structural colour. J. Opt. A Pure Appl. Opt. 2000, 2, 15. [Google Scholar] [CrossRef]
- Vukusic, P.; Stavenga, D.G. Physical methods for investigating structural colors in biological systems. J. R. Soc. Interface 2009, 6, S133–S148. [Google Scholar] [CrossRef] [Green Version]
- Haddock, S.H.D.; Moline, M.A.; Case, J.F. Bioluminescence in the Sea. Annu. Rev. Mar. Sci. 2010, 2, 443–493. [Google Scholar] [CrossRef] [Green Version]
- Wilson, T.; Hastings, J.W. Bioluminescence. Annu. Rev. Cell Dev. Biol. 1998, 14, 197–230. [Google Scholar] [CrossRef]
- Shang, L.; Zhang, W.; Xu, K.; Zhao, Y. Bio-inspired intelligent structural color materials. Mater. Horiz. 2019, 6, 945–958. [Google Scholar] [CrossRef]
- Kreit, E.; Mathger, L.M.; Hanlon, R.T.; Dennis, P.B.; Naik, R.R.; Forsythe, E.; Heikenfeld, J. Biological versus electronic adaptive coloration : How can one inform the other? J. R. Soc. Interface 2013, 10, 20120601. [Google Scholar] [CrossRef] [Green Version]
- Phan, L.; Kautz, R.; Leung, E.M.; Naughton, K.L.; Dyke, Y.V.; Gorodetsky, A.A. Dynamic materials inspired by cephalopods. Chem. Mater. 2016, 19, 6804–6816. [Google Scholar] [CrossRef]
- Chatterjee, A.; Norton-Baker, B.; Bagge, L.E.; Patel, P.; Gorodetsky, A.A. An introduction to color-changing systems from the cephalopod protein reflectin. Bioinspir. Biomim. 2018, 13, 045001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawelek, J.; Wong, G.; Sansone, M.; Morowitz, J. Molecular controls in mammalian pigmentation. Yale J. Biol. Med. 1973, 46, 430–443. [Google Scholar]
- Aspengren, S.; Hedberg, D.; Skold, H.N.; Wallin, M. New insights into melanosome transport in vertebrate pigment cells. Int. Rev. Cell Mol. Biol. 2009, 272, 245–302. [Google Scholar] [PubMed]
- Yang, S.; Zhou, J.; Li, D. Functions and diseases of the retinal pigment epithelium. Front. Parmacol. 2021, 12, 1976. [Google Scholar] [CrossRef]
- George, S.M.; Lu, F.; Rao, M.; Leach, L.L.; Gross, J.M. The retinal pigment epithelium: Development, injury responses, and regenerative potential in mammalian and non-mammalian systems. Prog. Retin. Eye Res. 2021, 85, 100969. [Google Scholar] [CrossRef]
- Ralph, C.L. The control of color in birds. Am. Zool. 1969, 9, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Galvan, I.; Solano, F. Bird integumentary melanins: Biosynthesis, forms, function and evolution. Int. J. Mol. Sci. 2016, 17, 520. [Google Scholar] [CrossRef] [Green Version]
- Rembold, H.; Rascher, J.; Eder, J.; Umebachi, Y. Partial structure of the papiliochrome, the yellow wing pigment of the papilionid butterflies. Z. Nat. C 1978, 33, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Koch, P.B.; Behnecke, B.; Weigmann-Lenz, M.; Ffrench-Constant, R.H. Insect pigmentation: Activities of beta-alanyldopamine synthase in wing color patterns of wild-type and melanic mutant swallowtail butterfly Papilio glaucus. Pigment Cell Res. 2000, 13, 54–58. [Google Scholar] [CrossRef]
- Stavenga, D.G.; Giraldo, M.A.; Leertouwer, H.L. Butterfly wing colors: Glass scales of Graphium sarpedon cause polarized iridescence and enhance blue/green pigment coloration of the wing membrane. J. Exp. Biol. 2010, 213, 1731–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilts, B.D.; Matsushita, A.; Arikawa, K.; Stavenga, D.G. Spectrally tuned structural and pigmentary coloration of birdwing butterfly wing scales. J. R. Soc. Interface 2015, 12, 20150717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackiston, D.; Briscoe, A.D.; Weiss, M.R. Color vision and learning in the monarch butterfly, Danaus plexippus (Nymphalidae). J. Exp. Biol. 2011, 214, 509–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, M. Morphological color changes in fish: Regulation of pigment cell density and morphology. Micros. Res. Tech. 2002, 58, 496–503. [Google Scholar] [CrossRef]
- Hirata, M.; Nakamura, K.; Kondo, S. Pigment cell distributions in different tissues of the zebrafish, with special reference to the striped pigment pattern. Dev. Dyn. 2005, 234, 293–300. [Google Scholar] [CrossRef]
- Pereira, D.M.; Valentao, P.; Andrade, P.B. Marine natural pigments: Chemistry, distribution and analysis. Dyes Pigm. 2014, 111, 124–134. [Google Scholar] [CrossRef]
- Mlodzinska, E. Survey of plant pigments: Molecular and environmental determinants of plant colors. Acta Biol. Crac. Ser. Bot. 2009, 51, 7–16. [Google Scholar]
- Sandmann, G. Carotenoid biosynthesis and biotechnological application. Arch. Biochem. Biophys. 2001, 385, 4–12. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.-M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef]
- Rossbach, S.; Subedi, R.C.; Ng, T.K.; Ooi, B.S.; Duarte, C.M. Iridocytes mediate photonic cooperation between giant clams (Tridacninae) and their photosynthetic symbionts. Front. Mar. Sci. 2020, 7, 465. [Google Scholar] [CrossRef]
- Gur, D.; Leshem, B.; Pierantoni, M.; Farstey, V.; Oron, D.; Weiner, S.; Addadi, L. Structural basis for the brilliant colors of the Sapphirinid Copepods. J. Am. Chem. Soc. 2015, 137, 8408–8411. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, S.; Yoshioka, S.; Miyazaki, J. Physics of structural colors. Rep. Prog. Phys. 2008, 71, 76401–76500. [Google Scholar] [CrossRef] [Green Version]
- Seago, A.E.; Brady, P.; Vigneron, J.P.; Schultz, T.D. Gold bugs and beyond: A review of iridescence and structural colour mechanisms in beetles (Coleoptera). J. R. Soc. Interface 2008, 6, S165–S184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shawkey, M.D.; Morehouse, N.I.; Vukusic, P. A protean palette: Colour materials and mixing in birds and butterflies. J. R. Soc. Interface 2009, 6, S221–S231. [Google Scholar] [CrossRef] [Green Version]
- Frohnhofer, H.G.; Krauss, J.; Maischein, H.-M.; Nusslein-Volhard, C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development 2013, 140, 2997–3007. [Google Scholar] [CrossRef] [Green Version]
- Vignolini, S.; Moyroud, E.; Glover, B.J.; Steiner, U. Analysing photonic structures in plants. J. R. Soc. Interface 2013, 10, 20130394. [Google Scholar] [CrossRef] [Green Version]
- Mäthger, L.M.; Bell, G.R.R.; Kuzirian, A.M.; Allen, J.J.; Hanlon, R.T. How does the blue-ringed octopus (Hapalochlaena lunulata) flash its blue rings? J. Exp. Biol. 2012, 215, 3752–3757. [Google Scholar] [CrossRef] [Green Version]
- Mäthger, L.M.; Denton, E.J.; Marshall, N.J.; Hanlon, R.T. Mechanisms and behavioural functions of structural coloration in cephalopods. J. R. Soc. Interface 2008, 6, S149–S163. [Google Scholar] [CrossRef] [Green Version]
- Hanlon, R.T.; Messenger, J.B. Cephalopod Behaviour; Cambridge University Press: New York, NY, USA, 2018; Available online: https://www.cambridge.org/core/books/cephalopod-behaviour/2D21474D460811C160EFDBA35796FAC0 (accessed on 11 May 2022).
- Stevens, M.; Merialaita, S. Animal Camouflage: Mechanisms and Functions; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Bryne, J.H. The Oxford Handbook of Invertebrate Neurobiology; Oxford University Press: Oxford, UK, 2018; Available online: https://www.oxfordhandbooks.com/view/10.1093/oxfordhb/9780190456757.001.0001/oxfordhb-9780190456757 (accessed on 11 May 2022).
- Cloney, R.A.; Brocco, S.L. Chromatophore organs, reflector cells, iridocytes and leucophores in cephalopods. Am. Zool. 1983, 23, 581–592. [Google Scholar] [CrossRef]
- Williams, T.L.; Senft, S.L.; Yeo, J.; Martin-Martinez, F.; Kuzirian, A.M.; Martin, C.A.; DiBona, C.W.; Chen, C.-T.; Dinneen, S.R.; Nguyen, H.T.; et al. Dynamic pigmentary and structural coloration within cephalopod chromatophore organs. Nat. Commun. 2019, 10, 1004. [Google Scholar] [CrossRef]
- Hanlon, R.T.; Cooper, K.M.; Budelmann, B.U.; Pappas, T.C. Physiological color change in squid iridophores. I. Behavior, morphology and pharmacology in Lolliguncula brevis. Cell Tissue Res. 1990, 259, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Mäthger, L.M.; Collins, T.F.T.; Lima, P.A. The role of muscarinic receptors and intracellular Ca2+ in the spectral reflectivity changes of squid iridophores. J. Exp. Biol. 2004, 207, 1759–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wardill, T.J.; Gonzalez-Bellido, P.T.; Crook, R.J.; Hanlon, R.T. Neural control of tuneable skin iridescence in squid. Proc. R. Soc. B 2012, 279, 4243–4252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, A.R.; DeMartini, D.G.; Izumi, M.; Sweeney, A.M.; Holt, A.L.; Morse, D.E. The role of protein assembly in dynamically tunable bio-optical tissues. Biomaterials 2010, 31, 793–801. [Google Scholar] [CrossRef] [PubMed]
- DeMartini, D.G.; Izumi, M.; Weaver, A.T.; Pandolfi, E.; Morse, D.E. Structures, organization, and function of reflectin proteins in dynamically tunable reflective cells. J. Biol. Chem. 2015, 290, 15238–15249. [Google Scholar] [CrossRef] [Green Version]
- DeMartini, D.G.; Krogstad, D.V.; Morse, D.E. Membrane invaginations facilitate reversible water flux driving tunable iridescence in a dynamic biophotonic system. Proc. Natl. Acad. Sci. USA 2013, 110, 2552–2556. [Google Scholar] [CrossRef] [Green Version]
- Cooper, K.M.; Hanlon, R.T.; Budelmann, B.U. Physiological color change in squid iridophores. II. Ultrastructural mechanisms in Lolliguncula brevis. Cell Tissue Res. 1990, 259, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Born, M.; Wolf, E. Electromagnetic Theory of Propagation, Interference and Diffraction of Light. In Principles of Optics; Cambridge University Press: London, UK, 2018; Available online: https://www.cambridge.org/core/books/principles-of-optics/D12868B8AE26B83D6D3C2193E94FFC32 (accessed on 11 May 2022).
- Land, M.F. The physics and biology of animal reflectors. Prog. Biophys. Mol. Biol. 1972, 24, 75–106. [Google Scholar] [CrossRef]
- DeMartini, D.G.; Ghoshal, A.; Pandolfi, E.; Weaver, A.T.; Baum, M.; Morse, D.E. Dynamic biophotonics: Female squid exhibit sexually dimorphic tunable leucophores and iridocytes. J. Exp. Biol. 2013, 216, 3733–3741. [Google Scholar] [CrossRef] [Green Version]
- Stavenga, D.G.; Leertouwer, H.L.; Osorio, D.C.; Wilts, B.D. High refractive index of melanin in shiny occipital feathers of a bird of paradise. Light Sci. Appl. 2015, 4, e243. [Google Scholar] [CrossRef]
- Leertouwer, H.L.; Wilts, B.D.; Stavenga, D.G. Refractive index and dispersion of butterfly chitin and bird keratin measured by polarizing interference microscopy. Opt. Express 2011, 24, 24061–24066. [Google Scholar] [CrossRef] [PubMed]
- Mäthger, L.M.; Senft, S.L.; Gao, M.; Karaveli, S.; Bell, G.R.R.; Zia, R.; Kuzirian, A.M.; Dennis, P.B.; Crookes-Goodson, W.J.; Naik, R.R.; et al. Bright white scattering from protein spheres in color changing, flexible cuttlefish skin. Adv. Funct. Mater. 2013, 23, 3980–3989. [Google Scholar] [CrossRef]
- Ghoshal, A.; DeMartinin, D.G.; Eck, E.; Morse, D.E. Expreimental determination of refractive index of condensed reflectin in squid iridocytes. J. R. Soc. Interface 2014, 11, 20140106. [Google Scholar] [CrossRef] [Green Version]
- Umerani, M.J.; Pratakshya, P.; Chatterjee, A.; Sanchez, J.A.C.; Kim, H.S.; Ilc, G.; Kovačič, M.; Magnan, C.; Marmiroli, B.; Sartori, B.; et al. Structure, self-assembly, and properties of a truncated reflectin variant. Proc. Natl. Acad. Sci. USA 2020, 117, 32891–32901. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.M.; Crookes-Goodson, W.J.; Naik, R.R. The self-organizing properties of squid reflectin protein. Nat. Mater. 2007, 6, 533–538. [Google Scholar] [CrossRef]
- Phan, L.; Iv, W.G.W.; Ordinario, D.D.; Karshalev, E.; Jocson, J.-M.; Burke, A.M.; Gorodetsky, A.A. Reconfigurable infrared camouflage coatings from a cephalopod protein. Adv. Mater. 2013, 25, 5621–5625. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Sanchez, J.A.C.; Yamauchi, T.; Taupin, V.; Couvrette, J.; Gorodetsky, A.A. Cephalopod-inspired optical engineering of human cells. Nat. Commun. 2020, 11, 2708. [Google Scholar] [CrossRef]
- Crookes, W.J.; Ding, L.L.; Huang, Q.L.; Kimbell, J.R.; Horwitz, J.; McFall-Ngai, M.J. Reflectins: The unusual proteins of squid reflective tissues. Science 2004, 303, 235–238. [Google Scholar] [CrossRef]
- Levenson, R.; Braken, C.; Bush, N.; Morse, D.E. Cyclable condensation and hierarchical assembly of metastable reflectin proteins, the drivers of tunable biophotonics. J. Biol. Chem. 2016, 291, 4058–4068. [Google Scholar] [CrossRef] [Green Version]
- Naughton, K.L.; Phan, L.; Leung, E.M.; Kautz, R.; Lin, Q.; Van Dyke, Y.; Marmiroli, B.; Sartori, B.; Arvai, A.; Li, S.; et al. Self-assembly of the cephalopod protein reflectin. Adv. Mater. 2016, 28, 8405–8412. [Google Scholar] [CrossRef]
- Ordinario, D.D.; Leung, E.M.; Phan, L.; Kautz, R.; Lee, W.K.; Naeim, M.; Kerr, J.P.; Aquino, M.J.; Sheehan, P.E.; Gorodetsky, A.A. Protochromic devices from a cephalopod structural protein. Adv. Opt. Mater. 2017, 5, 1600751. [Google Scholar] [CrossRef]
- Qin, G.; Dennis, P.B.; Zhang, Y.; Hu, X.; Bressner, J.E.; Sun, Z.; Goodson, W.; Naik, R.R.; Omenetto, F.G.; Kaplan, D.L. Recombinant reflectin-based optical materials. J. Polym. Sci. B Polym. Phys. 2013, 51, 254–264. [Google Scholar] [CrossRef]
- Wolde-Michael, E. Design and fabrication of recombinant reflectin-based multilayer reflectors: Bio-design engineering and photoisomerism induced wavelength modulation. Sci. Rep. 2021, 11, 14580. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.B.; Singh, K.M.; Vasudev, M.C.; Naik, R.R.; Crookes-Goodson, W.J. Research update: A minimal region of squid reflectin for vapor-induced light scattering. APL Mater. 2017, 5, 120701. [Google Scholar] [CrossRef] [Green Version]
- Guan, Z.; Cai, T.; Liu, Z.; Dou, Y.; Hu, X.; Zhang, P.; Sun, X.; Li, H.; Kuang, Y.; Zhai, Q.; et al. Origin of the reflectin gene and hierarchical assembly of its protein. Curr. Biol. 2017, 27, 2833–2842. [Google Scholar] [CrossRef] [Green Version]
- Hsiung, B.K.; Siddique, R.H.; Jiang, L.; Liu, Y.; Lu, Y.; Shawkey, M.D.; Blackledge, T.A. Tarantula-inspired noniridescent photonics with long-range order. Adv. Opt. Mater. 2017, 5, 1600599. [Google Scholar] [CrossRef]
- Zyla, G.; Kovalev, A.; Grafen, M.; Gurevich, E.; Esen, C.; Ostendorf, A.; Gorb, S. Generation of bioinspired structural colors via two-photon polymerization. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Zhang, H.; Bu, X.; Yip, S.P.; Liang, X.; Ho, J.C. Self-assembly of colloidal particles for fabrication of structural color materials toward advanced intelligent systems. Adv. Intell. Syst. 2020, 2, 1900085. [Google Scholar] [CrossRef]
- Zyla, G.; Kovalev, A.; Gurevich, E.L.; Esen, C.; Liu, Y.; Lu, Y.; Gorb, S.; Ostendorf, A. Structural colors with angle-insensitive optical properties generated by Morpho-inspired 2PP structures. Appl. Phys. A 2020, 126, 1–11. [Google Scholar] [CrossRef]
- Li, K.; Li, C.; Li, H.; Li, M.; Song, Y. Designable structural coloration by colloidal particle assembly: From nature to artificial manufacturing. iScience 2021, 24, 102121. [Google Scholar] [CrossRef]
- Lu, Y.; Pratakshya, P.; Chatterjee, A.; Jia, X.; Ordinario, D.D.; Phan, L.; Sanchez, J.A.C.; Kautz, R.; Tyagi, V.; Patel, P.; et al. Proton conduction in inkjet-printed reflectin films. APL Mater. 2020, 8, 101113. [Google Scholar] [CrossRef]
- Jeong, S.H. Analytical methods for formulation factors to enhance protein stability in solution. Arch. Pharm. Res. 2012, 35, 1871–1886. [Google Scholar] [CrossRef] [PubMed]
- Mikos, A.C.; Sarkar, M.; Wang, Y.; Pielak, G.J. Protein crowding tunes protein stability. J. Am. Chem. Soc. 2011, 133, 7116–7120. [Google Scholar] [CrossRef]
- Chi, E.Y.; Krishnan, S.; Randolph, T.W.; Carpenter, J.F. Physical stability of proteins in aqueous solution: Mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 2003, 20, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Schneider, C.P.; Trout, B.K. Complex interactions between molecular ions in solution and their effect on protein stability. J. Am. Chem. Soc. 2011, 133, 18713–18718. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Mantsch, H.H. The Use and Misuse of FTIR Spectroscopy in the Determination of Protein Structure. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 95–120. [Google Scholar] [CrossRef]
- Haris, P.I.; Severcan, F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B Enzym. 1999, 7, 207–221. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Benevides, J.M.; Overman, S.A.; Thomas, G.J., Jr. Raman spectroscopy of proteins. Curr. Protoc. Protein Sci. 2003, 33, 17–18. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Micsonai, A.; Bulyáki, E.; Kardos, J. BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy. In Structural Genomics. Methods in Molecular Biology; Chen, Y.W., You, C.P.B., Eds.; Humana: New York, NY, USA, 2021; Volume 2199. [Google Scholar]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.W. Protein structure from X-ray diffraction. J. Biol. Phys. 2003, 29, 341–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, M.S.; Martin, J.H.J. X-ray crystallography. J. Clin. Pathol. Mol. Pathol. 2000, 53, 8–14. [Google Scholar] [CrossRef]
- Lyumkis, D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 2019, 294, 5181–5197. [Google Scholar] [CrossRef] [Green Version]
- Turk, M.; Baumeister, W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 2020, 594, 3243–3261. [Google Scholar] [CrossRef]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef]
- Zölls, S.; Gregoritza, M.; Tantipolphan, R.; Wiggenhorn, M.; Winter, G.; Friess, W.; Hawe, A. How subvisible particles become invisible—Relevance of the refractive index for protein particle analysis. J. Pharm. Sci. 2013, 102, 1434–1446. [Google Scholar] [CrossRef]
- Malmsten, M. Ellipsometry studies of protein layers adsorbed at hydrophobic surfaces. J. Colloid Interface Sci. 1994, 166, 333–342. [Google Scholar] [CrossRef]
- Bucciarelli, A.; Mulloni, V.; Maniglio, D.; Pal, R.; Yadavalli, V.; Motta, A.; Quaranta, A. A comparative study of the refractive index of silk protein thin films towards biomaterial based optical devices. Opt. Mater. 2018, 78, 407–414. [Google Scholar] [CrossRef]
- Vörös, J. The density and refractive index of adsorbing protein layers. Biophys. J. 2004, 87, 553–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, T.; Kathman, A.; Koszelak, S.; McPherson, A. Determination of local refractive index for protein and virus crystals in solution by Mach-Zehnder interferometry. Anal. Biochem. 1995, 231, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Kim, Y.; Jung, J.; Ihee, H.; Park, Y. Measurements of complex refractive index change of photoactive yellow protein over a wide wavelength range using hyperspectral quantitative phase imaging. Sci. Rep. 2018, 8, 3064. [Google Scholar] [CrossRef] [PubMed]
- McMeekin, T.L.; Groves, M.L.; Hipp, N.J. Refractive Indices of Amino Acids, Proteins, and Related Substances. In Amino Acids and Serum Proteins, 2nd ed.; Stekol, J.A., Ed.; American Chemical Society: Washington, DC, USA, 1964; pp. 54–66. [Google Scholar]
- Perlmann, G.E.; Longsworth, L.G. The specific refractive increment of some purified proteins. J. Am. Chem. Soc. 1948, 70, 2719–2724. [Google Scholar] [CrossRef]
- Doty, P.; Geiduschek, E.P. The Proteins; Neurath, H., Bailey, K., Eds.; Academic Press: New York, NY, USA, 1953; Volume 1A, p. 393. [Google Scholar]
- Krivosudsky, O.; Draber, P.; Cifra, M. Resolving controversy of unusually high refractive index of a tubulin. Europhys. Lett. 2017, 117, 38003. [Google Scholar] [CrossRef] [Green Version]
- Khago, D.; Bierma, J.C.; Roskamp, K.W.; Kozlyuk, N.; Martin, R.W. Protein refractive index increment is determined by conformation as well as composition. J. Phys. Condens. Matter 2018, 30, 435101. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Brown, P.H.; Schuck, P. On the Distribution of Protein Refractive Index Increments. Biophys. J. 2011, 100, 2309–2317. [Google Scholar] [CrossRef] [Green Version]
- Barer, R.; Tkaczyk, S. Refractive index of concentrated protein solutions. Nature 1954, 173, 821–822. [Google Scholar] [CrossRef]
- Balasubramani, V.; Kus, A.; Tu, H.Y.; Cheng, C.J.; Baczewska, M.; Krauwze, W.; Kujawinska, M. Holographic tomography: Techniques and biomedical applications. Appl. Opt. 2021, 60, B65–B80. [Google Scholar] [CrossRef]
- Kim, D.; Lee, S.; Lee, M.; Oh, J.; Yang, S.A.; Park, Y. Holotomography: Refractive Index as an Intrinsic Imaging Contrast for 3-D Label-Free Live Cell Imaging. In Advanced Imaging and Bio Techniques for Convergence Science; Kim, J.K., Kim, J.K., Park, C.G., Eds.; Springer: Berlin/Heidelberg, Germany, 2021; pp. 211–238. [Google Scholar]
- Kim, K.; Park, W.S.; Na, S.; Kim, S.; Kim, T.; Heo, W.D.; Park, Y. Correlative three-dimensional fluorescence and refractive index tomography: Bridging the gap between molecular specificity and quantitative bioimaging. Biomed. Opt. Express 2017, 8, 5688–5697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-K.; Lee, B.-W.; Fujii, F.; Kim, J.K.; Pack, C.-G. Physicochemical properties of nucleoli in live cells analyzed by label-free optical diffraction tomography. Cells 2019, 8, 699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucic, V.; Leis, A.; Baumeister, W. Cryo-electron tomography of cells: Connecting structure and function. Histochem. Cell Biol. 2008, 130, 185–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asason, S.; Engel, B.D.; Baumeister, W. In Situ cryo-electron tomography: A post-reductionist approach to structural biology. J. Mol. Biol. 2016, 428, 332–343. [Google Scholar]
- Koning, R.I.; Koster, A.J.; Sharp, T.H. Advances in cryo-electron tomography for biology and medicine. Ann. Anat. 2018, 217, 82–96. [Google Scholar] [CrossRef]
- Gui, L.; Ebner, J.L.; Mileant, A.; Williams, J.A.; Lee, K.K. Visualization and sequencing of membrane remodeling leading to influenza virus fusion. J. Virol. 2016, 90, 6948–6962. [Google Scholar] [CrossRef] [Green Version]
- Gipson, P.; Fukuda, Y.; Danev, R.; Lai, Y.; Chen, D.-H.; Baumeister, W.; Brunger, A.T. Morphologies of synaptic protein membrane fusion interfaces. Proc. Natl. Acad. Sci. USA 2017, 114, 9110–9115. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.D.; Beaujeux, T.P.; Shaw, P.J. Protein aggregation in motor neurone disorders. Neuropathol. Appl. Neurobiol. 2003, 29, 529–545. [Google Scholar] [CrossRef]
- Vacquer-Alicea, J.; Diamond, M.I. Propagation of protein aggregation in neurodegenerative diseases. Annu. Rev. Biochem. 2019, 88, 785–810. [Google Scholar] [CrossRef]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef]
- Irvine, G.B.; El-Agnaf, O.M.; Shankar, G.M.; Walsh, D.M. Protein aggregation in the brain: The molecular basis for Alzheimer’s and Parkinson’s diseases. Mol. Med. 2008, 14, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Shastry, B.S. Neurodegnerative disorders of protein aggregation. Neurochem. Int. 2003, 43, 1–7. [Google Scholar] [CrossRef]

| Protein | Refractive Index | Description | References |
|---|---|---|---|
| Melanin | 1.7–1.8 | Bird of paradise melanin measured with Jamin-Lebedeff interference microscopy fit with the Cauchy equation | [54] |
| Keratin | 1.532 | Bird (Anas anas domestica) keratin measured with Jamin-Lebedeff interference microscopy fit with the Cauchy equation | [55] |
| 1.54–1.57 | Bird of paradise keratin measured with Jamin-Lebedeff interference microscopy fit with the Cauchy equation | [54] | |
| Chitin | 1.517 | Butterfly (Graphium Sarpedon) chitin measured with Jamin-Lebedeff interference microscopy fit with the Cauchy equation | [55] |
| Reflectin | 1.41 | Cuttlefish leucosomes | [56] |
| 1.44 | Condensed platelets in squid iridophores | [57] | |
| 1.40–1.47 | Structures formed in solution of a truncated reflectin variant | [58] | |
| 1.54–1.59 | Reflectin-based substrates | [59,60] | |
| 1.42–1.62 | Reflectin-based structures in engineered cells | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatterjee, A. At the Intersection of Natural Structural Coloration and Bioengineering. Biomimetics 2022, 7, 66. https://doi.org/10.3390/biomimetics7020066
Chatterjee A. At the Intersection of Natural Structural Coloration and Bioengineering. Biomimetics. 2022; 7(2):66. https://doi.org/10.3390/biomimetics7020066
Chicago/Turabian StyleChatterjee, Atrouli. 2022. "At the Intersection of Natural Structural Coloration and Bioengineering" Biomimetics 7, no. 2: 66. https://doi.org/10.3390/biomimetics7020066
APA StyleChatterjee, A. (2022). At the Intersection of Natural Structural Coloration and Bioengineering. Biomimetics, 7(2), 66. https://doi.org/10.3390/biomimetics7020066





