Microbial Cells as a Microrobots: From Drug Delivery to Advanced Biosensors
Abstract
:1. Introduction
2. Current Approaches in the Microbial Cell Based Microrobots Development
2.1. Chemotaxis-Based Microrobots
- The chemotaxis process is highly expensive for bacteria, for example, it requires around 3% of the total protein amount in Escherichia Coli [26];
- Bacterial cells do not move as a number of standalone agents using chemotaxis—they have a mechanism of cell-to-cell chemical communication by secreting and sensing small molecules in the environment, which is named quorum sensing [27];
- Both chemotaxis and quorum sensing take place in the liquid media, thus, diffusion is low and hydrodynamics can make such signals noisy [19]; and
- Even in the clonal population of the bacteria, chemotactic sensitivity can be different [28].
2.2. Phototaxis-Based Microrobots
2.3. Magnetotaxis-Based Microrobots
2.4. Comparison of the Different Motility Types
3. Synthetic Biology Approaches for Microbial Cell-Based Microrobots Design
4. Current and Future Applications
- The removal of toxic pollutants from the environment. As in the case of similar applications in medicine, microrobots can realize search-and-destroy behavior to remove toxic molecules. This requires modification of the chemotaxis receptors to react on the target toxic chemical as an attractant.
- HLM with microorganisms that provide self-healing of the material and/or additional functionality such as sensing and the production of useful chemicals, air treatment, etc. [109,110,111]. In the case of the application of a microfluidic network that can provide efficient microorganism transport, microrobots can be a living part of an HLM.
- -
- Crowd effect—the more cells via chemotaxis generate an output signal in some places that have a higher concentration of attractant. Thus, distribution and power of attractant sources theoretically can be displayed;
- -
- -
- Microrobot sensor networks—distribution of sensing duties between different groups of cells, each of them responsible for sensing its own group of parameters [61].
5. Discussion and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| DOX | Doxorubicin |
| HLM | Hybrid living materials |
| MRI | Magnetic resonance imaging |
| nnAAs | Non-canonical amino acids |
| RNA | Ribonucleic acid |
| XNA | Xeno-nucleic acid |
References
- Zhang, D.; Gorochowski, T.E.; Marucci, L.; Lee, H.-T.; Gil, B.; Li, B.; Hauert, S.; Yeatman, E. Advanced medical micro-robotics for early diagnosis and therapeutic interventions. Front. Robot. AI 2023, 9, 358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, W.; Xi, N.; Wang, Y.; Liu, L. Development and Future Challenges of Bio-Syncretic Robots. Engineering 2018, 4, 452–463. [Google Scholar] [CrossRef]
- Li, Z.; Balance, W.C.; Hossain, S.; Higueras-ruiz, D.R.; Nishikawa, K. Biohybrid robots: Recent progress, challenges, and perspectives. Bioinspir. Biomim. 2023, 18, 015001. [Google Scholar]
- Li, J.; De Ávila, B.E.-F.; Gao, W.; Zhang, L.; Wang, J. Micro/nanorobots for biomedicine: Delivery, surgery, sensing, and detoxification. Sci. Robot. 2017, 2, eaam6431. [Google Scholar] [CrossRef]
- Dabbagh, S.R.; Sarabi, M.R.; Birtek, M.T.; Seyfi, S.; Sitti, M.; Tasoglu, S. 3D-printed microrobots from design to translation. Nat. Commun. 2022, 13, 5875. [Google Scholar] [CrossRef]
- Peyer, K.E.; Zhang, L.; Nelson, B.J. Bio-inspired magnetic swimming microrobots for biomedical applications. Nanoscale 2013, 5, 1259–1272. [Google Scholar] [CrossRef]
- Dodampegama, S.; Mudugamuwa, A.; Konara, M.; Perera, N.; De Silva, D.; Roshan, U.; Amarasinghe, R.; Jayaweera, N.; Tamura, H. A Review on the Motion of Magnetically Actuated Bio-Inspired Microrobots. Appl. Sci. 2022, 12, 11542. [Google Scholar] [CrossRef]
- Wei, Y.; Wu, Z.; Dai, Z.; Zhou, B.; Xu, Q. Design of a Magnetic Soft Inchworm Millirobot Based on Pre-Strained Elastomer with Micropillars. Biomimetics 2023, 8, 22. [Google Scholar] [CrossRef]
- Sitti, M.; Wiersma, D.S. Pros and Cons: Magnetic versus Optical Microrobots. Adv. Mater. 2020, 32, 1906766. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, H.; Fu, R.; Wang, X.; Yu, J.; Zhang, S.; Huang, Q.; Sun, Y.; Fukuda, T. A review on microrobots driven by optical and magnetic fields. Lab Chip 2023, 23, 848–868. [Google Scholar] [CrossRef]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.K.; Zhao, X.; Tansi, B.M.; Méndez-Ortiz, W.J.; Córdova-Figueroa, U.M.; Golestanian, R.; Sen, A. Micromotors Powered by Enzyme Catalysis. Nano Lett. 2015, 15, 8311–8315. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, P.; Der, B.S.; Bhatia, S.; Roehner, N.; Silva, R.; Voigt, C.A.; Densmore, D. A Framework for Genetic Logic Synthesis. Proc. IEEE 2015, 103, 2196–2207. [Google Scholar] [CrossRef]
- Jones, T.S.; Oliveira, S.M.D.; Myers, C.J.; Voigt, C.A.; Densmore, D. Genetic circuit design automation with Cello 2.0. Nat. Protoc. 2022, 17, 1097–1113. [Google Scholar] [CrossRef]
- Schauer, O.; Mostaghaci, B.; Colin, R.; Hürtgen, D.; Kraus, D.; Sitti, M.; Sourjik, V. Motility and chemotaxis of bacteria-driven microswimmers fabricated using antigen 43-mediated biotin display. Sci. Rep. 2018, 8, 9801. [Google Scholar] [CrossRef] [Green Version]
- Akolpoglu, M.B.; Dogan, N.O.; Bozuyuk, U.; Ceylan, H.; Kizilel, S.; Sitti, M. High-Yield Production of Biohybrid Microalgae for On-Demand Cargo Delivery. Adv. Sci. 2020, 7, 2001256. [Google Scholar] [CrossRef]
- Weibel, D.B.; Garstecki, P.; Ryan, D.; DiLuzio, W.R.; Mayer, M.; Seto, J.E.; Whitesides, G.M. Microoxen: Microorganisms to move microscale loads. Proc. Natl. Acad. Sci. USA 2005, 102, 11963–11967. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Liu, A.; Diller, E.; Sitti, M. Chemotactic steering of bacteria propelled microbeads. Biomed. Microdevices 2012, 14, 1009–1017. [Google Scholar] [CrossRef]
- Micali, G.; Endres, R.G. Bacterial chemotaxis: Information processing, thermodynamics, and behavior. Curr. Opin. Microbiol. 2016, 30, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Arrieta, J.; Barreira, A.; Chioccioli, M.; Polin, M.; Tuval, I. Phototaxis beyond turning: Persistent accumulation and response acclimation of the microalga Chlamydomonas reinhardtii. Sci. Rep. 2017, 7, 3447. [Google Scholar] [CrossRef]
- Faivre, D.; Schüler, D. Magnetotactic Bacteria and Magnetosomes. Chem. Rev. 2008, 108, 4875–4898. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Zhao, S.; Jiang, X.; Sun, Y.; Zhao, S.; Gao, J.; Borleis, J.; Willard, S.; Tang, M.; Cai, H.; et al. A large-scale screen reveals genes that mediate electrotaxis in Dictyostelium discoideum. Sci. Signal. 2015, 8, ra50. [Google Scholar] [CrossRef] [Green Version]
- Chong, P.; Erable, B.; Bergel, A. How bacteria use electric fields to reach surfaces. Biofilm 2021, 3, 100048. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.D.; Wolanin, P.M.; Stock, J.B. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 2006, 9, 187–192. [Google Scholar] [CrossRef]
- Cogan, N.G.; Gunn, J.S.; Wozniak, D.J. Biofilms and infectious diseases: Biology to mathematics and back again. FEMS Microbiol. Lett. 2011, 322, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.C. The Rotary Motor of Bacterial Flagella. Annu. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salek, M.M.; Carrara, F.; Fernandez, V.; Guasto, J.S.; Stocker, R. Bacterial chemotaxis in a microfluidic T-maze reveals strong phenotypic heterogeneity in chemotactic sensitivity. Nat. Commun. 2019, 10, 1877. [Google Scholar] [CrossRef] [Green Version]
- Tindall, M.J.; Maini, P.K.; Porter, S.L.; Armitage, J.P. Overview of Mathematical Approaches Used to Model Bacterial Chemotaxis II: Bacterial Populations. Bull. Math. Biol. 2008, 70, 1570–1607. [Google Scholar] [CrossRef] [Green Version]
- D’Acunto, B.; Frunzo, L.; Klapper, I.; Mattei, M.R.; Stoodley, P. Mathematical modeling of dispersal phenomenon in biofilms. Math. Biosci. 2019, 307, 70–87. [Google Scholar] [CrossRef] [Green Version]
- Acemel, R.D.; Govantes, F.; Cuetos, A. Computer simulation study of early bacterial biofilm development. Sci. Rep. 2018, 8, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Guo, S. Dynamics for a two-species competitive Keller–Segel chemotaxis system with a free boundary. J. Math. Anal. Appl. 2021, 502, 125259. [Google Scholar] [CrossRef]
- Bubba, F.; Lorenzi, T.; Macfarlane, F.R. From a discrete model of chemotaxis with volume-filling to a generalized Patlak–Keller–Segel model. Proc. R. Soc. A 2020, 476, 20190871. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, G.; Erhardt, A.H.; Eswaramoorthy, I.; Krishnan, B. Existence of weak solutions to the Keller–Segel chemotaxis system with additional cross-diffusion. Nonlinear Anal. Real World Appl. 2020, 54, 103090. [Google Scholar] [CrossRef]
- Izumi, S.; Azuma, S.I.; Sugie, T. Multi-Robot Control Inspired by Bacterial Chemotaxis: Coverage and Rendezvous via Networking of Chemotaxis Controllers. IEEE Access 2020, 8, 124172–124184. [Google Scholar] [CrossRef]
- Yang, B.; Ding, Y.; Jin, Y.; Hao, K. Self-organized swarm robot for target search and trapping inspired by bacterial chemotaxis. Robot. Auton. Syst. 2015, 72, 83–92. [Google Scholar] [CrossRef]
- Shechter, E.; Martel, S. Principles of motion control of bacterial micro-robots using oxygen gradients. In Proceedings of the IEEE/ASME International Conference on Advanced Intelligent Mechatronics, AIM 2010, Montreal, QC, Canada, 6–9 July 2010; pp. 848–853. [Google Scholar]
- Higashi, K.; Miki, N. A self-swimming microbial robot using microfabricated nanofibrous hydrogel. Sens. Actuators B Chem. 2014, 202, 301–306. [Google Scholar] [CrossRef]
- Kuo, C.H.; Chen, J.H.; Liou, B.K.; Lee, C.K. Utilization of acetate buffer to improve bacterial cellulose production by Gluconacetobacter xylinus. Food Hydrocoll. 2016, 53, 98–103. [Google Scholar] [CrossRef]
- Schaller, K.; David, R.; Uhl, R. How Chlamydomonas keeps track of the light once it has reached the right phototactic orientation. Biophys. J. 1997, 73, 1562–1572. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, S.K.; Baskaran, A.; Sharma, P. Reentrant Efficiency of Phototaxis in Chlamydomonas reinhardtii Cells. Biophys. J. 2019, 117, 1508–1513. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, S.P.; Manuell, A.L.; Chen, S.; Wu, J.; Tran, M.; Siefker, D.; Muto, M.; Marin-Navarro, J. Chlamydomonas reinhardtii chloroplasts as protein factories. Curr. Opin. Biotechnol. 2007, 18, 126–133. [Google Scholar] [CrossRef]
- Redekop, P.; Rothhausen, N.; Rothhausen, N.; Melzer, M.; Mosebach, L.; Dülger, E.; Bovdilova, A.; Caffarri, S.; Hippler, M.; Jahns, P. PsbS contributes to photoprotection in Chlamydomonas reinhardtii independently of energy dissipation. Biochim. Biophys. Acta-Bioenerg. 2020, 1861, 148183. [Google Scholar] [CrossRef] [PubMed]
- Giometto, A.; Altermatt, F.; Maritan, A.; Stocker, R.; Rinaldo, A. Generalized receptor law governs phototaxis in the phytoplankton Euglena gracilis. Proc. Natl. Acad. Sci. USA 2015, 112, 7045–7050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.; Barzyk, A.; Bertin, E.; Peyla, P.; Rafai, S. Photofocusing: Light and flow of phototactic microswimmer suspension. Phys. Rev. E 2016, 93, 051101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, S.; Jiao, N.; Tung, S.; Liu, L. Controlled regular locomotion of algae cell microrobots. Biomed. Microdevices 2016, 18, 47. [Google Scholar] [CrossRef]
- Sukhinov, D.V.; Gorin, K.V.; Romanov, A.O.; Gotovtsev, P.M.; Sergeeva, Y.E. Increased C-phycocyanin extract purity by flocculation of Arthrospira platensis with chitosan. Algal Res. 2021, 58, 102393. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, Y.; Yin, Q.; Liu, G.; Liu, H.; Huang, Y.; Li, B. Phycocyanin: A Potential Drug for Cancer Treatment. J. Cancer 2017, 8, 3416–3429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Zhuang, J.; Li, Z.; Gong, H.; de Ávila, B.E.-F.; Duan, Y.; Zhang, Q.; Zhou, J.; Yin, L.; Karshalev, E.; et al. Nanoparticle-modified microrobots for in vivo antibiotic delivery to treat acute bacterial pneumonia. Nat. Mater. 2022, 21, 1324–1332. [Google Scholar] [CrossRef]
- Choi, H.; Kim, B.; Jeong, S.H.; Kim, T.Y.; Kim, D.-P.; Oh, Y.-K.; Hahn, S.K. Microalgae-Based Biohybrid Microrobot for Accelerated Diabetic Wound Healing. Small 2022, 19, 2204617. [Google Scholar] [CrossRef]
- Martel, S. Flagellated bacterial nanorobots for medical interventions in the human body. In Surgical Robotics: Systems Applications and Visions; Springer: New York, NY, USA, 2011; pp. 397–416. ISBN 9781441911254. [Google Scholar]
- Yasa, O.; Erkoc, P.; Alapan, Y.; Sitti, M. Microalga-Powered Microswimmers toward Active Cargo Delivery. Adv. Mater. 2018, 30, 1804130. [Google Scholar] [CrossRef]
- Klumpp, S.; Kiani, B.; Vach, P.; Faivre, D. Navigation with magnetic nanoparticles: Magnetotactic bacteria and magnetic micro-robots. Phys. Scr. 2015, 2015, 014044. [Google Scholar] [CrossRef]
- Martel, S.; Tremblay, C.C.; Ngakeng, S.; Langlois, G. Controlled manipulation and actuation of micro-objects with magnetotactic bacteria. Appl. Phys. Lett. 2006, 89, 233904. [Google Scholar] [CrossRef] [Green Version]
- Alapan, Y.; Yasa, O.; Schauer, O.; Giltinan, J.; Tabak, A.F.; Sourjik, V.; Sitti, M. Soft erythrocyte-based bacterial microswimmers for cargo delivery. Sci. Robot. 2018, 3, eaar4423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Soto, F.; Liu, S.; Yin, Q.; Purcell, E.; Zeng, Y.; Hsu, E.C.; Akin, D.; Sinclair, B.; Stoyanova, T.; et al. Volbots: Volvox Microalgae-Based Robots for Multimode Precision Imaging and Therapy. Adv. Funct. Mater. 2022, 32, 2201800. [Google Scholar] [CrossRef]
- Bi, S.; Sourjik, V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 2018, 45, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Carroll, B.L.; Nishikino, T.; Guo, W.; Zhu, S.; Kojima, S.; Homma, M.; Liu, J. The flagellar motor of Vibrio alginolyticus undergoes major structural remodeling during rotational switching. Elife 2020, 9, e61446. [Google Scholar] [CrossRef]
- Hicks, M.; Bachmann, T.T.; Wang, B. Synthetic Biology Enables Programmable Cell-Based Biosensors. ChemPhysChem 2020, 21, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Del Vecchio, D.; Qian, Y.; Murray, R.M.; Sontag, E.D. Future systems and control research in synthetic biology. Annu. Rev. Control. 2018, 45, 5–17. [Google Scholar] [CrossRef]
- Gotovtsev, P.M.; Konova, I.A. Synthetic Biology as a Bridge to Integration of Bio Objects into Internet of Things. In Proceedings of the 2019 International Conference on Sensing and Instrumentation in IoT Era (ISSI), Lisbon, Portugal, 29–30 August 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–6. [Google Scholar]
- Del Valle, I.; Fulk, E.M.; Kalvapalle, P.; Silberg, J.J.; Masiello, C.A.; Stadler, L.B. Translating New Synthetic Biology Advances for Biosensing Into the Earth and Environmental Sciences. Front. Microbiol. 2021, 11, 618373. [Google Scholar] [CrossRef]
- Lapinaite, A.; Knott, G.J.; Palumbo, C.M.; Lin-Shiao, E.; Richter, M.F.; Zhao, K.T.; Beal, P.A.; Liu, D.R.; Doudna, J.A. DNA capture by a CRISPR-Cas9–guided adenine base editor. Science 2020, 369, 566–571. [Google Scholar] [CrossRef]
- Nielsen, A.K.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Densmore, D.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352, 53–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, J.C.; Glass, J.I.; Hutchison, C.A.; Vashee, S. Synthetic chromosomes, genomes, viruses, and cells. Cell 2022, 185, 2708–2724. [Google Scholar] [CrossRef] [PubMed]
- Mazzolini, R.; Rodríguez-Arce, I.; Fernández-Barat, L.; Piñero-Lambea, C.; Garrido, V.; Rebollada-Merino, A.; Motos, A.; Torres, A.; Grilló, M.J.; Serrano, L.; et al. Engineered live bacteria suppress Pseudomonas aeruginosa infection in mouse lung and dissolve endotracheal-tube biofilms. Nat. Biotechnol. 2023, 1–10. [Google Scholar] [CrossRef] [PubMed]
- De Bhowmick, G.; Koduru, L.; Sen, R. Metabolic pathway engineering towards enhancing microalgal lipid biosynthesis for biofuel application—A review. Renew. Sustain. Energy Rev. 2015, 50, 1239–1253. [Google Scholar] [CrossRef]
- Hidalgo Martinez, D.; Betterle, N.; Melis, A. Phycocyanin Fusion Constructs for Heterologous Protein Expression Accumulate as Functional Heterohexameric Complexes in Cyanobacteria. ACS Synth. Biol. 2022, 11, 1152–1166. [Google Scholar] [CrossRef]
- Naduthodi, M.I.S.; Claassens, N.J.; D’Adamo, S.; van der Oost, J.; Barbosa, M.J. Synthetic Biology Approaches To Enhance Microalgal Productivity. Trends Biotechnol. 2021, 39, 1019–1036. [Google Scholar] [CrossRef]
- Sexton, J.T.; Tabor, J.J. Multiplexing cell-cell communication. Mol. Syst. Biol. 2020, 16, e9618. [Google Scholar] [CrossRef]
- Nakano, T.; Suda, T.; Okaie, Y.; Moore, M.J.; Vasilakos, A.V. Molecular Communication Among Biological Nanomachines: A layered architecture and research Issues. IEEE Trans. Nanobiosci. 2014, 13, 169–197. [Google Scholar] [CrossRef]
- Zhou, X.; Franklin, R.A.; Adler, M.; Jacox, J.B.; Bailis, W.; Shyer, J.A.; Flavell, R.A.; Mayo, A.; Alon, U.; Medzhitov, R. Circuit Design Features of a Stable Two-Cell System. Cell 2018, 172, 744–757.e17. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, J.; Yin, P.; Ortiz, M.E.; Subsoontorn, P.; Endy, D. Amplifying Genetic Logic Gates. Science 2013, 340, 599–603. [Google Scholar] [CrossRef]
- Frei, T.; Khammash, M. Adaptive circuits in synthetic biology. Curr. Opin. Syst. Biol. 2021, 28, 100399. [Google Scholar] [CrossRef]
- Bi, S.; Pollard, A.M.; Yang, Y.; Jin, F.; Sourjik, V. Engineering Hybrid Chemotaxis Receptors in Bacteria. ACS Synth. Biol. 2016, 5, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Duran-Nebreda, S.; Solé, R.V. Toward Synthetic Spatial Patterns in Engineered Cell Populations with Chemotaxis. ACS Synth. Biol. 2016, 5, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Fernandes e Silva, E.; Figueira, F.D.S.; Lettnin, A.P.; Carrett-Dias, M.; Filgueira, D.D.M.V.B.; Kalil, S.; Trindade, G.S.; Votto, A.P.D.S. C-Phycocyanin: Cellular targets, mechanisms of action and multi drug resistance in cancer. Pharmacol. Rep. 2018, 70, 75–80. [Google Scholar] [CrossRef]
- Jiang, W.Z.; Weeks, D.P. A gene-within-a-gene Cas9/sgRNA hybrid construct enables gene editing and gene replacement strategies in Chlamydomonas reinhardtii. Algal Res. 2017, 26, 474–480. [Google Scholar] [CrossRef]
- Shin, S.E.; Lim, J.M.; Koh, H.G.; Kim, E.K.; Kang, N.K.; Jeon, S.; Kwon, S.; Shin, W.S.; Lee, B.; Hwangbo, K.; et al. CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci. Rep. 2016, 6, 27810. [Google Scholar] [CrossRef]
- Navarro, F.; Baulcombe, D.C. miRNA-Mediated Regulation of Synthetic Gene Circuits in the Green Alga Chlamydomonas reinhardtii. ACS Synth. Biol. 2019, 8, 358–370. [Google Scholar] [CrossRef]
- Yamano, T.; Iguchi, H.; Fukuzawa, H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 2013, 115, 691–694. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Patena, W.; Fauser, F.; Jinkerson, R.E.; Saroussi, S.; Meyer, M.T.; Ivanova, N.; Robertson, J.M.; Yue, R.; Zhang, R.; et al. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 2019, 51, 627–635. [Google Scholar] [CrossRef]
- Mogas-Díez, S.; Gonzalez-Flo, E.; Macía, J. 2D printed multicellular devices performing digital and analogue computation. Nat. Commun. 2021, 12, 1679. [Google Scholar] [CrossRef]
- Gotovtsev, P.M.; Kirillova, D.A.; Vasilov, R.G. Biocomputers: Problems They Solve, State of the Art, and Prospects. Nanotechnol. Russ. 2020, 15, 3–12. [Google Scholar] [CrossRef]
- Colin, R.; Sourjik, V. Emergent properties of bacterial chemotaxis pathway. Curr. Opin. Microbiol. 2017, 39, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Kassinger, S.J.; van Hoek, M.L. Biofilm architecture: An emerging synthetic biology target. Synth. Syst. Biotechnol. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Bagdonas, H.; Fogarty, C.A.; Fadda, E.; Agirre, J. The case for post-predictional modifications in the AlphaFold Protein Structure Database. Nat. Struct. Mol. Biol. 2021, 28, 869–870. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, W. Synthetic biology: Recent progress, biosafety and biosecurity concerns, and possible solutions. J. Biosaf. Biosecurity 2019, 1, 22–30. [Google Scholar] [CrossRef]
- Schmidt, M. Xenobiology: A new form of life as the ultimate biosafety tool. Bioessays 2010, 32, 322–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acevedo-Rocha, C.G.; Budisa, N. Xenomicrobiology: A roadmap for genetic code engineering. Microb. Biotechnol. 2016, 9, 666–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, M.; Pei, L.; Budisa, N. Xenobiology: State-of-the-art, ethics, and philosophy of new-to-nature organisms. In Advances in Biochemical Engineering/Biotechnology; Springer Science and Business Media Deutschland GmbH: Cham, Switzerland, 2018; Volume 162, pp. 301–316. [Google Scholar]
- Maiti, M.; Maiti, M.; Knies, C.; Dumbre, S.; Lescrinier, E.; Rosemeyer, H.; Ceulemans, A.; Herdewijn, P. Xylonucleic acid: Synthesis, structure, and orthogonal pairing properties. Nucleic Acids Res. 2015, 43, 7189–7200. [Google Scholar] [CrossRef] [Green Version]
- Hoshika, S.; Leal, N.A.; Kim, M.-J.; Kim, M.-S.; Karalkar, N.B.; Kim, H.-J.; Bates, A.M.; Watkins, N.E.; Santa Lucia, H.A.; Meyer, A.J.; et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 2019, 363, 884–887. [Google Scholar] [CrossRef]
- Whitford, C.M.; Dymek, S.; Kerkhoff, D.; März, C.; Schmidt, O.; Edich, M.; Droste, J.; Pucker, B.; Rückert, C.; Kalinowski, J. Auxotrophy to Xeno-DNA: An exploration of combinatorial mechanisms for a high-fidelity biosafety system for synthetic biology applications. J. Biol. Eng. 2018, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Shetty, R.; Endy, D.; Knight, T. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008, 2, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhari, V.; Hanson, M. GoldBricks: An improved cloning strategy that combines features of Golden Gate and BioBricks for better efficiency and usability. Synth. Biol. 2021, 6, ysab032. [Google Scholar] [CrossRef]
- Karr, J.R.; Sanghvi, J.C.; Macklin, D.N.; Gutschow, M.V.; Jacobs, J.M.; Bolival, B.; Assad-Garcia, N.; Glass, J.I.; Covert, M.W. A Whole-Cell Computational Model Predicts Phenotype from Genotype. Cell 2012, 150, 389–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornburg, Z.R.; Bianchi, D.M.; Brier, T.A.; Gilbert, B.R.; Earnest, T.M.; Melo, M.C.R.; Safronova, N.; Sáenz, J.P.; Cook, A.T.; Wise, K.S.; et al. Fundamental behaviors emerge from simulations of a living minimal cell. Cell 2022, 185, 345–360.e28. [Google Scholar] [CrossRef]
- Frei, T.; Chang, C.-H.; Filo, M.; Arampatzis, A.; Khammash, M. A genetic mammalian proportional–integral feedback control circuit for robust and precise gene regulation. Proc. Natl. Acad. Sci. USA 2022, 119, e2122132119. [Google Scholar] [CrossRef] [PubMed]
- Grunberg, T.W.; Del Vecchio, D. Modular Analysis and Design of Biological Circuits. Curr. Opin. Biotechnol. 2020, 63, 41–47. [Google Scholar] [CrossRef]
- Sørensen, S.J.; Burmølle, M.; Hansen, L.H. Making bio-sense of toxicity: New developments in whole-cell biosensors. Curr. Opin. Biotechnol. 2006, 17, 11–16. [Google Scholar] [CrossRef]
- Woo, S.G.; Moon, S.J.; Kim, S.K.; Kim, T.H.; Lim, H.S.; Yeon, G.H.; Sung, B.H.; Lee, C.H.; Lee, S.G.; Hwang, J.H.; et al. A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens. Bioelectron. 2020, 168, 112523. [Google Scholar] [CrossRef] [PubMed]
- Riangrungroj, P.; Bever, C.S.; Hammock, B.D.; Polizzi, K.M. A label-free optical whole-cell Escherichia coli biosensor for the detection of pyrethroid insecticide exposure. Sci. Rep. 2019, 9, 12466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotovtsev, P.M.; Dyakov, A.V. Biotechnology and Internet of Things for green smart city application. In Proceedings of the 2016 IEEE 3rd World Forum on Internet of Things (WF-IoT), Reston, VA, USA, 12–14 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 542–546. [Google Scholar]
- Chen, S.Y.; Wei, W.; Yin, B.-C.; Tong, Y.; Lu, J.; Ye, B.C. Development of a Highly Sensitive Whole-Cell Biosensor for Arsenite Detection through Engineered Promoter Modifications. ACS Synth. Biol. 2019, 8, 2295–2302. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, M.; Rühs, P.A.; Coulter, F.; Kilcher, S.; Studart, A.R. 3D printing of bacteria into functional complex materials. Sci. Adv. 2017, 3, eaao6804. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.S.H.; Bader, C.; Sharma, S.; Kolb, D.; Tang, T.C.; Hosny, A.; Moser, F.; Weaver, J.C.; Voigt, C.A.; Oxman, N. Hybrid Living Materials: Digital Design and Fabrication of 3D Multimaterial Structures with Programmable Biohybrid Surfaces. Adv. Funct. Mater. 2020, 30, 1907401. [Google Scholar] [CrossRef]
- Gotovtsev, P. How IoT Can Integrate Biotechnological Approaches for City Applications—Review of Recent Advancements, Issues, and Perspectives. Appl. Sci. 2020, 10, 3990. [Google Scholar] [CrossRef]
- Rodrigues, D.; Barbosa, A.; Rebelo, R.; Kwon, I.; Reis, R.; Correlo, V. Skin-integrated wearable systems and implantable biosensors: A comprehensive review. Biosensors 2020, 10, 79. [Google Scholar] [CrossRef]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef]
- Hutchison, C.A.; Chuang, R.-Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef] [Green Version]
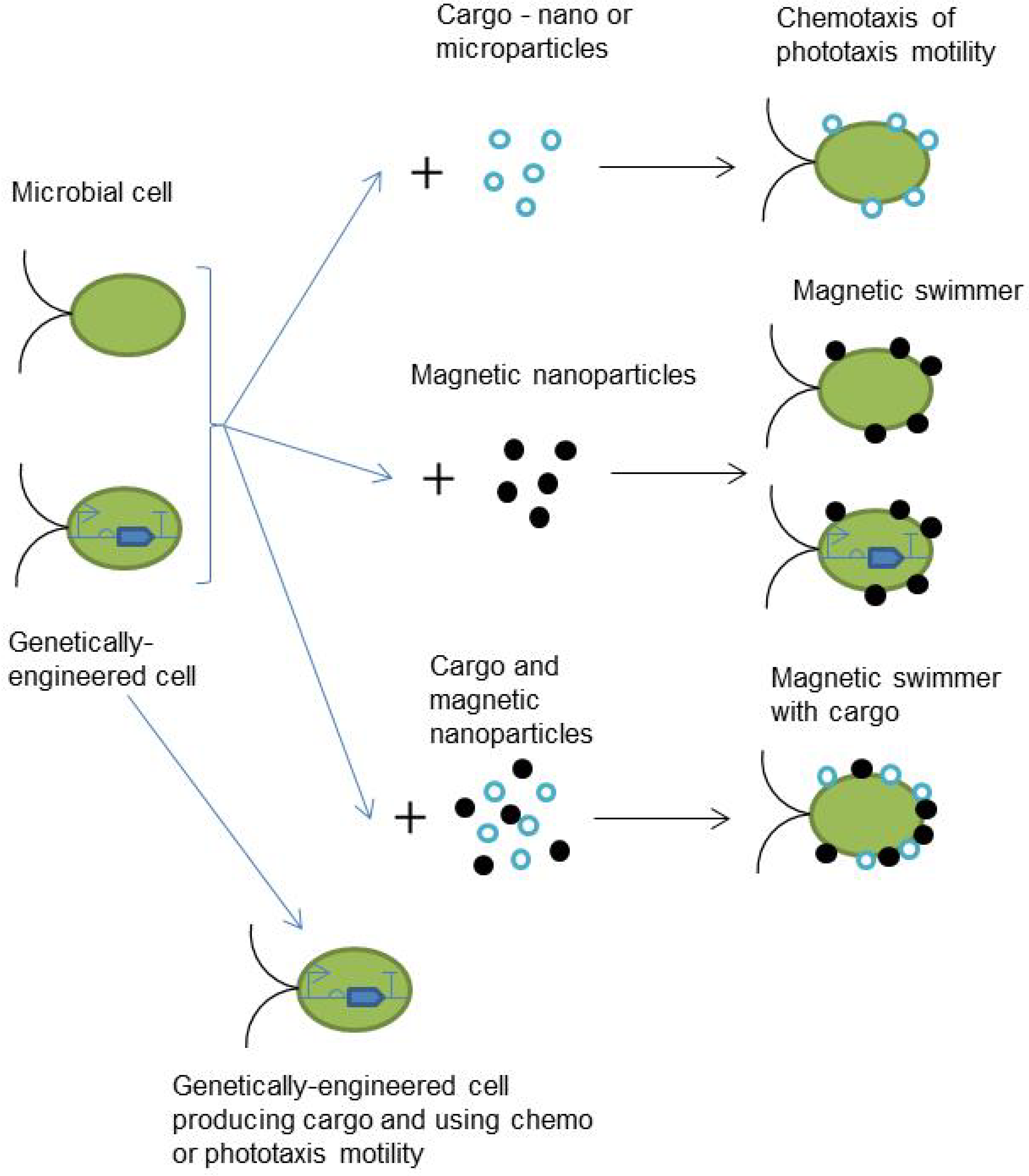
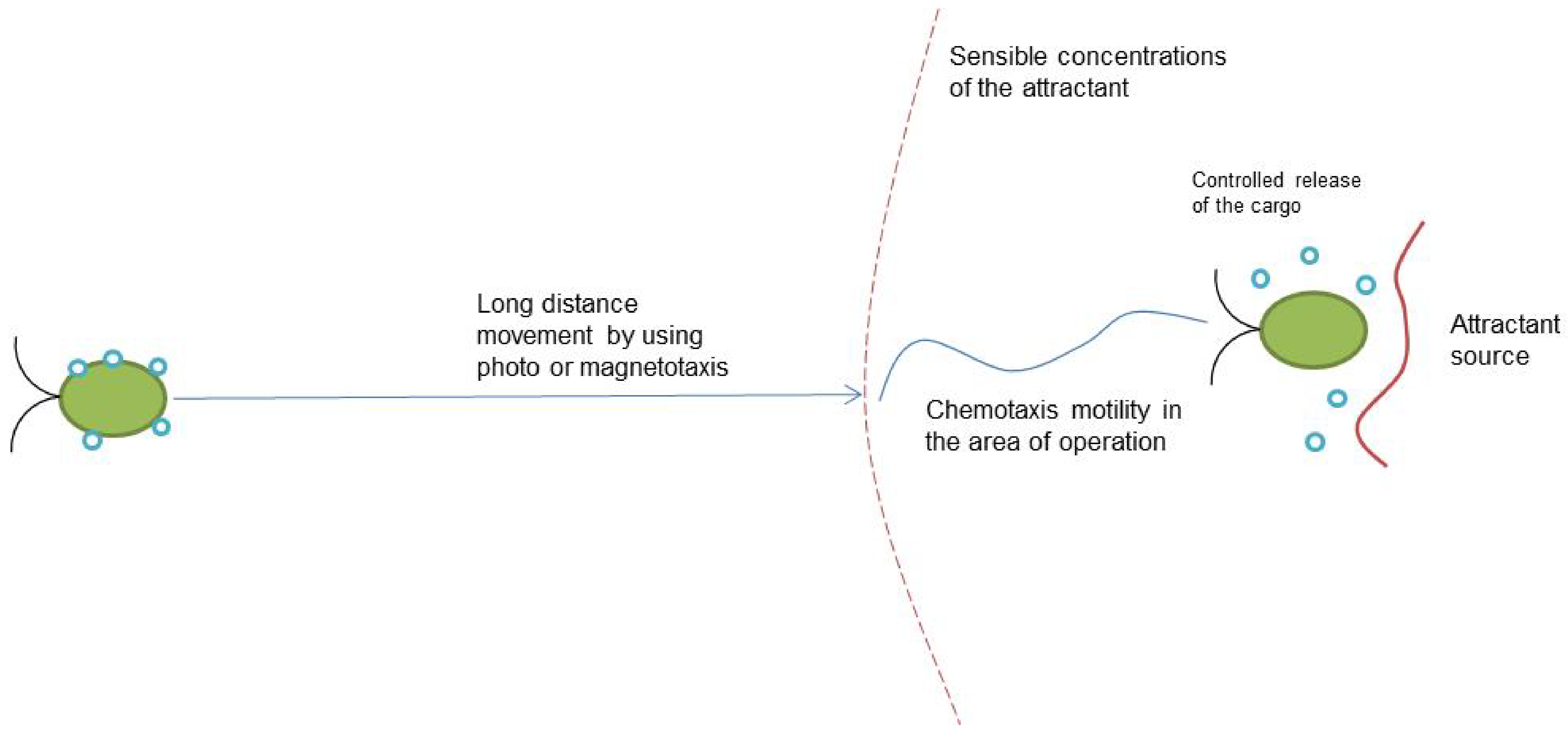
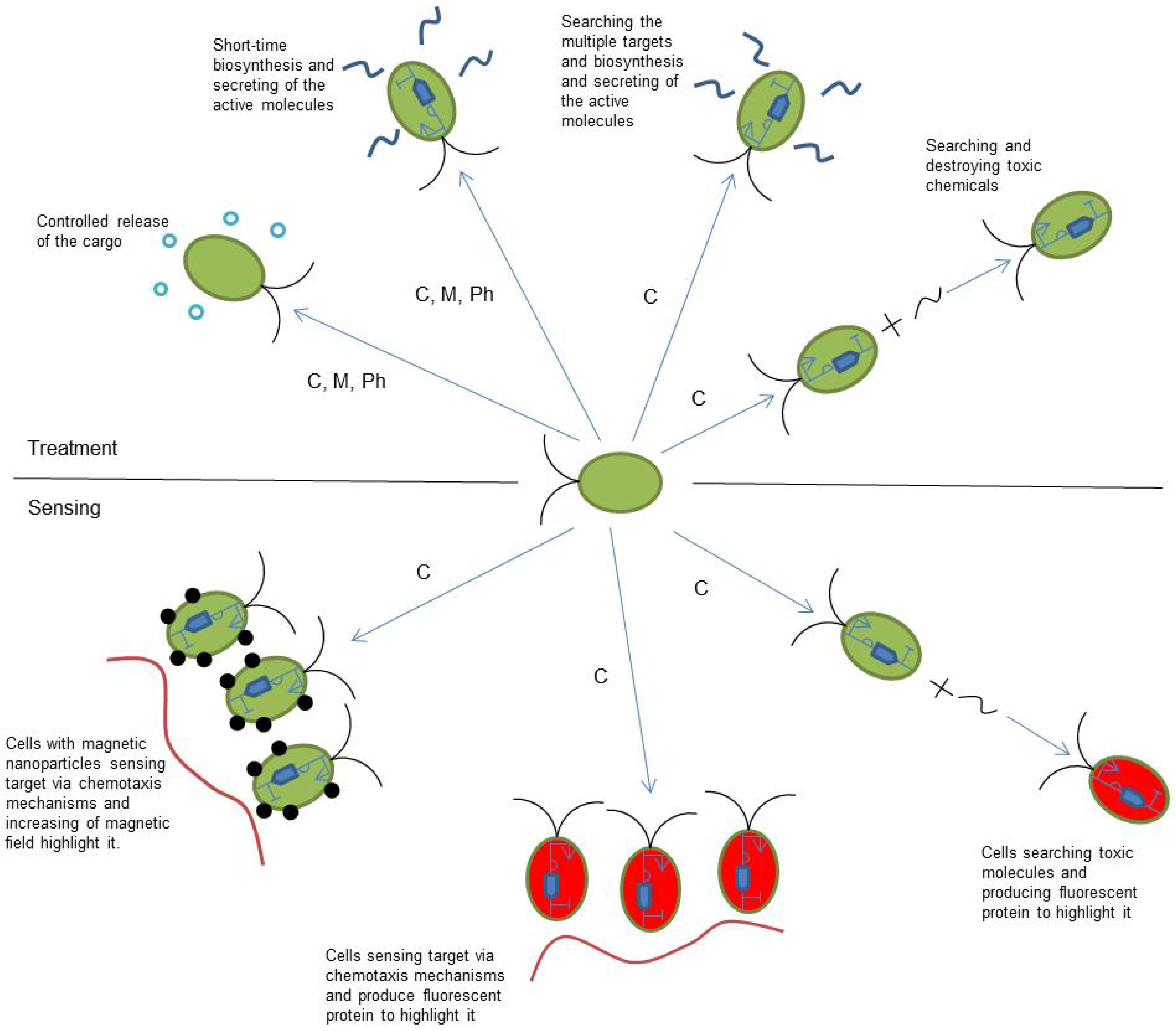
| Motility Type | Speed without Cargo | External Stimulus | Advantages | Limitations | References |
|---|---|---|---|---|---|
| Chemotaxis | Around 20–50 μm/s | Attractant, but it can be an internal chemical synthesized inside the organism where the microrobot is applied | Can be realized without external stimulus. Well-known and can be discussed as a target for genetic engineering | Lower speed, more difficult behavior in comparison with other types of motility. | [15,38] |
| Phototaxis | Can be larger than 100 μm/s | Light needed as an external stimulus | Phototrophic microorganisms also produce oxygen that is helpful to prevent hypoxia. Can be to the light and out of light depending on the strain and light intensity. | Cannot work without light. | [16,41,49,50] |
| Magnetotaxis | Dependent on the strain and magnetic field parameters | A magnetic field is necessary | Can be compatible with MRI. Possible to induce by adding nanoparticles to the cells. | An external magnetic field is necessary. Artificial magnetotaxis is not inherited through the generations of the cells. | [52,54] |
| Synthetic Biology Approach | References | Possible Applications for Microrobots | Comment |
|---|---|---|---|
| Adding biosynthesis of new for the microorganism’s chemicals | [67,68,69] | Production of the necessary chemical that can be a drug against target illness | Replacing cargo with biosynthesis leads to the saving of this ability in throw-out generations. |
| Computations in the cells, genetic logic circuits, and based on those methods of cell-to-cell communications | [70,71,72] | Enhancement of quorum sensing and efficiency of chemotaxis | The more microrobots with some toxic anticancer cargo reach the target, the less negative impact they will have on the whole organism. |
| [14,64,73,74] | Development of analysis of the received signal and generation of the answer based on the provided computation | Offers the possibility to make the behavior of the microrobot more complex and can add some additional chemical sensors to enhance efficiency in reaching the target. | |
| Engineering of motility related mechanisms and sensors | [75,76] | Engineering receptors for the new attractant related to the targets, or modification of the chemotaxis pathway to fit it with new receptors. | Increasing the efficiency of chemotaxis, development of synthetic chemotaxis pathways related to the microrobot’s target. |
| Synthetic Biology Tool | References | Possible Applications for Microrobotic Engineering |
|---|---|---|
| DNA assembly and DNA synthesis | [97] | Biosynthesis of chemicals for treatment, cell-to-cell communications, enhanced sensing, novel sensing molecules for chemotaxis |
| Genome editing | [78,79,97] | All applications where manipulation with genome required |
| Genetic circuits | [13,14] | Computation in cells, cell-to-cell communications, triggers, and switches |
| XNA assembly and integration | [94,95] | Safety, control of cell population |
| Biological parts/biobricks | [98,99] | Fast development of synthetic genetic circuits |
| Intracellular processes and cell behavior through mathematical modeling and simulations | [100,101] | Modeling intracellular processes and the behavior of developed microrobots, and the simulation of its application |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gotovtsev, P. Microbial Cells as a Microrobots: From Drug Delivery to Advanced Biosensors. Biomimetics 2023, 8, 109. https://doi.org/10.3390/biomimetics8010109
Gotovtsev P. Microbial Cells as a Microrobots: From Drug Delivery to Advanced Biosensors. Biomimetics. 2023; 8(1):109. https://doi.org/10.3390/biomimetics8010109
Chicago/Turabian StyleGotovtsev, Pavel. 2023. "Microbial Cells as a Microrobots: From Drug Delivery to Advanced Biosensors" Biomimetics 8, no. 1: 109. https://doi.org/10.3390/biomimetics8010109
APA StyleGotovtsev, P. (2023). Microbial Cells as a Microrobots: From Drug Delivery to Advanced Biosensors. Biomimetics, 8(1), 109. https://doi.org/10.3390/biomimetics8010109





