Gut-on-a-Chip Research for Drug Development: Implications of Chip Design on Preclinical Oral Bioavailability or Intestinal Disease Studies
Abstract
:1. Introduction
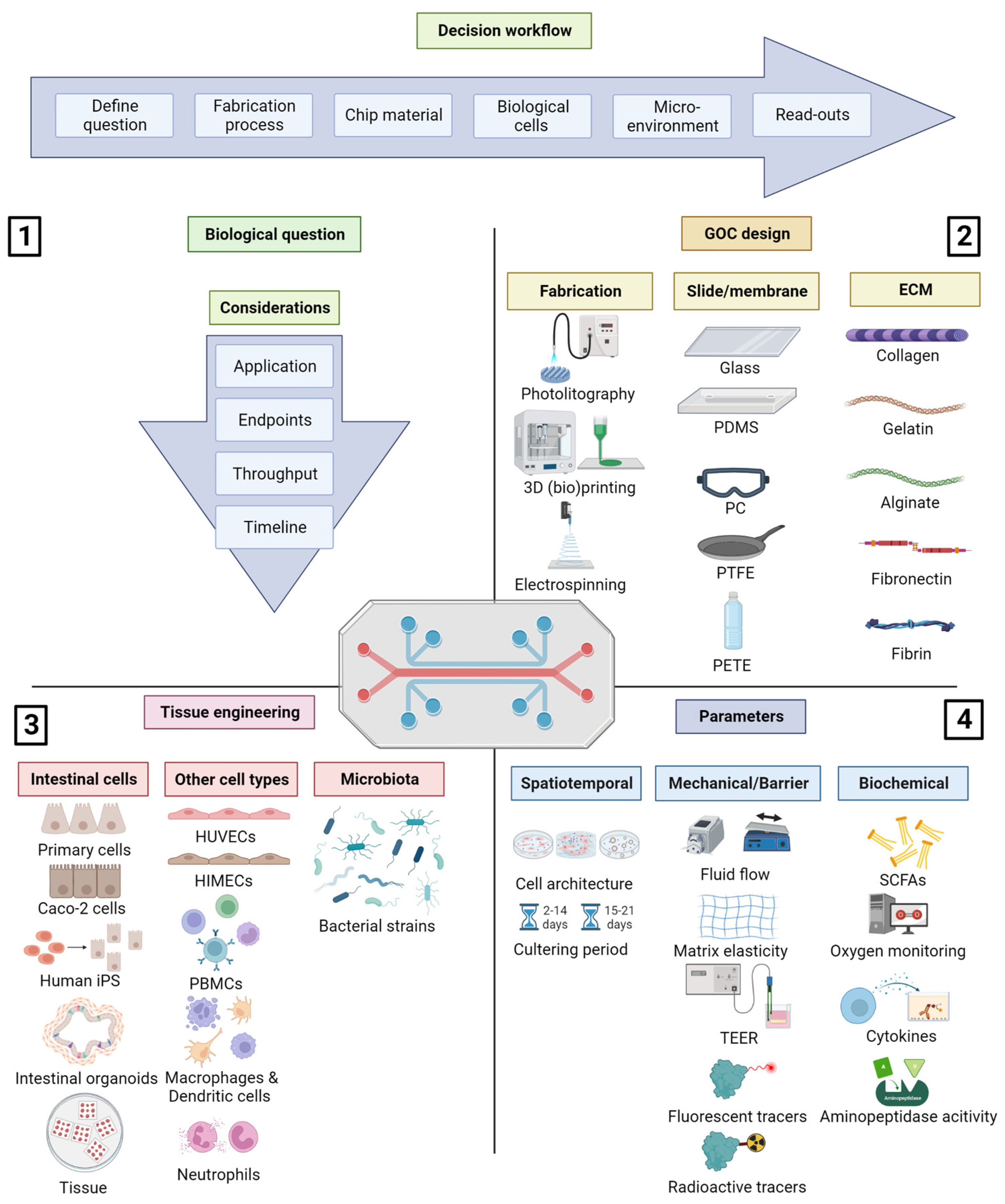
2. The Four Components Influencing GOC Design
2.1. The Biological Research Question Determines the Type of GOC to Use
2.2. Fabrication and Materials for the Individual Chip Components
2.2.1. Fabrication Techniques
2.2.2. Chip Slide Material
2.2.3. Membrane Material
2.2.4. Extracellular Matrix (ECM)
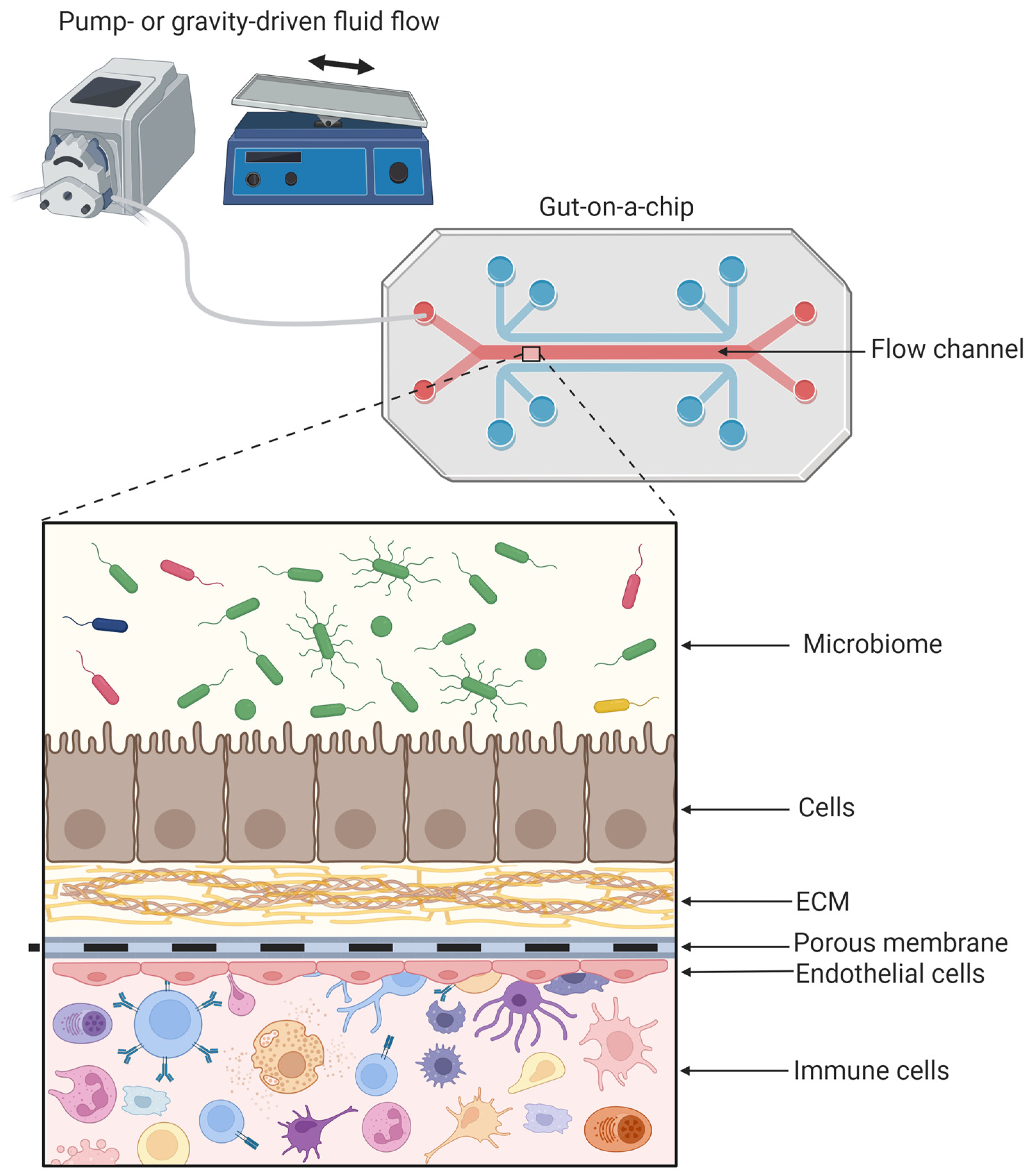
2.3. Tissue Engineering in the GOC
2.3.1. Source of the Intestinal Cells
2.3.2. Co-Culturing with Additional Cells: Endothelial and Immune Cells
2.3.3. The Gut Microbiome
2.4. Environmental Factors and Read-Out Parameters Important for the GOC Design
2.4.1. Mechanical Cues Important in the GOC
2.4.2. Measurement of the Intestinal Barrier Function
2.4.3. Control and Regulation of Physiochemical Parameters Oxygen and pH
2.4.4. Biochemical Cues
3. GOC for Preclinical Drug Development Research
3.1. GOC for ADME Research
3.2. GOC to Study Gut Health and Disease
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Okumura, R.; Takeda, K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef] [PubMed]
- Bowcutt, R.; Forman, R.; Glymenaki, M.; Carding, S.R.; Else, K.J.; Cruickshank, S.M. Heterogeneity across the murine small and large intestine. World J. Gastroenterol. 2014, 20, 15216–15232. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S.; Gordon, J.I.; Glimcher, L.H. Homeostasis and Inflammation in the Intestine. Cell 2010, 140, 859–870. [Google Scholar] [CrossRef]
- Rousset, M. The human colon carcinoma cell lines HT-29 and Caco-2: Two in vitro models for the study of intestinal differentiation. Biochimie 1986, 68, 1035–1040. [Google Scholar] [CrossRef]
- Hidalgo, I.J.; Raub, T.J.; Borchardt, R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989, 96, 736–749. [Google Scholar] [CrossRef]
- Rahman, S.; Ghiboub, M.; Donkers, J.M.; van de Steeg, E.; van Tol, E.A.F.; Hakvoort, T.B.M.; de Jonge, W.J. The progress of intestinal epithelial models from cell lines to gut-on-chip. Int. J. Mol. Sci. 2021, 22, 13472. [Google Scholar] [CrossRef] [PubMed]
- Vaessen, S.F.C.; van Lipzig, M.M.H.; Pieters, R.H.H.; Krul, C.A.M.; Wortelboer, H.M.; van de Steeg, E. Regional expression levels of drug transporters and metabolizing enzymes along the pig and human intestinal tract and comparison with Caco-2 cells. Drug Metab. Dispos. 2017, 45, 353–360. [Google Scholar] [CrossRef]
- Milani, N.; Parrott, N.; Ortiz Franyuti, D.; Godoy, P.; Galetin, A.; Gertz, M.; Fowler, S. Application of a gut-liver-on-a-chip device and mechanistic modelling to the quantitative in vitro pharmacokinetic study of mycophenolate mofetil. Lab Chip 2022, 22, 2853–2868. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.E.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.G. 2D-and 3D-based intestinal stem cell cultures for personalized medicine. Cells 2018, 7, 225. [Google Scholar] [CrossRef]
- Kwon, O.; Jung, K.B.; Lee, K.R.; Son, Y.S.; Lee, H.; Kim, J.J.; Kim, K.; Lee, S.; Song, Y.K.; Jung, J.; et al. The development of a functional human small intestinal epithelium model for drug absorption. Sci. Adv. 2021, 7, eabh1586. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Inui, T.; Yokota, J.; Kawakami, K.; Morinaga, G.; Takatani, M.; Hirayama, D.; Nomoto, R.; Ito, K.; Cui, Y.; et al. Monolayer platform using human biopsy-derived duodenal organoids for pharmaceutical research. Mol. Ther. Methods Clin. Dev. 2021, 22, 263–278. [Google Scholar] [CrossRef]
- Westerhout, J.; Wortelboer, H.; Verhoeckx, K. Ussing Chamber. In The Impact of Food Bioactives on Health; Springer International Publishing: Cham, Switzerland, 2015; pp. 263–273. ISBN 9783319161044. [Google Scholar]
- Herrmann, J.R.; Turner, J.R. Beyond Ussing’s chambers: Contemporary thoughts on integration of transepithelial transport. Am. J. Physiol. Physiol. 2016, 310, C423–C431. [Google Scholar] [CrossRef] [PubMed]
- Streekstra, E.J.; Kiss, M.; van den Heuvel, J.; Nicolaï, J.; van den Broek, P.; Botden, S.M.B.I.; Stommel, M.W.J.; van Rijssel, L.; Ungell, A.L.; van de Steeg, E.; et al. A proof of concept using the Ussing chamber methodology to study pediatric intestinal drug transport and age-dependent differences in absorption. Clin. Transl. Sci. 2022, 15, 2392–2402. [Google Scholar] [CrossRef]
- Alam, M.A.; Al-Jenoobi, F.I.; Al-Mohizea, A.M. Everted gut sac model as a tool in pharmaceutical research: Limitations and applications. J. Pharm. Pharmacol. 2012, 64, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Stevens, L.J.; van Lipzig, M.M.H.; Erpelinck, S.L.A.; Pronk, A.; van Gorp, J.; Wortelboer, H.M.; van de Steeg, E. A higher throughput and physiologically relevant two-compartmental human ex vivo intestinal tissue system for studying gastrointestinal processes. Eur. J. Pharm. Sci. 2019, 137, 104989. [Google Scholar] [CrossRef]
- Westerhout, J.; Van De Steeg, E.; Grossouw, D.; Zeijdner, E.E.; Krul, C.A.M.; Verwei, M.; Wortelboer, H.M. A new approach to predict human intestinal absorption using porcine intestinal tissue and biorelevant matrices. Eur. J. Pharm. Sci. 2014, 63, 167–177. [Google Scholar] [CrossRef]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y.; et al. Organ-On-A-Chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies. Adv. Healthc. Mater. 2018, 7, 1700506. [Google Scholar] [CrossRef]
- Donkers, J.M.; Amirabadi, H.E.; van de Steeg, E. Intestine-on-a-chip: Next level in vitro research model of the human intestine. Curr. Opin. Toxicol. 2020, 25, 6–14. [Google Scholar] [CrossRef]
- Sung, J.H.; Wang, Y.I.; Narasimhan Sriram, N.; Jackson, M.; Long, C.; Hickman, J.J.; Shuler, M.L. Recent Advances in Body-on-a-Chip Systems. Anal. Chem. 2019, 91, 330–351. [Google Scholar] [CrossRef]
- Bovard, D.; Iskandar, A.; Luettich, K.; Hoeng, J.; Peitsch, M.C. Organs-on-a-chip: A new paradigm for toxicological assessment and preclinical drug development. Toxicol. Res. Appl. 2017, 1, 239784731772635. [Google Scholar] [CrossRef]
- Fois, C.A.M.; Le, T.Y.L.; Schindeler, A.; Naficy, S.; McClure, D.D.; Read, M.N.; Valtchev, P.; Khademhosseini, A.; Dehghani, F. Models of the Gut for Analyzing the Impact of Food and Drugs. Adv. Healthc. Mater. 2019, 8, 1900968. [Google Scholar] [CrossRef]
- Kimura, H.; Yamamoto, T.; Sakai, H.; Sakai, Y.; Fujii, T. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 2008, 8, 741–746. [Google Scholar] [CrossRef]
- Eslami Amirabadi, H.; Donkers, J.; Wierenga, E.; Ingenhut, B.; Pieters, L.; Stevens, L.; Donkers, T.; Westerhout, J.; Masereeuw, R.; Bobeldijk, I.; et al. Intestinal Explant Barrier Chip: Long-term intestinal absorption screening in a novel microphysiological system using tissue explants. Lab Chip 2022, 22, 326–342. [Google Scholar] [CrossRef] [PubMed]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cmgh 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Chi, M.; Yi, B.; Oh, S.; Park, D.J.; Sung, J.H.; Park, S. A microfluidic cell culture device (μFCCD) to culture epithelial cells with physiological and morphological properties that mimic those of the human intestine. Biomed. Microdevices 2015, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, V.; Corrado, B.; Sbrescia, S.; Sibilio, S.; Urciuolo, F.; Netti, P.A.; Imparato, G. Intestine-on-chip device increases ECM remodeling inducing faster epithelial cell differentiation. Biotechnol. Bioeng. 2020, 117, 556–566. [Google Scholar] [CrossRef]
- Tarricone, G.; Carmagnola, I.; Chiono, V. Tissue-Engineered Models of the Human Brain: State-of-the-Art Analysis and Challenges. J. Funct. Biomater. 2022, 13, 146. [Google Scholar] [CrossRef]
- Kellner, K.; Liebsch, G.; Klimant, I.; Wolfbeis, O.S.; Blunk, T.; Schulz, M.B.; Göpferich, A. Determination of oxygen gradients in engineered tissue using a fluorescent sensor. Biotechnol. Bioeng. 2002, 80, 73–83. [Google Scholar] [CrossRef]
- Abbott, R.D.; Kaplan, D.L. Strategies for improving the physiological relevance of human engineered tissues. Trends Biotechnol. 2015, 33, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Dawson, A.; Dyer, C.; Macfie, J.; Davies, J.; Karsai, L.; Greenman, J.; Jacobsen, M. A microfluidic chip based model for the study of full thickness human intestinal tissue using dual flow. Biomicrofluidics 2016, 10, 064101. [Google Scholar] [CrossRef]
- Sung, J.H.; Yu, J.; Luo, D.; Shuler, M.L.; March, J.C. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011, 11, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.Y.; Lee, D.; Han, J.; Nguyen, N.T.; Park, S.; Sung, J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 2017, 19, 37. [Google Scholar] [CrossRef] [PubMed]
- Gencturk, E.; Mutlu, S.; Ulgen, K.O. Advances in microfluidic devices made from thermoplastics used in cell biology and analyses. Biomicrofluidics 2017, 11, 051502. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.B.; Ng, S.H.; Li, K.H.H.; Yoon, Y.J. 3D printed microfluidics for biological applications. Lab Chip 2015, 15, 3627–3637. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.P.; Matsu-Ura, T.; Rosselot, A.E.; Broda, T.R.; Wells, J.M.; Hong, C.I. An integrated microfluidic bubble pocket for long-term perfused three-dimensional intestine-on-a-chip model. Biomicrofluidics 2021, 15, 014110. [Google Scholar] [CrossRef] [PubMed]
- Creff, J.; Courson, R.; Mangeat, T.; Foncy, J.; Souleille, S.; Thibault, C.; Besson, A.; Malaquin, L. Fabrication of 3D scaffolds reproducing intestinal epithelium topography by high-resolution 3D stereolithography. Biomaterials 2019, 221, 119404. [Google Scholar] [CrossRef]
- Lee, M.; Dunn, J.C.Y.; Wu, B.M. Scaffold fabrication by indirect three-dimensional printing. Biomaterials 2005, 26, 4281–4289. [Google Scholar] [CrossRef]
- Kim, W.J.; Kim, G.H. An innovative cell-printed microscale collagen model for mimicking intestinal villus epithelium. Chem. Eng. J. 2018, 334, 2308–2318. [Google Scholar] [CrossRef]
- Chuchuy, J.; Rogal, J.; Ngo, T.; Stadelmann, K.; Antkowiak, L.; Achberger, K.; Liebau, S.; Schenke-Layland, K.; Loskill, P. Integration of electrospun membranes into low-absorption thermoplastic organ-on-chip. ACS Biomater. Sci. Eng. 2021, 7, 3006–3017. [Google Scholar] [CrossRef]
- Ding, C.; Chen, X.; Kang, Q.; Yan, X. Biomedical Application of Functional Materials in Organ-on-a-Chip. Front. Bioeng. Biotechnol. 2020, 8, 823. [Google Scholar] [CrossRef]
- Mahler, G.J.; Esch, M.B.; Glahn, R.P.; Shuler, M.L. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009, 104, 193–205. [Google Scholar] [CrossRef]
- Kotz, F.; Plewa, K.; Bauer, W.; Schneider, N.; Keller, N.; Nargang, T.; Helmer, D.; Sachsenheimer, K.; Schäfer, M.; Worgull, M.; et al. Liquid Glass: A Facile Soft Replication Method for Structuring Glass. Adv. Mater. 2016, 28, 4646–4650. [Google Scholar] [CrossRef] [PubMed]
- Schulze, T.; Mattern, K.; Früh, E.; Hecht, L.; Rustenbeck, I.; Dietzel, A. A 3D microfluidic perfusion system made from glass for multiparametric analysis of stimulus-secretioncoupling in pancreatic islets. Biomed. Microdevices 2017, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.W. Polymer microfluidics: Simple, low-cost fabrication process bridging academic lab research to commercialized production. Micromachines 2016, 7, 225. [Google Scholar] [CrossRef] [PubMed]
- Auner, A.W.; Tasneem, K.M.; Markov, D.A.; McCawley, L.J.; Hutson, M.S. Chemical-PDMS binding kinetics and implications for bioavailability in microfluidic devices. Lab Chip 2019, 19, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Berthier, E.; Young, E.W.K.; Beebe, D. Engineers are from PDMS-land, biologists are from polystyrenia. Lab Chip 2012, 12, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.; Gruner, D.; Richter, A.; Loskill, P. Membrane integration into PDMS-free microfluidic platforms for organ-on-chip and analytical chemistry applications. Lab Chip 2021, 21, 1866–1885. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Onoe, H.; Osaki, T.; Kawano, R.; Takeuchi, S. Parylene-coating in PDMS microfluidic channels prevents the absorption of fluorescent dyes. Sens. Actuators B Chem. 2010, 150, 478–482. [Google Scholar] [CrossRef]
- Ren, K.; Zhao, Y.; Su, J.; Ryan, D.; Wu, H. Convenient method for modifying poly(dimethylsiloxane) to be airtight and resistive against absorption of small molecules. Anal. Chem. 2010, 82, 5965–5971. [Google Scholar] [CrossRef]
- van Meer, B.J.; de Vries, H.; Firth, K.S.A.; van Weerd, J.; Tertoolen, L.G.J.; Karperien, H.B.J.; Jonkheijm, P.; Denning, C.; IJzerman, A.P.; Mummery, C.L. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem. Biophys. Res. Commun. 2017, 482, 323–328. [Google Scholar] [CrossRef]
- Sollier, E.; Murray, C.; Maoddi, P.; Di Carlo, D. Rapid prototyping polymers for microfluidic devices and high pressure injections. Lab Chip 2011, 11, 3752–3765. [Google Scholar] [CrossRef] [PubMed]
- Domansky, K.; Sliz, J.D.; Wen, N.; Hinojosa, C.; Thompson, G.; Fraser, J.P.; Hamkins-Indik, T.; Hamilton, G.A.; Levner, D.; Ingber, D.E. SEBS elastomers for fabrication of microfluidic devices with reduced drug absorption by injection molding and extrusion. Microfluid. Nanofluidics 2017, 21, 107. [Google Scholar] [CrossRef]
- Sano, E.; Mori, C.; Matsuoka, N.; Ozaki, Y.; Yagi, K.; Wada, A.; Tashima, K.; Yamasaki, S.; Tanabe, K.; Yano, K.; et al. Tetrafluoroethylene-propylene elastomer for fabrication of microfluidic organs-on-chips resistant to drug absorption. Micromachines 2019, 10, 793. [Google Scholar] [CrossRef]
- Thomas, D.P.; Zhang, J.; Nguyen, N.-T.; Ta, H.T. Microfluidic Gut-on-a-Chip: Fundamentals and Challenges. Biosensors 2023, 13, 136. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Hauser, R.; Marcus, M.; Olea, N.; Welshons, W.V. Human exposure to bisphenol A (BPA). Reprod. Toxicol. 2007, 24, 139–177. [Google Scholar] [CrossRef] [PubMed]
- Donkers, J.M.; Höppener, E.M.; Grigoriev, I.; Will, L.; Melgert, B.N.; van der Zaan, B.; van de Steeg, E.; Kooter, I.M. Advanced epithelial lung and gut barrier models demonstrate passage of microplastic particles. Microplast. Nanoplast. 2022, 2, 6. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Karamanos, N.K.; Theocharis, A.D.; Piperigkou, Z.; Manou, D.; Passi, A.; Skandalis, S.S.; Vynios, D.H.; Orian-Rousseau, V.; Ricard-Blum, S.; Schmelzer, C.E.H.; et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021, 288, 6850–6912. [Google Scholar] [CrossRef]
- Kyburz, K.A.; Anseth, K.S. Synthetic Mimics of the Extracellular Matrix: How Simple is Complex Enough? Ann. Biomed. Eng. 2015, 43, 489–500. [Google Scholar] [CrossRef]
- Unal, A.Z.; West, J.L. Synthetic ECM: Bioactive Synthetic Hydrogels for 3D Tissue Engineering. Bioconjug. Chem. 2020, 31, 2253–2271. [Google Scholar] [CrossRef]
- Naomi, R.; Bahari, H.; Ridzuan, P.M.; Othman, F. Natural-based biomaterial for skin wound healing (Gelatin vs. collagen): Expert review. Polymers 2021, 13, 2319. [Google Scholar] [CrossRef]
- Shaikh, F.M.; Callanan, A.; Kavanagh, E.G.; Burke, P.E.; Grace, P.A.; McGloughlin, T.M. Fibrin: A natural biodegradable scaffold in vascular tissue engineering. Cells Tissues Organs 2008, 188, 333–346. [Google Scholar] [CrossRef]
- Antoine, E.E.; Vlachos, P.P.; Rylander, M.N. Review of Collagen I Hydrogels for Bioengineered Tissue Microenvironments: Characterization of Mechanics, Structure, and Transport. Tissue Eng. Part B Rev. 2014, 20, 683–696. [Google Scholar] [CrossRef]
- Kreger, S.T.; Bell, B.J.; Bailey, J.; Stites, E.; Kuske, J.; Waisner, B.; Voytik-Harbin, S.L. Polymerization and matrix physical properties as important design considerations for soluble collagen formulations. Biopolymers 2010, 93, 690–707. [Google Scholar] [CrossRef]
- Xiang, Y.; Wen, H.; Yu, Y.; Li, M.; Fu, X.; Huang, S. Gut-on-chip: Recreating human intestine in vitro. J. Tissue Eng. 2020, 11, 2041731420965318. [Google Scholar] [CrossRef]
- Richardson, A.; Schwerdtfeger, L.A.; Eaton, D.; McLean, I.; Henry, C.S.; Tobet, S.A. A microfluidic organotypic device for culture of mammalian intestines: Ex vivo. Anal. Methods 2020, 12, 297–303. [Google Scholar] [CrossRef]
- Meunier, V.; Bourrié, M.; Berger, Y.; Fabre, G. The human intestinal epithelial cell line Caco-2; pharmacological and pharmacokinetic applications. Cell Biol. Toxicol. 1995, 11, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.; Zihler Berner, A.; Chervet, N.; Chassard, C.; Lacroix, C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 2013, 94, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Han, L.; Zhang, Y.; Yu, Y.; Liu, J. Optimization of Caco-2 and HT29 co-culture in vitro cell models for permeability studies. Int. J. Food Sci. Nutr. 2015, 66, 680–685. [Google Scholar] [CrossRef]
- Richter, M.; Piwocka, O.; Musielak, M.; Piotrowski, I.; Suchorska, W.M.; Trzeciak, T. From Donor to the Lab: A Fascinating Journey of Primary Cell Lines. Front. Cell Dev. Biol. 2021, 9, 711381. [Google Scholar] [CrossRef]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef]
- Yin, J.; Sunuwar, L.; Kasendra, M.; Yu, H.; Tse, C.-M.; Talbot, C.; Boronina, T.N.; Cole, R.N.; Karalis, K.; Donowitz, M. Fluid Shear Stress Enhances Differentiation of Jejunal Human Enteroids in Intestine-Chip. Am. J. Physiol. Liver Physiol. 2020, 320, G258–G271. [Google Scholar] [CrossRef]
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; De Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Apostolou, A.; Panchakshari, R.A.; Banerjee, A.; Manatakis, D.V.; Paraskevopoulou, M.D.; Luc, R.; AbuAli, G.; Dimitriou, A.; Lucchesi, C.; Kulkarni, G.; et al. A Micro-engineered Human Colon Intestine-Chip Platform to Study Leaky Barrier. bioRXiv 2020, 8, 271759. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.J.; Yoon, J.Y.; Kemmitt, J.; Wright, C.; Schneider, K.; Sphabmixay, P.; Hernandez-Gordillo, V.; Holcomb, S.J.; Bhushan, B.; et al. Primary Human Colonic Mucosal Barrier Crosstalk with Super Oxygen-Sensitive Faecalibacterium prausnitzii in Continuous Culture. Med 2021, 2, 74–98.e9. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Benda, C.; Dunzinger, S.; Huang, Y.; Ho, J.C.; Yang, J.; Wang, Y.; Zhang, Y.; Zhuang, Q.; Li, Y.; et al. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012, 7, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Rowe, R.G.; Daley, G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019, 20, 377–388. [Google Scholar] [CrossRef]
- Naumovska, E.; Aalderink, G.; Valencia, C.W.; Kosim, K.; Nicolas, A.; Brown, S.; Vulto, P.; Erdmann, K.S.; Kurek, D. Direct on-chip differentiation of intestinal tubules from induced pluripotent stem cells. Int. J. Mol. Sci. 2020, 21, 4964. [Google Scholar] [CrossRef] [PubMed]
- Lanik, W.E.; Luke, C.J.; Nolan, L.S.; Gong, Q.; Frazer, L.C.; Rimer, J.M.; Gale, S.E.; Luc, R.; Bidani, S.S.; Sibbald, C.A.; et al. Microfluidic device facilitates in vitro modeling of human neonatal necrotizing enterocolitis-on-a-chip. JCI Insight 2023, 8, e146496. [Google Scholar] [CrossRef]
- Zhao, W.; Yao, Y.; Zhang, T.; Lu, H.; Zhang, X.; Zhao, L.; Chen, X.; Zhu, J.; Sui, G.; Zhao, W. Primary exploration of host–microorganism interaction and enteritis treatment with an embedded membrane microfluidic chip of the human intestinal–vascular microsystem. Front. Bioeng. Biotechnol. 2022, 10, 35647. [Google Scholar] [CrossRef]
- Kasendra, M.; Luc, R.; Yin, J.; Manatakis, D.V.; Kulkarni, G.; Lucchesi, C.; Sliz, J.; Apostolou, A.; Sunuwar, L.; Obrigewitch, J.; et al. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. Elife 2020, 9, e50135. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Maurer, M.; Gresnigt, M.S.; Last, A.; Wollny, T.; Berlinghof, F.; Pospich, R.; Cseresnyes, Z.; Medyukhina, A.; Graf, K.; Gröger, M.; et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019, 220, 119396. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef]
- Seiler, K.M.; Bajinting, A.; Alvarado, D.M.; Traore, M.A.; Binkley, M.M.; Goo, W.H.; Lanik, W.E.; Ou, J.; Ismail, U.; Iticovici, M.; et al. Patient-derived small intestinal myofibroblasts direct perfused, physiologically responsive capillary development in a microfluidic Gut-on-a-Chip Model. Sci. Rep. 2020, 10, 3842. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Luo, R.; Wang, Y.; Deng, P.; Song, T.; Zhang, M.; Wang, P.; Zhang, X.; Cui, K.; Tao, T.; et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci. Bull. 2021, 66, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, M.; Mitrofanova, O.; Broguiere, N.; Geraldo, S.; Dutta, D.; Tabata, Y.; Elci, B.; Brandenberg, N.; Kolotuev, I.; Gjorevski, N.; et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020, 585, 574–578. [Google Scholar] [CrossRef]
- Jing, B.; Wang, Z.A.; Zhang, C.; Deng, Q.; Wei, J.; Luo, Y.; Zhang, X.; Li, J.; Du, Y. Establishment and Application of Peristaltic Human Gut-Vessel Microsystem for Studying Host–Microbial Interaction. Front. Bioeng. Biotechnol. 2020, 8, 272. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Yuan Hsin, H.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Gijzen, L.; Marescotti, D.; Raineri, E.; Nicolas, A.; Lanz, H.L.; Guerrera, D.; van Vught, R.; Joore, J.; Vulto, P.; Peitsch, M.C.; et al. An Intestine-on-a-Chip Model of Plug-and-Play Modularity to Study Inflammatory Processes. SLAS Technol. 2020, 25, 585–597. [Google Scholar] [CrossRef]
- Shin, W.; Kim, H.J. Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. USA 2018, 115, E10539–E10547. [Google Scholar] [CrossRef]
- Gjorevski, N.; Avignon, B.; Gérard, R.; Cabon, L.; Roth, A.B.; Bscheider, M.; Moisan, A. Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab Chip 2020, 20, 3365–3374. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota-masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Chi, M.; Yi, B.; Kim, S.H.; Oh, S.; Kim, Y.; Park, S.; Sung, J.H. Three-dimensional intestinal villi epithelium enhances protection of human intestinal cells from bacterial infection by inducing mucin expression. Integr. Biol. 2014, 6, 1122–1131. [Google Scholar] [CrossRef]
- Grassart, A.; Malardé, V.; Gobba, S.; Sartori-Rupp, A.; Kerns, J.; Karalis, K.; Marteyn, B.; Sansonetti, P.; Sauvonnet, N. Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection. Cell Host Microbe 2019, 26, 435–444.e4. [Google Scholar] [CrossRef]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadaghian Sadabad, M.; Pinheiro, I.; Possemiers, S.; Van Den Abbeele, P.; Derycke, L.; Bracke, M.; Pieters, J.; et al. The HMITM module: A new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 2014, 14, 133. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef]
- Shin, W.; Wu, A.; Massidda, M.W.; Foster, C.; Thomas, N.; Lee, D.W.; Koh, H.; Ju, Y.; Kim, J.; Kim, H.J. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front. Bioeng. Biotechnol. 2019, 7, 13. [Google Scholar] [CrossRef]
- Shin, Y.C.; Shin, W.; Koh, D.; Wu, A.; Ambrosini, Y.M.; Min, S.; Gail Eckhardt, S.; Declan Fleming, R.Y.; Kim, S.; Park, S.; et al. Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal interface-on-a-chip. Micromachines 2020, 11, 663. [Google Scholar] [CrossRef]
- De Gregorio, V.; Sgambato, C.; Urciuolo, F.; Vecchione, R.; Netti, P.A.; Imparato, G. Immunoresponsive microbiota-gut-on-chip reproduces barrier dysfunction, stromal reshaping and probiotics translocation under inflammation. Biomaterials 2022, 286, 121573. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dang, T.; Baste, J.; Anil Joshi, A.; Bhushan, A. A novel standalone microfluidic device for local control of oxygen tension for intestinal-bacteria interactions. FASEB J. 2021, 35, e21291. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, R.; Zheng, X.; Hou, W.; Wu, X.; Zhao, H.; Wang, G.; Tian, T. Establishment of a gut-on-a-chip device with controllable oxygen gradients to study the contribution of Bifidobacterium bifidum to inflammatory bowel disease. Biomater. Sci. 2023, 11, 2504–2517. [Google Scholar] [CrossRef]
- Trietsch, S.J.; Naumovska, E.; Kurek, D.; Setyawati, M.C.; Vormann, M.K.; Wilschut, K.J.; Lanz, H.L.; Nicolas, A.; Ng, C.P.; Joore, J.; et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 2017, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Fois, C.A.M.; Schindeler, A.; Valtchev, P.; Dehghani, F. Dynamic flow and shear stress as key parameters for intestinal cells morphology and polarization in an organ-on-a-chip model. Biomed. Microdevices 2021, 23, 55. [Google Scholar] [CrossRef]
- Delon, L.C.; Guo, Z.; Oszmiana, A.; Chien, C.C.; Gibson, R.; Prestidge, C.; Thierry, B. A systematic investigation of the effect of the fluid shear stress on Caco-2 cells towards the optimization of epithelial organ-on-chip models. Biomaterials 2019, 225, 119521. [Google Scholar] [CrossRef]
- Jochems, P.G.M.; van Bergenhenegouwen, J.; van Genderen, A.M.; Eis, S.T.; Wilod Versprille, L.J.F.; Wichers, H.J.; Jeurink, P.V.; Garssen, J.; Masereeuw, R. Development and validation of bioengineered intestinal tubules for translational research aimed at safety and efficacy testing of drugs and nutrients. Toxicol. Vitr. 2019, 60, 1–11. [Google Scholar] [CrossRef]
- Langerak, N.; Ahmed, H.M.M.; Li, Y.; Middel, I.R.; Eslami Amirabadi, H.; Malda, J.; Masereeuw, R.; van Roij, R. A Theoretical and Experimental Study to Optimize Cell Differentiation in a Novel Intestinal Chip. Front. Bioeng. Biotechnol. 2020, 8, 763. [Google Scholar] [CrossRef] [PubMed]
- Pompili, S.; Latella, G.; Gaudio, E.; Sferra, R.; Vetuschi, A. The Charming World of the Extracellular Matrix: A Dynamic and Protective Network of the Intestinal Wall. Front. Med. 2021, 8, 610189. [Google Scholar] [CrossRef]
- Bordeleau, F.; Mason, B.N.; Lollis, E.M.; Mazzola, M.; Zanotelli, M.R.; Somasegar, S.; Califano, J.P.; Montague, C.; LaValley, D.J.; Huynh, J.; et al. Matrix stiffening promotes a tumor vasculature phenotype. Proc. Natl. Acad. Sci. USA 2017, 114, 492–497. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER Measurement Techniques for In Vitro Barrier Model Systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef]
- Wang, L.; Han, J.; Su, W.; Li, A.; Zhang, W.; Li, H.; Hu, H.; Song, W.; Xu, C.; Chen, J. Gut-on-a-chip for exploring the transport mechanism of Hg(II). Microsyst. Nanoeng. 2023, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Bossink, E.G.B.M.; Zakharova, M.; De Bruijn, D.S.; Odijk, M.; Segerink, L.I. Measuring barrier function in organ-on-chips with cleanroom-free integration of multiplexable electrodes. Lab Chip 2021, 21, 2040–2049. [Google Scholar] [CrossRef]
- Hoffmann, P.; Burmester, M.; Langeheine, M.; Brehm, R.; Empl, M.T.; Seeger, B.; Breves, G. Caco-2/HT29-MTX co-cultured cells as a model for studying physiological properties and toxin-induced effects on intestinal cells. PLoS ONE 2021, 16, e0257824. [Google Scholar] [CrossRef] [PubMed]
- Manjakkal, L.; Szwagierczak, D.; Dahiya, R. Metal oxides based electrochemical pH sensors: Current progress and future perspectives. Prog. Mater. Sci. 2020, 109, 100635. [Google Scholar] [CrossRef]
- Rajapaksha, R.D.A.A.; Hashim, U.; Gopinath, S.C.B.; Fernando, C.A.N. Sensitive pH detection on gold interdigitated electrodes as an electrochemical sensor. Microsyst. Technol. 2018, 24, 1965–1974. [Google Scholar] [CrossRef]
- McCracken, K.W.; Aihara, E.; Martin, B.; Crawford, C.M.; Broda, T.; Treguier, J.; Zhang, X.; Shannon, J.M.; Montrose, M.H.; Wells, J.M. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature 2017, 541, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Zachos, N.C.; Kovbasnjuk, O.; Foulke-Abel, J.; In, J.; Blutt, S.E.; De Jonge, H.R.; Estes, M.K.; Donowitz, M. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J. Biol. Chem. 2016, 291, 3759–3766. [Google Scholar] [CrossRef] [PubMed]
- Hight, M.R.; Nolting, D.D.; McKinley, E.T.; Lander, A.D.; Wyatt, S.K.; Gonyea, M.; Zhao, P.; Manning, H.C. Multispectral fluorescence imaging to assess pH in biological specimens. J. Biomed. Opt. 2011, 16, 016007. [Google Scholar] [CrossRef] [PubMed]
- Philippova, O.E.; Hourdet, D.; Audebert, R.; Khokhlov, A.R. pH-responsive gels of hydrophobically modified poly(acrylic acid). Macromolecules 1997, 30, 8278–8285. [Google Scholar] [CrossRef]
- Gil, E.S.; Hudson, S.M. Stimuli-reponsive polymers and their bioconjugates. Prog. Polym. Sci. 2004, 29, 1173–1222. [Google Scholar] [CrossRef]
- Mohamed, M.A.; Fallahi, A.; El-Sokkary, A.M.A.; Salehi, S.; Akl, M.A.; Jafari, A.; Tamayol, A.; Fenniri, H.; Khademhosseini, A.; Andreadis, S.T.; et al. Stimuli-responsive hydrogels for manipulation of cell microenvironment: From chemistry to biofabrication technology. Prog. Polym. Sci. 2019, 98, 101147. [Google Scholar] [CrossRef]
- Lee, K.; Silva, E.A.; Mooney, D.J. Growth factor delivery-based tissue engineering: General approaches and a review of recent developments. J. R. Soc. Interface 2011, 8, 153–170. [Google Scholar] [CrossRef]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Ramadan, Q.; Jing, L. Characterization of tight junction disruption and immune response modulation in a miniaturized Caco-2/U937 coculture-based in vitro model of the human intestinal barrier. Biomed. Microdevices 2016, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Trapecar, M.; Communal, C.; Velazquez, J.; Maass, C.A.; Huang, Y.J.; Schneider, K.; Wright, C.W.; Butty, V.; Eng, G.; Yilmaz, O.; et al. Gut-Liver Physiomimetics Reveal Paradoxical Modulation of IBD-Related Inflammation by Short-Chain Fatty Acids. Cell Syst. 2020, 10, 223–239.e9. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, Q.; Jafarpoorchekab, H.; Huang, C.; Silacci, P.; Carrara, S.; Koklü, G.; Ghaye, J.; Ramsden, J.; Ruffert, C.; Vergeres, G.; et al. NutriChip: Nutrition analysis meets microfluidics. Lab Chip 2013, 13, 196–203. [Google Scholar] [CrossRef]
- de Haan, P.; Santbergen, M.J.C.; van der Zande, M.; Bouwmeester, H.; Nielen, M.W.F.; Verpoorte, E. A versatile, compartmentalised gut-on-a-chip system for pharmacological and toxicological analyses. Sci. Rep. 2021, 11, 4920. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Liu, H.; Lin, J.M.; Wang, Y.; Jiang, Y. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip 2013, 13, 978–985. [Google Scholar] [CrossRef]
- Santbergen, M.J.C.; van der Zande, M.; Gerssen, A.; Bouwmeester, H.; Nielen, M.W.F. Dynamic in vitro intestinal barrier model coupled to chip-based liquid chromatography mass spectrometry for oral bioavailability studies. Anal. Bioanal. Chem. 2020, 412, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.R.; Huey-Jen Hsu, S.; Chou, D.A.; Wang, G.J.; Chang, C.C. Surface plasmon-enhanced fluorescence and surface-enhanced Raman scattering dual-readout chip constructed with silver nanowires: Label-free clinical detection of direct-bilirubin. Biosens. Bioelectron. 2022, 213, 114440. [Google Scholar] [CrossRef]
- Persichetti, G.; Grimaldi, I.A.; Testa, G.; Bernini, R. Multifunctional optofluidic lab-on-chip platform for Raman and fluorescence spectroscopic microfluidic analysis. Lab Chip 2017, 17, 2631–2639. [Google Scholar] [CrossRef]
- Doogue, M.P.; Polasek, T.M. The ABCD of clinical pharmacokinetics. Ther. Adv. Drug Saf. 2013, 4, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.B.; Marques, M.C.; Hacke, A.; Loubet Filho, P.S.; Cazarin, C.B.B.; Mariutti, L.R.B. Trust your gut: Bioavailability and bioaccessibility of dietary compounds. Curr. Res. Food Sci. 2022, 5, 228–233. [Google Scholar] [CrossRef]
- Guo, Y.; Deng, P.; Chen, W.; Li, Z. Modeling pharmacokinetic profiles for assessment of anti-cancer drug on a microfluidic system. Micromachines 2020, 11, 551. [Google Scholar] [CrossRef]
- Satoh, T.; Sugiura, S.; Shin, K.; Onuki-Nagasaki, R.; Ishida, S.; Kikuchi, K.; Kakiki, M.; Kanamori, T. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab Chip 2018, 18, 115–125. [Google Scholar] [CrossRef]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Sensoy, I. A review on the food digestion in the digestive tract and the used in vitro models. Curr. Res. Food Sci. 2021, 4, 308–319. [Google Scholar] [CrossRef]
- Imura, Y.; Yoshimura, E.; Sato, K. Micro total bioassay system for oral drugs: Evaluation of gastrointestinal degradation, intestinal absorption, hepatic metabolism, and bioactivity. Anal. Sci. 2012, 28, 197–200. [Google Scholar] [CrossRef]
- Marin, T.M.; de Carvalho Indolfo, N.; Rocco, S.A.; Basei, F.L.; de Carvalho, M.; de Almeida Gonçalves, K.; Pagani, E. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem. Biol. Interact. 2019, 299, 59–76. [Google Scholar] [CrossRef]
- Choe, A.; Ha, S.K.; Choi, I.; Choi, N.; Sung, J.H. Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed. Microdevices 2017, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Tsamandouras, N.; Chen, W.L.K.; Edington, C.D.; Stokes, C.L.; Griffith, L.G.; Cirit, M. Integrated Gut and Liver Microphysiological Systems for Quantitative In Vitro Pharmacokinetic Studies. AAPS J. 2017, 19, 1499–1512. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Ikeda, T.; Nakayama, H.; Sakai, Y.; Fujii, T. An On-Chip Small Intestine–Liver Model for Pharmacokinetic Studies. J. Lab. Autom. 2015, 20, 265–273. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef]
- Mayorga, C.; Palomares, F.; Cañas, J.A.; Pérez-Sánchez, N.; Núñez, R.; Torres, M.J.; Gómez, F. New insights in therapy for food allergy. Foods 2021, 10, 1037. [Google Scholar] [CrossRef] [PubMed]
- Moerkens, R.; Mooiweer, J.; Withoff, S.; Wijmenga, C. Celiac disease-on-chip: Modeling a multifactorial disease in vitro. United Eur. Gastroenterol. J. 2019, 7, 467–476. [Google Scholar] [CrossRef]
- Vergères, G.; Bogicevic, B.; Buri, C.; Carrara, S.; Chollet, M.; Corbino-Giunta, L.; Egger, L.; Gille, D.; Kopf-Bolanz, K.; Laederach, K.; et al. The NutriChip project-translating technology into nutritional knowledge. Br. J. Nutr. 2012, 108, 762–768. [Google Scholar] [CrossRef]
- Martin, T.D.; Chan, S.S.M.; Hart, A.R. Environmental Factors in the Relapse and Recurrence of Inflammatory Bowel Disease: A Review of the Literature. Dig. Dis. Sci. 2015, 60, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Beaurivage, C.; Naumovska, E.; Chang, Y.X.; Elstak, E.D.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; et al. Development of a gut-on-a-chip model for high throughput disease modeling and drug discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef] [PubMed]
- Jing, B.; Xia, K.; Zhang, C.; Jiao, S.; Zhu, L.; Wei, J.; Wang, Z.A.; Chen, N.; Tu, P.; Li, J.; et al. Chitosan Oligosaccharides Regulate the Occurrence and Development of Enteritis in a Human Gut-On-a-Chip. Front. Cell Dev. Biol. 2022, 10, 877892. [Google Scholar] [CrossRef]
- Beaurivage, C.; Kanapeckaite, A.; Loomans, C.; Erdmann, K.S.; Stallen, J.; Janssen, R.A.J. Development of a human primary gut-on-a-chip to model inflammatory processes. Sci. Rep. 2020, 10, 21475. [Google Scholar] [CrossRef]
- Brennan, A.M. Development of synthetic biotics as treatment for human diseases. Synth. Biol. 2022, 7, ysac001. [Google Scholar] [CrossRef]
- Nelson, M.T.; Charbonneau, M.R.; Coia, H.G.; Castillo, M.J.; Holt, C.; Greenwood, E.S.; Robinson, P.J.; Merrill, E.A.; Lubkowicz, D.; Mauzy, C.A. Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip. Nat. Commun. 2021, 12, 2805. [Google Scholar] [CrossRef]
- Bein, A.; Kim, S.; Goyal, G.; Cao, W.; Fadel, C.; Naziripour, A.; Sharma, S.; Swenor, B.; LoGrande, N.; Nurani, A.; et al. Enteric Coronavirus Infection and Treatment Modeled With an Immunocompetent Human Intestine-On-A-Chip. Front. Pharmacol. 2021, 12, 718484. [Google Scholar] [CrossRef]
- Ly, K.L.; Luo, X.; Raub, C.B. Oral mucositis on a chip: Modeling induction by chemo- and radiation treatments and recovery. Biofabrication 2023, 15, 015007. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, C.; Rahimi, B.; Padova, D.; Rooholghodos, S.A.; Bienek, D.R.; Luo, X.; Kaufman, G.; Raub, C.B. Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 2018, 12, 054106. [Google Scholar] [CrossRef]
- Lee, E.; Kim, Y.; Salipante, P.; Kotula, A.P.; Lipshutz, S.; Graves, D.T.; Alimperti, S. Mechanical Regulation of Oral Epithelial Barrier Function. Bioengineering 2023, 10, 517. [Google Scholar] [CrossRef]
- Jin, L.; Kou, N.; An, F.; Gao, Z.; Tian, T.; Hui, J.; Chen, C.; Ma, G.; Mao, H.; Liu, H. Analyzing Human Periodontal Soft Tissue Inflammation and Drug Responses In Vitro Using Epithelium-Capillary Interface On-a-Chip. Biosensors 2022, 12, 345. [Google Scholar] [CrossRef]
- Makkar, H.; Zhou, Y.; Tan, K.S.; Lim, C.T.; Sriram, G. Modeling Crevicular Fluid Flow and Host-Oral Microbiome Interactions in a Gingival Crevice-on-Chip. Adv. Healthc. Mater. 2023, 12, 2202376. [Google Scholar] [CrossRef] [PubMed]
- Ewart, L.; Roth, A. Opportunities and challenges with microphysiological systems: A pharma end-user perspective. Nat. Rev. Drug Discov. 2021, 20, 327–328. [Google Scholar] [CrossRef]
- Clapp, N.; Amour, A.; Rowan, W.C.; Candarlioglu, P.L. Organ-on-chip applications in drug discovery: An end user perspective. Biochem. Soc. Trans. 2021, 49, 1881–1890. [Google Scholar] [CrossRef] [PubMed]
- Vulto, P.; Joore, J. Adoption of organ-on-chip platforms by the pharmaceutical industry. Nat. Rev. Drug Discov. 2021, 20, 961–962. [Google Scholar] [CrossRef]
- Roth, A. Human microphysiological systems for drug development. Science 2021, 373, 1304–1306. [Google Scholar] [CrossRef]
- Franzen, N.; van Harten, W.H.; Retèl, V.P.; Loskill, P.; van den Eijnden-van Raaij, J.; IJzerman, M. Impact of organ-on-a-chip technology on pharmaceutical R&D costs. Drug Discov. Today 2019, 24, 1720–1724. [Google Scholar] [CrossRef]
- Mesa+ Institute. Translational Organ-on-Chip Platform. Available online: https://www.utwente.nl/en/mesaplus/research/centres-of-expertise/oocct/top/ (accessed on 12 April 2023).
- Mastrangeli, M.; Millet, S.; van den Eijnden-van Raaij, J. Organ-on-chip in development: Towards a roadmap for organs-on-chip. ALTEX 2019, 36, 650–668. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donkers, J.M.; van der Vaart, J.I.; van de Steeg, E. Gut-on-a-Chip Research for Drug Development: Implications of Chip Design on Preclinical Oral Bioavailability or Intestinal Disease Studies. Biomimetics 2023, 8, 226. https://doi.org/10.3390/biomimetics8020226
Donkers JM, van der Vaart JI, van de Steeg E. Gut-on-a-Chip Research for Drug Development: Implications of Chip Design on Preclinical Oral Bioavailability or Intestinal Disease Studies. Biomimetics. 2023; 8(2):226. https://doi.org/10.3390/biomimetics8020226
Chicago/Turabian StyleDonkers, Joanne M., Jamie I. van der Vaart, and Evita van de Steeg. 2023. "Gut-on-a-Chip Research for Drug Development: Implications of Chip Design on Preclinical Oral Bioavailability or Intestinal Disease Studies" Biomimetics 8, no. 2: 226. https://doi.org/10.3390/biomimetics8020226
APA StyleDonkers, J. M., van der Vaart, J. I., & van de Steeg, E. (2023). Gut-on-a-Chip Research for Drug Development: Implications of Chip Design on Preclinical Oral Bioavailability or Intestinal Disease Studies. Biomimetics, 8(2), 226. https://doi.org/10.3390/biomimetics8020226





