RiPa-Net: Recognition of Rice Paddy Diseases with Duo-Layers of CNNs Fostered by Feature Transformation and Selection
Abstract
:1. Introduction
- Introducing an efficient and reliable pipeline to detect and classify nine paddy diseases based on three lightweight CNNs instead of a single DL model.
- Acquiring deep features from dual distinct deep layers of each CNN rather than obtaining deep features from one layer.
- Relying on spatial–spectral–temporal deep features as a replacement for only spatial features by adopting DTCWT to analyze and reduce deep features acquired from the first deep layer of each CNN and then concatenating it with deep features of the second layer.
- Employing PCA and DCT transformation methods that merge deep features of the three CNNs and reduce the dimension of deep features, thus reducing the training complexity of the recognition models.
- Blending deep features of the three CNNs to perform classification rather than depending on the deep features of a single CNN.
- Presenting a feature selection process to choose only the persuasive features and ignore unnecessary features, thus decreasing the classification complexity.
2. Previous Work on Paddy Disease Recognition
3. Materials and Methods
3.1. Transformation and Reduction Methods
3.1.1. Dual-Tree Complex Wavelet Transform
3.1.2. Principal Component Analysis
3.1.3. Discrete Cosine Transform
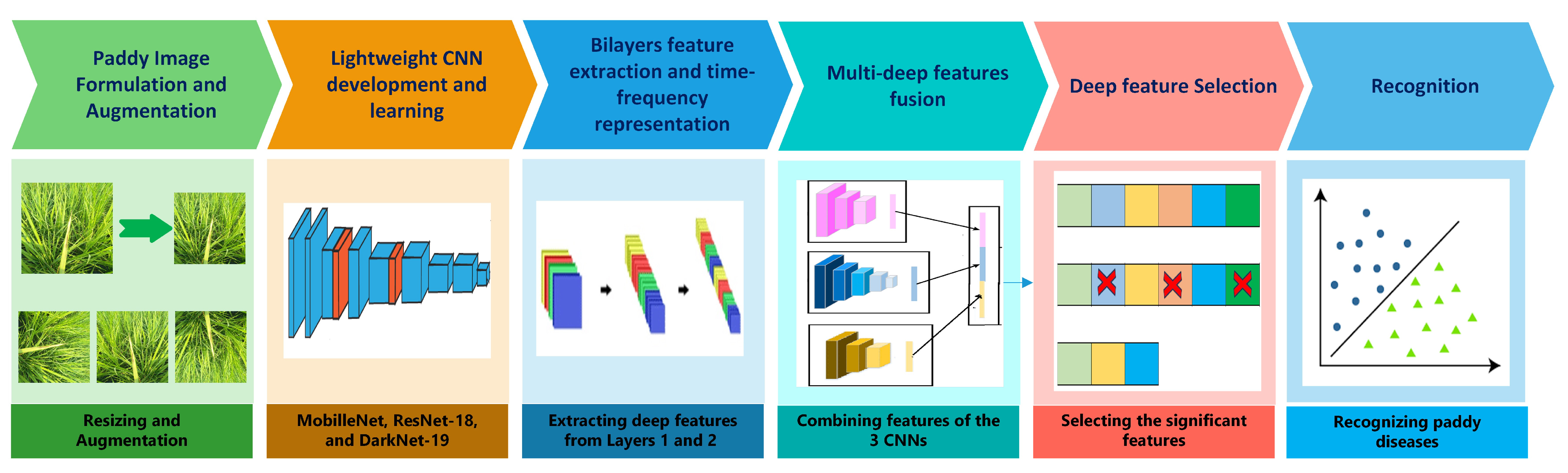
3.2. Suggested RiPa-Net Pipeline
| Algorithm 1. The steps of the proposed RiPa-Net pipeline. | |
| Input: Paddy RGB Images Output: Recognized paddy diseases | |
| 1. | Begin RiPa-Net: |
| 2. | Resize all images to fit the input size of the CNNs: 224 × 224 × 3 for MobileNet and ResNet-18 and 256 × 256 × 3 for DarkNet-19. |
| 3. | Augment images to avoid overfitting and boost CNNs performance: rotation, flipping, shearing, and scaling. |
| 4. | Create lightweight CNN models including MobileNet, ResNet-18, and DarkNet-19 |
| 5. | Set some CNN hyperparameters: learning rate (0.0001), mini-batch (10), epochs (20), and validation frequency (778). |
| 6. | Start: CNN learning process: |
| 7. | After the learning process is finished, End of the learning process. |
| 8. | For each CNN: |
| 9. | Extract deep features from layer 1 and layer 2. |
| 10. | Apply DTCWT to layer 1 features to obtain spatial–spectral–temporal deep features. |
| 11. | End For |
| 12. | Fuse Layer 1 deep features of the three CNNs: using PCA and DCT to fuse and reduce feature space dimensionality. |
| 13. | Concatenate deep features of the previous step with deep features of layer 2 of the three CNNs. |
| 14. | Apply mRMR feature selection to select the most significant features. |
| 15. | Construct classifiers: linear SVM, quadratic SVM, and cubic SVM. |
| 16. | Test classifiers: recognize paddy disease using the testing set (use 5-fold cross-validation). |
| 17. | End RiPa-Net |
3.2.1. Paddy Image Formulation and Augmentation
3.2.2. Lightweight CNN Development and Learning
3.2.3. Bilayers Feature Extraction and Time-Frequency Representation
3.2.4. Multi-Deep Features Fusion
3.2.5. Deep Feature Selection
3.2.6. Recognition
4. Pipeline Setting
4.1. Rice-Paddy Disease Dataset
4.2. Parameter Fine-Tuning
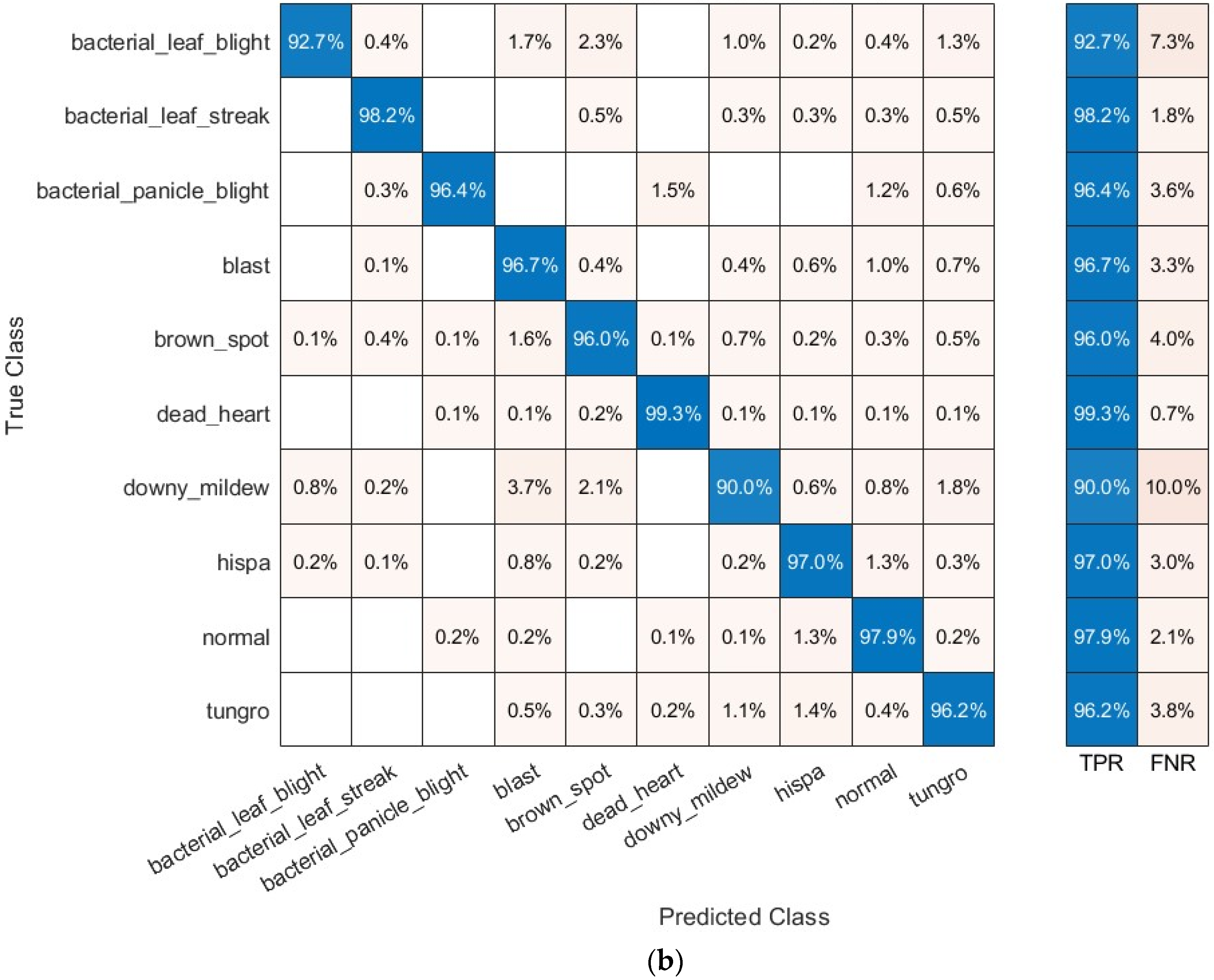
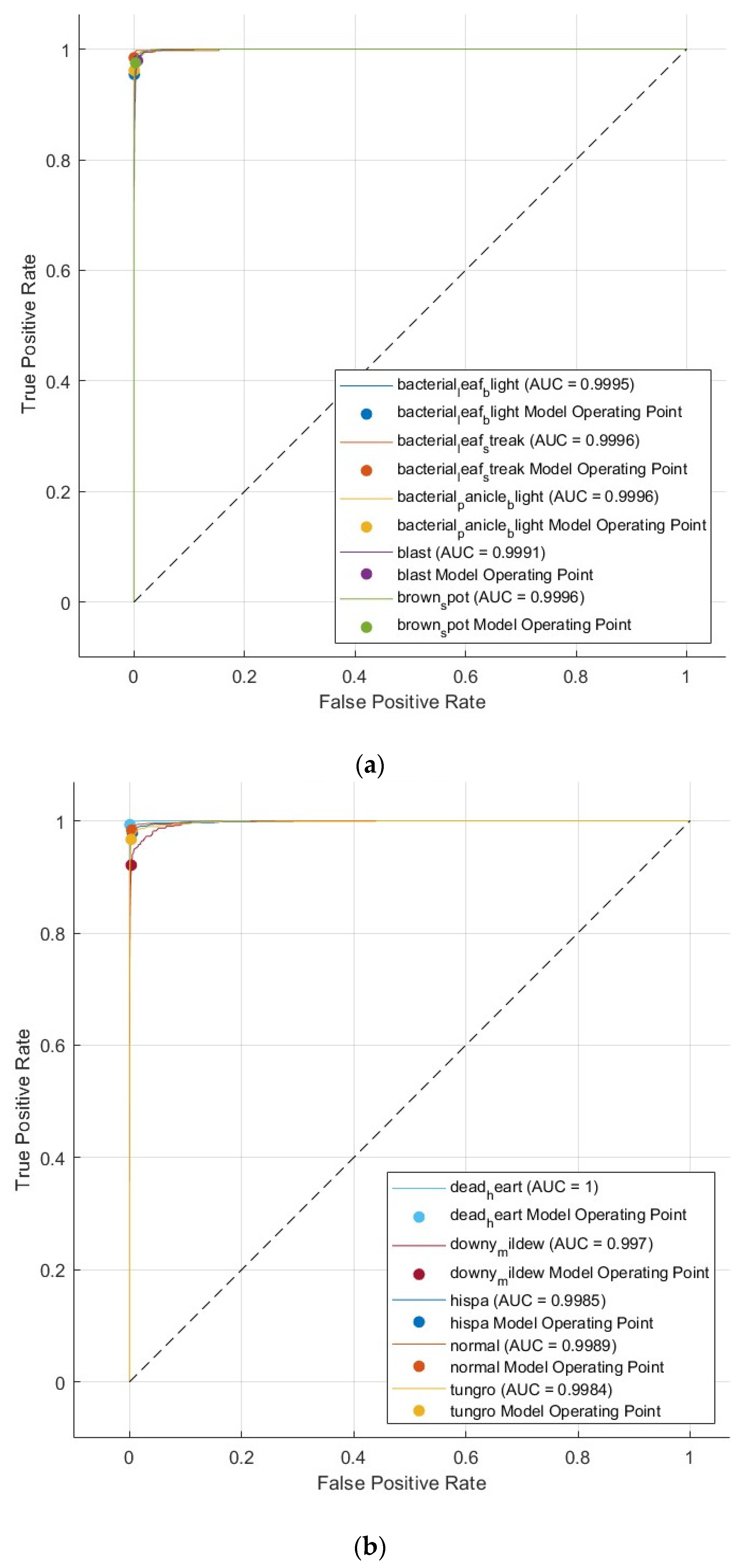
4.3. Assessment Measures
5. Results
5.1. Results of the First Scenario
5.2. Results of the Second Scenario
5.3. Results of the Third Scenario
6. Discussion
6.1. Comparative Evaluation
6.2. Shortcomings and Upcoming Directions
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Temniranrat, P.; Kiratiratanapruk, K.; Kitvimonrat, A.; Sinthupinyo, W.; Patarapuwadol, S. A System for Automatic Rice Disease Detection from Rice Paddy Images Serviced via a Chatbot. Comput. Electron. Agric. 2021, 185, 106156. [Google Scholar] [CrossRef]
- Rehman, A.U.; Farooq, M.; Rashid, A.; Nadeem, F.; Stuerz, S.; Asch, F.; Bell, R.W.; Siddique, K.H. Boron Nutrition of Rice in Different Production Systems. A Review. Agron. Sustain. Dev. 2018, 38, 25. [Google Scholar] [CrossRef]
- Shrivastava, V.K.; Pradhan, M.K.; Minz, S.; Thakur, M.P. Rice Plant Disease Classification Using Transfer Learning of Deep Convolution Neural Network. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 3, 631–635. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A Deep Learning-Based Approach for Automated Yellow Rust Disease Detection from High-Resolution Hyperspectral UAV Images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef]
- Tholkapiyan, M.; Aruna Devi, B.; Bhatt, D.; Saravana Kumar, E.; Kirubakaran, S.; Kumar, R. Performance Analysis of Rice Plant Diseases Identification and Classification Methodology. Wirel. Pers. Commun. 2023, 130, 1317–1341. [Google Scholar] [CrossRef]
- Attallah, O. GabROP: Gabor Wavelets-Based CAD for Retinopathy of Prematurity Diagnosis via Convolutional Neural Networks. Diagnostics 2023, 13, 171. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. A Computer-Aided Diagnostic Framework for Coronavirus Diagnosis Using Texture-Based Radiomics Images. Digit. Health 2022, 8, 20552076221092544. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. RADIC: A Tool for Diagnosing COVID-19 from Chest CT and X-Ray Scans Using Deep Learning and Quad-Radiomics. Chemom. Intell. Lab. Syst. 2023, 233, 104750. [Google Scholar] [CrossRef] [PubMed]
- Aurna, N.F.; Yousuf, M.A.; Taher, K.A.; Azad, A.K.M.; Moni, M.A. A Classification of MRI Brain Tumor Based on Two Stage Feature Level Ensemble of Deep CNN Models. Comput. Biol. Med. 2022, 146, 105539. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. Deep Learning-Based CAD System for COVID-19 Diagnosis via Spectral-Temporal Images. In Proceedings of the 2022 the 12th International Conference on Information Communication and Management, London, UK, 13–15 July 2022; pp. 25–33. [Google Scholar]
- Attallah, O. MB-AI-His: Histopathological Diagnosis of Pediatric Medulloblastoma and Its Subtypes via AI. Diagnostics 2021, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. An Intelligent ECG-Based Tool for Diagnosing COVID-19 via Ensemble Deep Learning Techniques. Biosensors 2022, 12, 299. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Chugh, N.; Balyan, A. Identification of ADHD Disorder in Children Using EEG Based on Visual Attention Task by Ensemble Deep Learning. In Proceedings of the International Conference on Data Science and Applications: ICDSA 2022, Kolkata, India, 26–27 March 2022; Springer: Berlin/Heidelberg, Germany, 2023; Volume 2, pp. 243–259. [Google Scholar]
- Attallah, O. Cervical Cancer Diagnosis Based on Multi-Domain Features Using Deep Learning Enhanced by Handcrafted Descriptors. Appl. Sci. 2023, 13, 1916. [Google Scholar] [CrossRef]
- Gong, W.; Chen, H.; Zhang, Z.; Zhang, M.; Wang, R.; Guan, C.; Wang, Q. A Novel Deep Learning Method for Intelligent Fault Diagnosis of Rotating Machinery Based on Improved CNN-SVM and Multichannel Data Fusion. Sensors 2019, 19, 1693. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Santhi, B.; Sridhar, S.; Vinolia, K.; Swaminathan, P. Nuclear Power Plant Thermocouple Sensor-Fault Detection and Classification Using Deep Learning and Generalized Likelihood Ratio Test. IEEE Trans. Nucl. Sci. 2017, 64, 1526–1534. [Google Scholar] [CrossRef]
- Attalah, O. Multitask Deep Learning-Based Pipeline for Gas Leakage Detection via E-Nose and Thermal Imaging Multimodal Fusion. Chemosensors 2023, 11, 364. [Google Scholar] [CrossRef]
- Rahate, A.; Mandaokar, S.; Chandel, P.; Walambe, R.; Ramanna, S.; Kotecha, K. Employing Multimodal Co-Learning to Evaluate the Robustness of Sensor Fusion for Industry 5.0 Tasks. Soft Comput. 2022, 27, 4139–4155. [Google Scholar] [CrossRef]
- Attallah, O. Tomato Leaf Disease Classification via Compact Convolutional Neural Networks with Transfer Learning and Feature Selection. Horticulturae 2023, 9, 149. [Google Scholar] [CrossRef]
- Javidan, S.M.; Banakar, A.; Vakilian, K.A.; Ampatzidis, Y. Diagnosis of Grape Leaf Diseases Using Automatic K-Means Clustering and Machine Learning. Smart Agric. Technol. 2023, 3, 100081. [Google Scholar] [CrossRef]
- Munjal, D.; Singh, L.; Pandey, M.; Lakra, S. A Systematic Review on the Detection and Classification of Plant Diseases Using Machine Learning. Int. J. Softw. Innov. 2023, 11, 1–25. [Google Scholar] [CrossRef]
- Thakur, P.S.; Sheorey, T.; Ojha, A. VGG-ICNN: A Lightweight CNN Model for Crop Disease Identification. Multimed. Tools Appl. 2023, 82, 497–520. [Google Scholar] [CrossRef]
- Tabbakh, A.; Barpanda, S.S. A Deep Features Extraction Model Based on the Transfer Learning Model and Vision Transformer “TLMViT” for Plant Disease Classification. IEEE Access 2023, 11, 45377–45392. [Google Scholar] [CrossRef]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Deep Feature Based Rice Leaf Disease Identification Using Support Vector Machine. Comput. Electron. Agric. 2020, 175, 105527. [Google Scholar] [CrossRef]
- Adeel, A.; Khan, M.A.; Akram, T.; Sharif, A.; Yasmin, M.; Saba, T.; Javed, K. Entropy-Controlled Deep Features Selection Framework for Grape Leaf Diseases Recognition. Expert Syst. 2022, 39, e12569. [Google Scholar] [CrossRef]
- Peng, Y.; Zhao, S.; Liu, J. Fused-Deep-Features Based Grape Leaf Disease Diagnosis. Agronomy 2021, 11, 2234. [Google Scholar] [CrossRef]
- Koklu, M.; Unlersen, M.F.; Ozkan, I.A.; Aslan, M.F.; Sabanci, K. A CNN-SVM Study Based on Selected Deep Features for Grapevine Leaves Classification. Measurement 2022, 188, 110425. [Google Scholar] [CrossRef]
- Farooqui, N.A.; Mishra, A.K.; Mehra, R. Concatenated Deep Features with Modified LSTM for Enhanced Crop Disease Classification. Int. J. Intell. Robot. Appl. 2022, 7, 510–534. [Google Scholar] [CrossRef]
- Attallah, O. CerCan·Net: Cervical Cancer Classification Model via Multi-Layer Feature Ensembles of Lightweight CNNs and Transfer Learning. Expert Syst. Appl. 2023, 229 Pt B, 120624. [Google Scholar] [CrossRef]
- Attallah, O. MonDiaL-CAD: Monkeypox Diagnosis via Selected Hybrid CNNs Unified with Feature Selection and Ensemble Learning. Digit. Health 2023, 9, 20552076231180056. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. ECG-BiCoNet: An ECG-Based Pipeline for COVID-19 Diagnosis Using Bi-Layers of Deep Features Integration. Comput. Biol. Med. 2022, 142, 105210. [Google Scholar] [CrossRef]
- Attallah, O.; Zaghlool, S. AI-Based Pipeline for Classifying Pediatric Medulloblastoma Using Histopathological and Textural Images. Life 2022, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Khullar, V.; Goyal, N. Contemporary and Futuristic Intelligent Technologies for Rice Leaf Disease Detection. In Proceedings of the 2022 10th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 13–14 October 2022; pp. 1–6. [Google Scholar]
- Maheswaran, S.; Sathesh, S.; Rithika, P.; Shafiq, I.M.; Nandita, S.; Gomathi, R.D. Detection and Classification of Paddy Leaf Diseases Using Deep Learning (CNN). In Computer, Communication, and Signal Processing, Proceedings of the 6th IFIP TC 5 International Conference, ICCCSP 2022, Chennai, India, 24–25 February 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 60–74. [Google Scholar]
- Upadhyay, S.K.; Kumar, A. Early-Stage Brown Spot Disease Recognition in Paddy Using Image Processing and Deep Learning Techniques. Trait. Signal 2021, 38, 1755–1766. [Google Scholar] [CrossRef]
- Ni, H.; Shi, Z.; Karungaru, S.; Lv, S.; Li, X.; Wang, X.; Zhang, J. Classification of Typical Pests and Diseases of Rice Based on the ECA Attention Mechanism. Agriculture 2023, 13, 1066. [Google Scholar] [CrossRef]
- Haridasan, A.; Thomas, J.; Raj, E.D. Deep Learning System for Paddy Plant Disease Detection and Classification. Environ. Monit. Assess. 2023, 195, 120. [Google Scholar] [CrossRef]
- Akyol, K. Handling Hypercolumn Deep Features in Machine Learning for Rice Leaf Disease Classification. Multimed. Tools Appl. 2022, 82, 19503–19520. [Google Scholar] [CrossRef]
- Yakkundimath, R.; Saunshi, G.; Anami, B.; Palaiah, S. Classification of Rice Diseases Using Convolutional Neural Network Models. J. Inst. Eng. (India) Ser. B 2022, 103, 1047–1059. [Google Scholar] [CrossRef]
- Lamba, S.; Baliyan, A.; Kukreja, V. A Novel GCL Hybrid Classification Model for Paddy Diseases. Int. J. Inf. Technol. 2023, 15, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Sethy, P.K.; Behera, S.K.; Kannan, N.; Narayanan, S.; Pandey, C. Smart Paddy Field Monitoring System Using Deep Learning and IoT. Concurr. Eng. 2021, 29, 16–24. [Google Scholar] [CrossRef]
- Saleem, M.A.; Aamir, M.; Ibrahim, R.; Senan, N.; Alyas, T. An Optimized Convolution Neural Network Architecture for Paddy Disease Classification. Comput. Mater. Contin. 2022, 71, 6053–6067. [Google Scholar]
- Attallah, O. CoMB-Deep: Composite Deep Learning-Based Pipeline for Classifying Childhood Medulloblastoma and Its Classes. Front. Neuroinform. 2021, 15, 663592. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, R.; Biswas, M.; Sen, S.; de Souza Júnior, L.A.; Papa, J.P.; Sarkar, R. A Fuzzy Distance-Based Ensemble of Deep Models for Cervical Cancer Detection. Comput. Methods Programs Biomed. 2022, 219, 106776. [Google Scholar] [CrossRef] [PubMed]
- Mallat, S.G. A Theory for Multiresolution Signal Decomposition: The Wavelet Representation. IEEE Trans. Pattern Anal. Mach. Intell. 1989, 11, 674–693. [Google Scholar] [CrossRef]
- Mallat, S. A Wavelet Tour of Signal Processing, 2nd ed.; New York Academic: New York, NY, USA, 1999. [Google Scholar]
- Cohen, A.; Daubechies, I.; Jawerth, B.; Vial, P. Multiresolution Analysis, Wavelets and Fast Algorithms on an Interval. C. R. Acad. Sci. Sér. 1 Math. 1993, 316, 417–421. [Google Scholar]
- Cohen, A.; Daubechies, I.; Feauveau, J.-C. Biorthogonal Bases of Compactly Supported Wavelets. Commun. Pure Appl. Math. 1992, 45, 485–560. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Dunteman, G.H. Principal Components Analysis; SAGE: Newcastle upon Tyne, UK, 1989; Volume 69. [Google Scholar]
- Ahmed, N.; Natarajan, T.; Rao, K.R. Discrete Cosine Transform. IEEE Trans. Comput. 1974, 100, 90–93. [Google Scholar] [CrossRef]
- Strang, G. The Discrete Cosine Transform. SIAM Rev. 1999, 41, 135–147. [Google Scholar] [CrossRef]
- Narasimha, M.; Peterson, A. On the Computation of the Discrete Cosine Transform. IEEE Trans. Commun. 1978, 26, 934–936. [Google Scholar] [CrossRef]
- Ochoa-Dominguez, H.; Rao, K.R. Discrete Cosine Transform; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Attallah, O.; Samir, A. A Wavelet-Based Deep Learning Pipeline for Efficient COVID-19 Diagnosis via CT Slices. Appl. Soft Comput. 2022, 128, 109401. [Google Scholar] [CrossRef]
- Jogin, M.; Madhulika, M.S.; Divya, G.D.; Meghana, R.K.; Apoorva, S. Feature Extraction Using Convolution Neural Networks (CNN) and Deep Learning. In Proceedings of the 2018 3rd IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT), Bangalore, India, 18–19 May 2018; pp. 2319–2323. [Google Scholar]
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature Selection in Machine Learning: A New Perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum Redundancy Maximum Relevance Feature Selection Approach for Temporal Gene Expression Data. BMC Bioinform. 2017, 18, 9. [Google Scholar] [CrossRef]
- Thai, L.H.; Hai, T.S.; Thuy, N.T. Image Classification Using Support Vector Machine and Artificial Neural Network. Int. J. Inf. Technol. Comput. Sci. 2012, 4, 32–38. [Google Scholar] [CrossRef]
- Attallah, O. A Deep Learning-Based Diagnostic Tool for Identifying Various Diseases via Facial Images. Digit. Health 2022, 8, 20552076221124430. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. DIAROP: Automated Deep Learning-Based Diagnostic Tool for Retinopathy of Prematurity. Diagnostics 2021, 11, 2034. [Google Scholar] [CrossRef] [PubMed]
- Murugan, D. Paddy Doctor: A Visual Image Dataset for Paddy Disease Classification. arXiv 2022, arXiv:2205.11108. [Google Scholar]
- Keskar, N.S.; Mudigere, D.; Nocedal, J.; Smelyanskiy, M.; Tang, P.T.P. On Large-Batch Training for Deep Learning: Generalization Gap and Sharp Minima. arXiv 2016, arXiv:1609.04836. [Google Scholar]
- Li, M.; Zhang, T.; Chen, Y.; Smola, A.J. Efficient Mini-Batch Training for Stochastic Optimization. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 661–670. [Google Scholar]
- Ullah, A.; Muhammad, K.; Ding, W.; Palade, V.; Haq, I.U.; Baik, S.W. Efficient Activity Recognition Using Lightweight CNN and DS-GRU Network for Surveillance Applications. Appl. Soft Comput. 2021, 103, 107102. [Google Scholar] [CrossRef]
- Nayem, Z.H.; Jahan, I.; Rakib, A.A.; Mia, S. Detection and Identification of Rice Pests Using Memory Efficient Convolutional Neural Network. In Proceedings of the 2023 International Conference on Computer, Electrical & Communication Engineering (ICCECE), Kolkata, India, 20–21 January 2023; pp. 1–6. [Google Scholar]
- Thai, H.-T.; Le, K.-H.; Nguyen, N.L.-T. FormerLeaf: An Efficient Vision Transformer for Cassava Leaf Disease Detection. Comput. Electron. Agric. 2023, 204, 107518. [Google Scholar] [CrossRef]
- Wang, F.; Rao, Y.; Luo, Q.; Jin, X.; Jiang, Z.; Zhang, W.; Li, S. Practical Cucumber Leaf Disease Recognition Using Improved Swin Transformer and Small Sample Size. Comput. Electron. Agric. 2022, 199, 107163. [Google Scholar] [CrossRef]
- Guo, Y.; Lan, Y.; Chen, X. CST: Convolutional Swin Transformer for Detecting the Degree and Types of Plant Diseases. Comput. Electron. Agric. 2022, 202, 107407. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]


| Augmentation Technique | Range |
|---|---|
| Flip horizontally and vertically | −25 to 25 |
| Scaling | 1 to 2 |
| Shearing vertically | −30 to 30 |
| Rotation horizontally and vertically | 25 to 25 |
| Model | Layer 1 | Layer 1 (After DTCWT) | Layer 2 |
|---|---|---|---|
| ResNet-18 | 512 | 256 | 10 |
| DarkNet-19 | 660 | 330 | 10 |
| MobileNet | 1280 | 640 | 10 |
| Paddy Disease Category | Sum of Photos |
|---|---|
| Blast | 1738 |
| Tungro | 1088 |
| Dead heart | 1442 |
| Bacterial leaf blight | 479 |
| Bacterial panicle blight | 337 |
| Bacterial leaf streak | 380 |
| Hispa | 1594 |
| Brown spot | 965 |
| Downy mildew | 620 |
| Normal | 1764 |
| ResNet-18 | MobileNet | DarkNet-19 | ||||
|---|---|---|---|---|---|---|
| Layer 1 | Layer 2 | Layer 1 | Layer 2 | Layer 1 | Layer 2 | |
| LSVM | 89.7 | 92.5 | 91.6 | 93.4 | 96.0 | 95.9 |
| Q-SVM | 92.6 | 93 | 93.1 | 93.4 | 96.4 | 96.3 |
| C-SVM | 93.4 | 92.7 | 94.0 | 92.9 | 96.3 | 95.6 |
| PCA Layer 1 Fused Features + Layer 2 Features | ||||||
| Number of Features | ||||||
| 50 | 100 | 150 | 200 | 250 | 300 | |
| LSVM | 57.4 | 58.4 | 81.1 | 96.4 | 97.0 | 97.1 |
| Q-SVM | 78.1 | 84.2 | 91.2 | 96.7 | 97.2 | 97.2 |
| C-SVM | 78.6 | 85.3 | 93.3 | 96.9 | 97.4 | 97.2 |
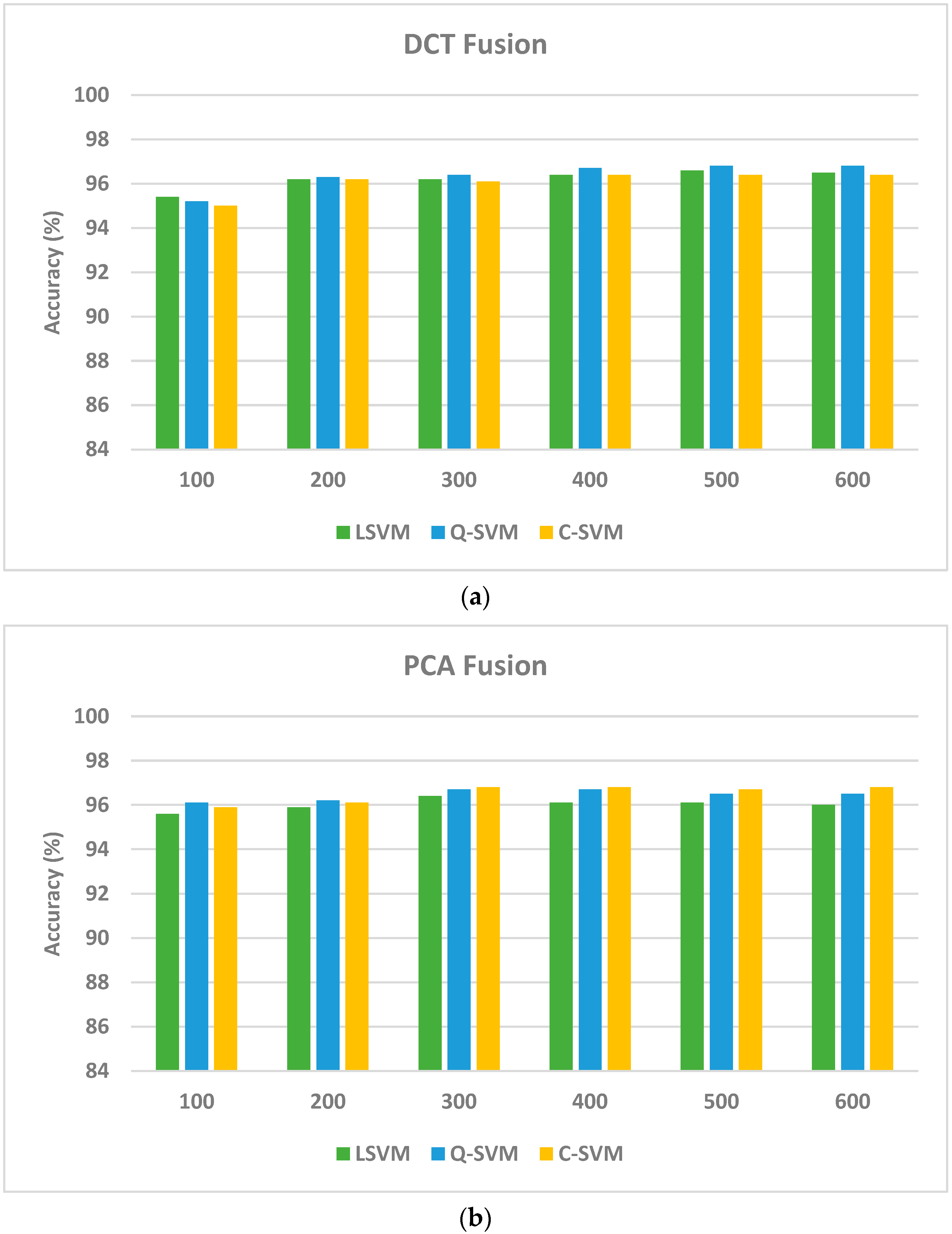
| DCT Layer 1 Fused Features + Layer 2 Features | ||||||
| Number of Features | ||||||
| 50 | 100 | 150 | 200 | 250 | 300 | |
| LSVM | 96.7 | 97.2 | 97.2 | 97.3 | 97.2 | 97.3 |
| Q-SVM | 97.0 | 97.4 | 97.5 | 97.3 | 97.5 | 97.5 |
| C-SVM | 97.0 | 97.4 | 97.3 | 97.5 | 97.5 | 97.5 |
| PCA Layer 1 Fused Features + Layer 2 Features | ||||||
| Accuracy | Sensitivity | Specificity | Precision | F1-Score | MCC | |
| Five-Fold Cross-Validation | ||||||
| LSVM | 0.9700 | 0.9564 | 0.9959 | 0.9634 | 0.9598 | 0.9557 |
| Q-SVM | 0.9720 | 0.9682 | 0.9968 | 0.9720 | 0.9700 | 0.96692 |
| C-SVM | 0.9740 | 0.9687 | 0.9970 | 0.9738 | 0.9712 | 0.9682 |
| Hold-Out Test Set | ||||||
| LSVM | 0.9702 | 0.9661 | 0.9966 | 0.9688 | 0.9674 | 0.9641 |
| Q-SVM | 0.9747 | 0.9721 | 0.9971 | 0.9750 | 0.9734 | 0.9706 |
| C-SVM | 0.9747 | 0.9724 | 0.9971 | 0.9750 | 0.9736 | 0.9707 |
| DCT Layer 1 Fused Features + Layer 2 Features | ||||||
| Accuracy | Sensitivity | Specificity | Precision | F1-Score | MCC | |
| Five-Fold Cross-Validation | ||||||
| LSVM | 0.9730 | 0.9672 | 0.9970 | 0.9721 | 0.9656 | 0.9667 |
| Q-SVM | 0.9750 | 0.9697 | 0.9971 | 0.9742 | 0.9719 | 0.9691 |
| C-SVM | 0.9750 | 0.9707 | 0.9971 | 0.9739 | 0.9722 | 0.9694 |
| Hold-Out Test Set | ||||||
| LSVM | 0.9702 | 0.9662 | 0.9966 | 0.9683 | 0.9671 | 0.9638 |
| Q-SVM | 0.9699 | 0.9644 | 0.9966 | 0.9681 | 0.9662 | 0.9628 |
| C-SVM | 0.9699 | 0.9657 | 0.9966 | 0.9676 | 0.9666 | 0.9632 |
| Reference | # Paddy Diseases | Model | Accuracy | F1 Score | Sensitivity | Precision |
|---|---|---|---|---|---|---|
| [36] | 6 | RepVGG | 0.9706 | 0.9709 | 0.9708 | 0.9713 |
| [66] | 10 | Custom CNN | 0.8888 | - | - | - |
| [67] | 10 | FormerLeaf: a customized vision transformer model | 0.9502 | 0.9616 | 0.9725 | 0.9210 |
| [68] | 10 | Swin Transformer | 0.9434 | 0.9343 | 0.9430 | 0.9252 |
| [69] | 10 | Convolutional Swin | 0.9565 | 0.9536 | 0.9615 | 0.9536 |
| [70] | 10 | Xception | 0.9251 | 0.9155 | 0.9180 | 0.9130 |
| [71] | 10 | MobileNet | 0.8987 | 0.9197 | 0.9348 | 0.9051 |
| [72] | 10 | ResNet-50 | 0.9113 | 0.8933 | 0.9082 | 0.8905 |
| Proposed RiPa-Net Five-fold cross-validation | 10 | MobileNet + ResNet-18 + DarkNet + DCT | 0.9750 | 0.9722 | 0.9707 | 0.9739 |
| Proposed RiPa-Net Hold-out | MobileNet + ResNet-18 + DarkNet + PCA | 0.9740 | 0.9666 | 0.9625 | 0.9680 | |
| MobileNet + ResNet-18 + DarkNet + DCT + CSVM | 0.9699 | 0.9666 | 0.9657 | 0.9676 | ||
| MobileNet + ResNet-18 + DarkNet + PCA + CSVM | 0.9747 | 0.9736 | 0.9724 | 0.9750 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attallah, O. RiPa-Net: Recognition of Rice Paddy Diseases with Duo-Layers of CNNs Fostered by Feature Transformation and Selection. Biomimetics 2023, 8, 417. https://doi.org/10.3390/biomimetics8050417
Attallah O. RiPa-Net: Recognition of Rice Paddy Diseases with Duo-Layers of CNNs Fostered by Feature Transformation and Selection. Biomimetics. 2023; 8(5):417. https://doi.org/10.3390/biomimetics8050417
Chicago/Turabian StyleAttallah, Omneya. 2023. "RiPa-Net: Recognition of Rice Paddy Diseases with Duo-Layers of CNNs Fostered by Feature Transformation and Selection" Biomimetics 8, no. 5: 417. https://doi.org/10.3390/biomimetics8050417
APA StyleAttallah, O. (2023). RiPa-Net: Recognition of Rice Paddy Diseases with Duo-Layers of CNNs Fostered by Feature Transformation and Selection. Biomimetics, 8(5), 417. https://doi.org/10.3390/biomimetics8050417








