Biosynthesis Strategy of Gold Nanoparticles and Biofabrication of a Novel Amoxicillin Gold Nanodrug to Overcome the Resistance of Multidrug-Resistant Bacterial Pathogens MRSA and E. coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Juniperus excelsa Extract
2.2. Biosynthesis of AuNPs
2.3. Factors Affecting the Synthesis of AuNPs
2.3.1. Effect of HAuCl43H2O
2.3.2. Effect of J. excelsa Leaf Extract
2.3.3. Effects of pH Levels
2.3.4. Effects of Temperatures
2.3.5. Effect of Incubation Time
2.4. Conjugation of Amoxicillin with AuNPs
2.5. Characterization of AuNPs and Amoxi-AuNPs
2.5.1. UV–Vis Spectroscopy
2.5.2. TEM Analysis
2.5.3. X-ray Diffraction (XRD) Analysis
2.5.4. FT-IR Analysis
2.6. Determination of Conjugating Efficiency of Amoxi-TPP-AuNPs
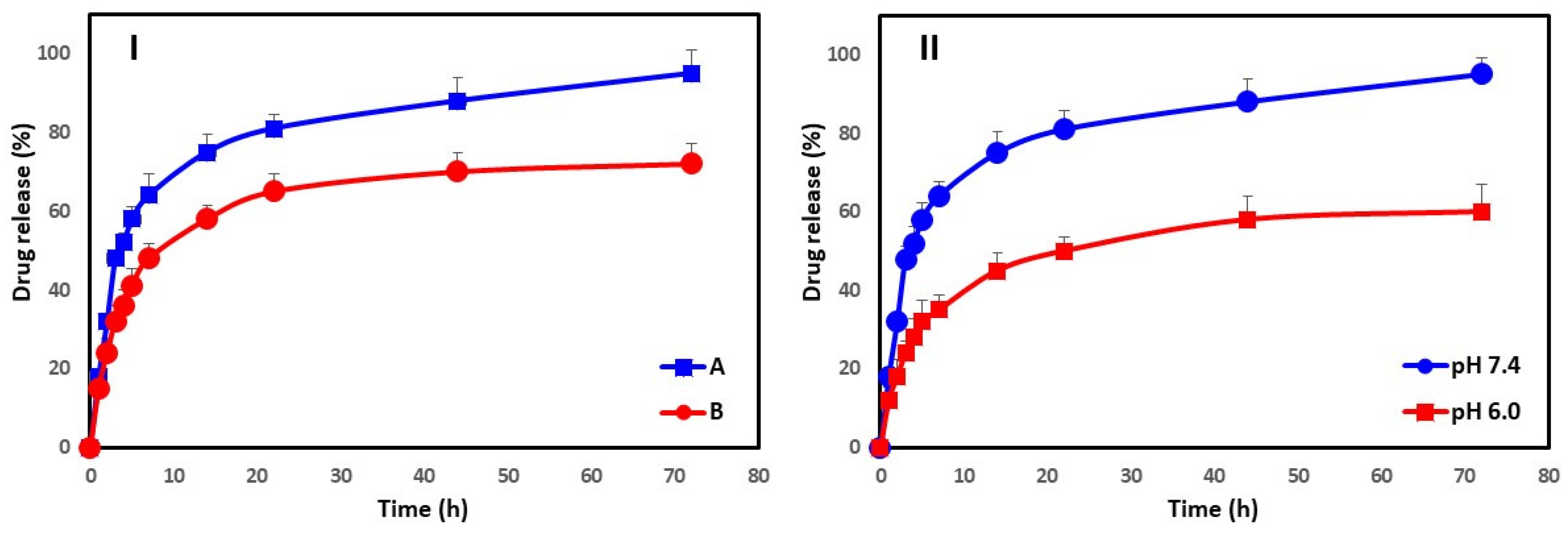
2.7. In Vitro Drug Release Kinetics
2.8. Isolation and Characterization of Clinical Isolates
2.8.1. Collection of Clinical Isolates and Growth Conditions
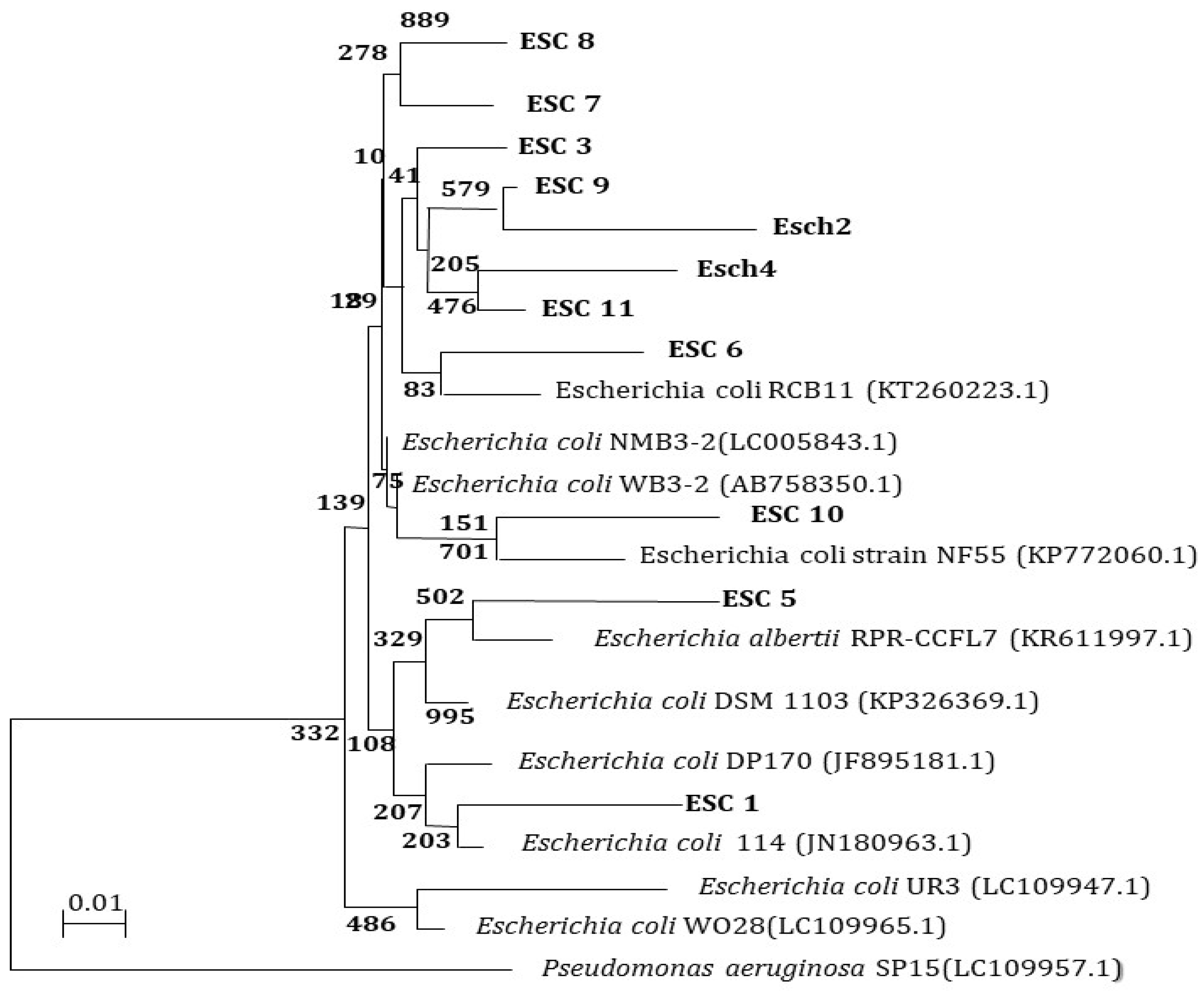
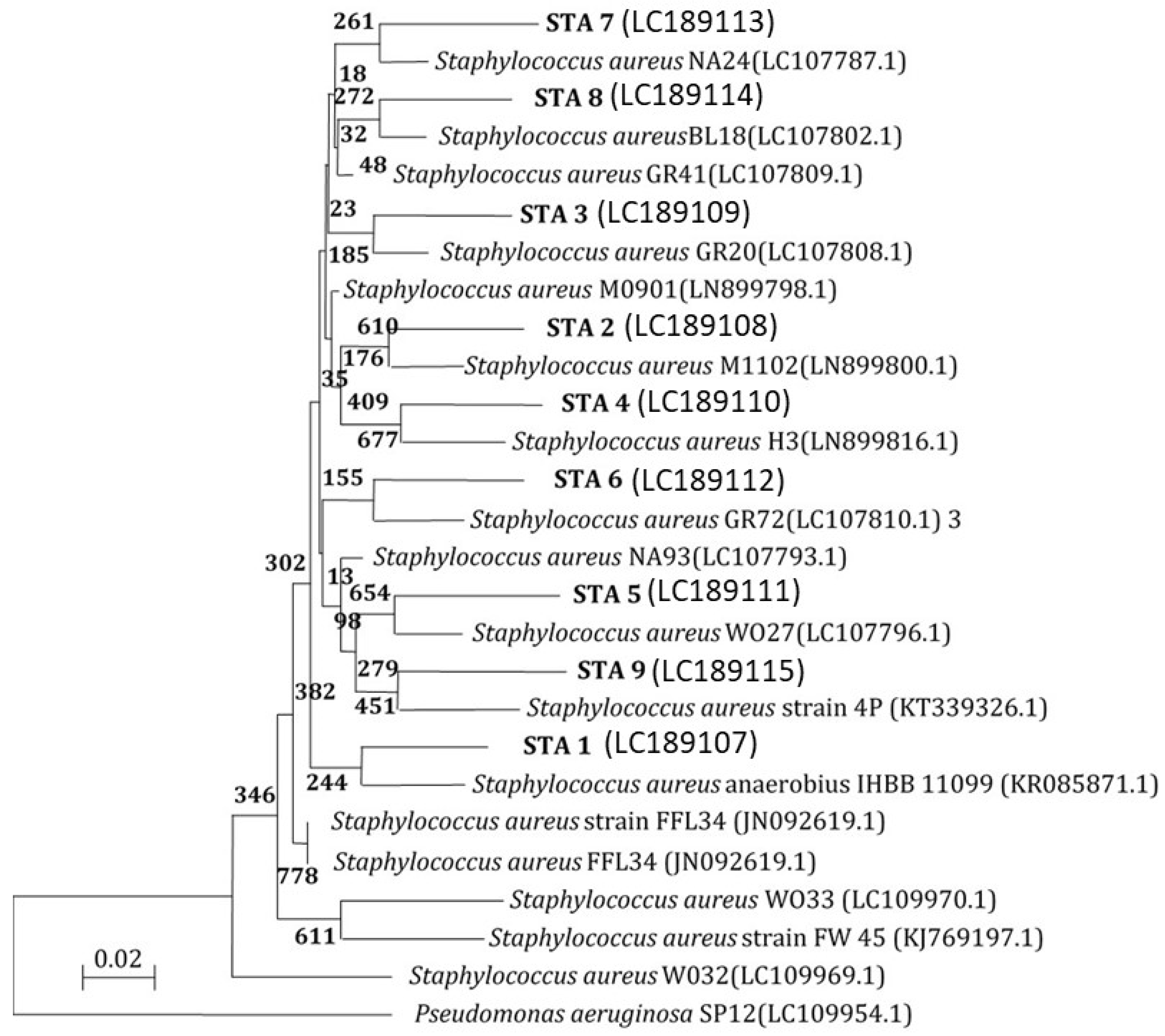
2.8.2. Molecular Typing
Molecular Characterization Using the 16S rRNA Gene
PCR of 16S rRNA Genes
16S rRNA Gene Analysis
2.9. Antibiotic Susceptibility Testing
Disk Diffusion Method
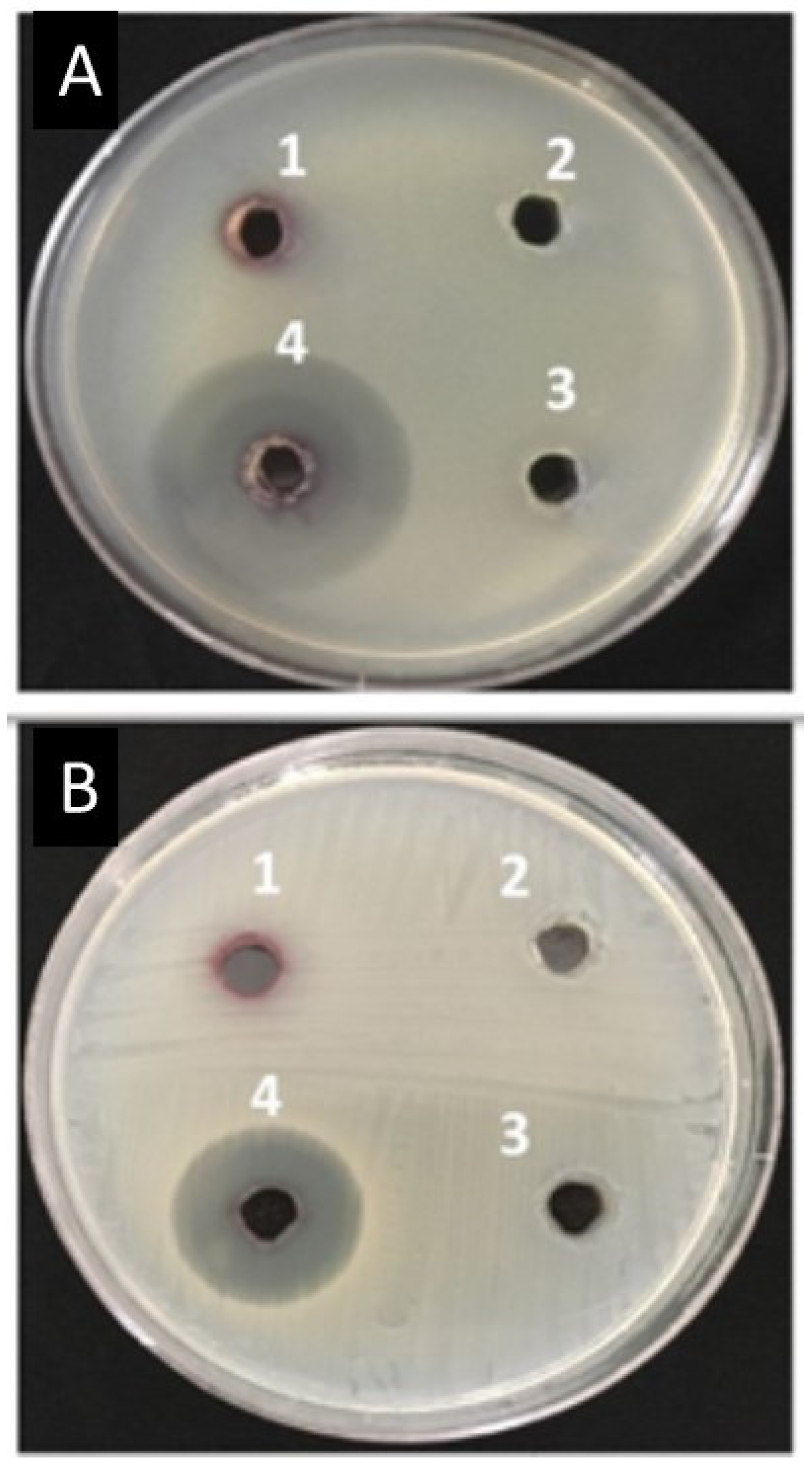
2.10. Antibacterial Activity of Amoxi-TPP-AuNPs Using the Well Agar Diffusion Method
2.11. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of Amoxi-TPP-AuNPs
2.12. Amoxi-TPP-AuNP Time Kill Test
3. Results
3.1. Biosynthesis of AuNPs
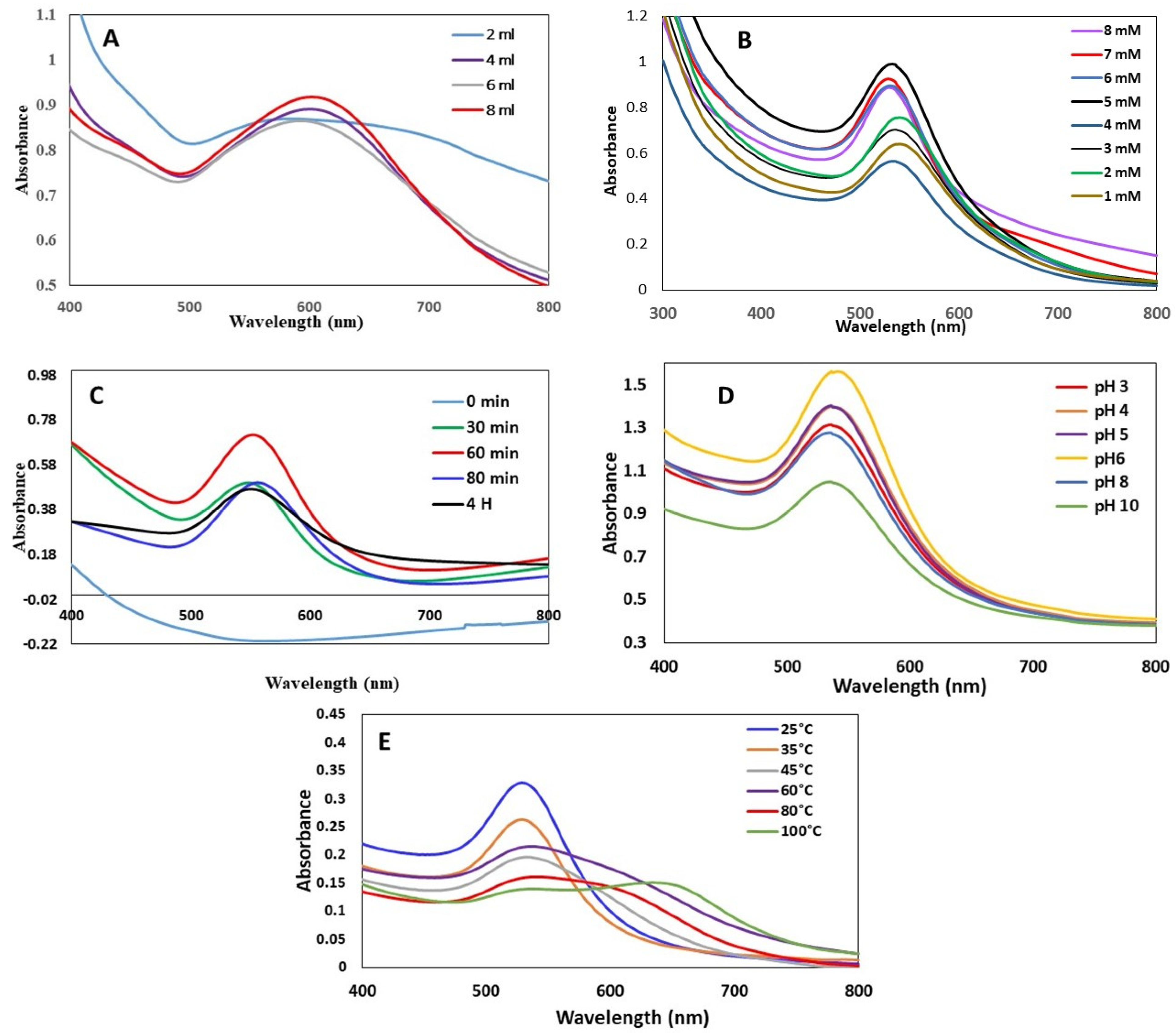
3.1.1. Effect of J. excelsa Extract Volume on AuNP Formation
3.1.2. Effect of HAuCl43H2O Concentration on AuNP Formation
3.1.3. Effect of Time on AuNP Formation
3.1.4. Effect of pH on the AuNP Formation
3.1.5. Effect of Temperature on the AuNP Formation
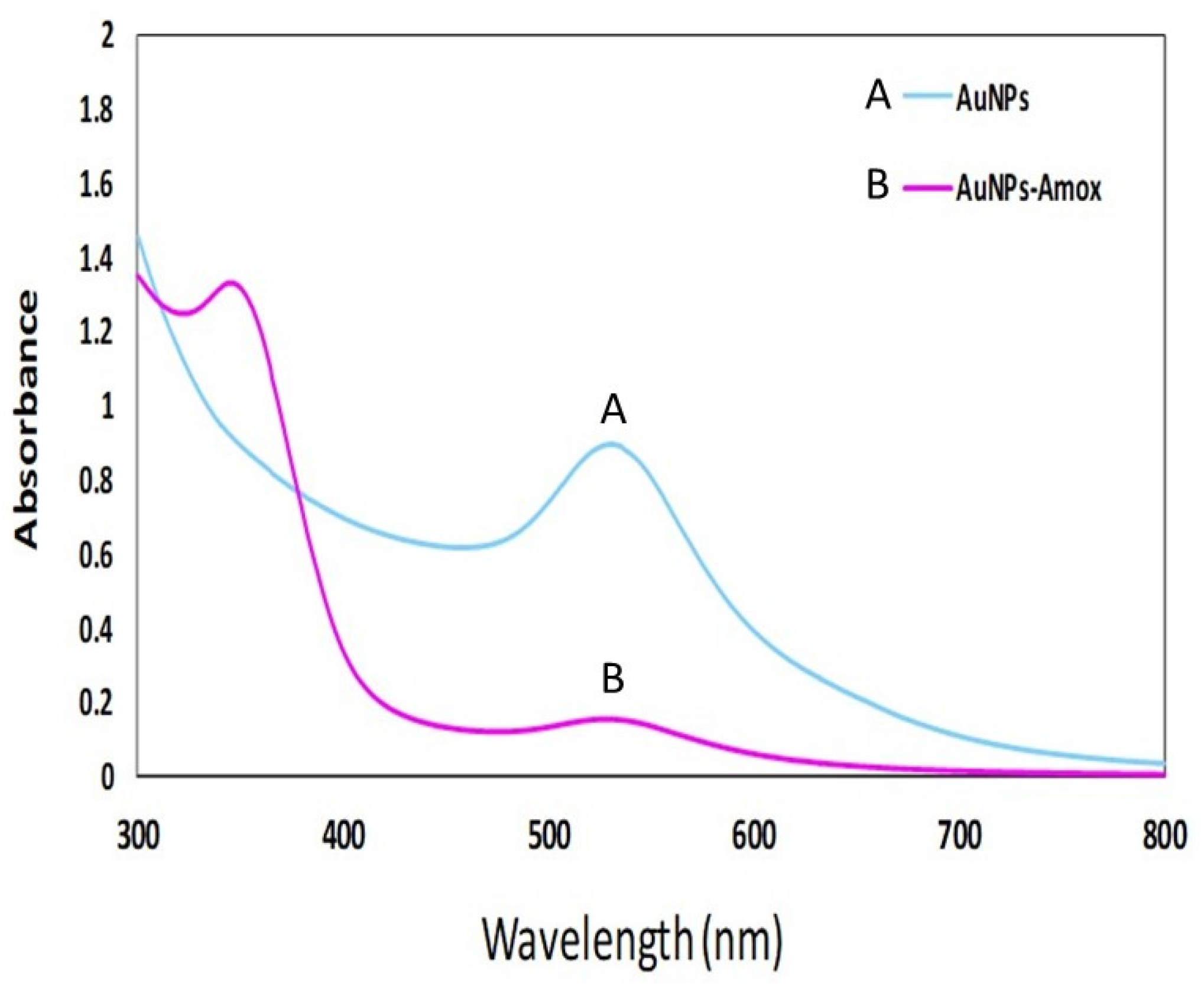
3.2. Bio-Fabrication of Amoxi-TPP-AuNPs
3.3. AuNP and Amoxi-TPP-AuNP Characterization
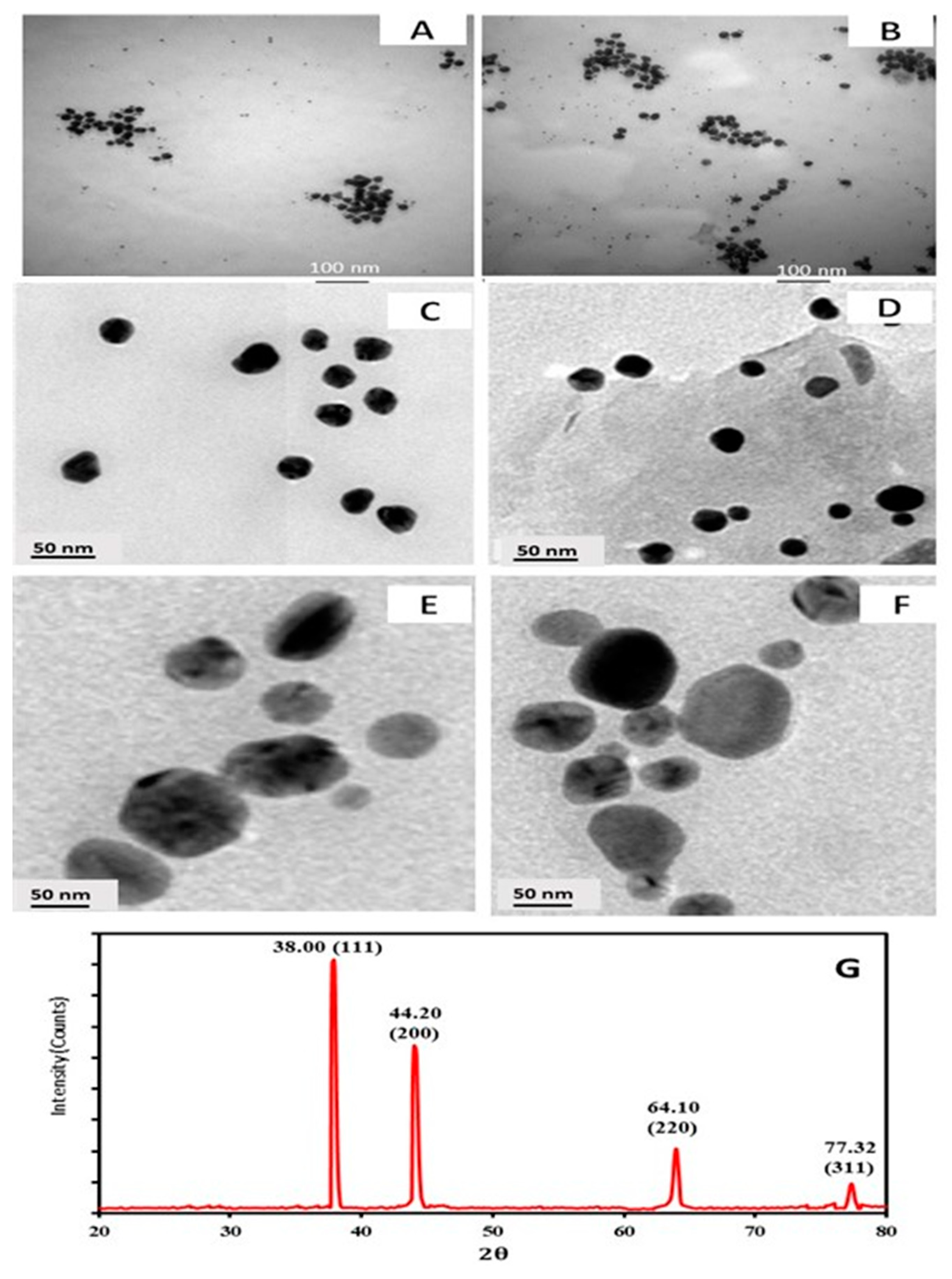
3.3.1. TEM with High Resolution
3.3.2. XRD Analysis
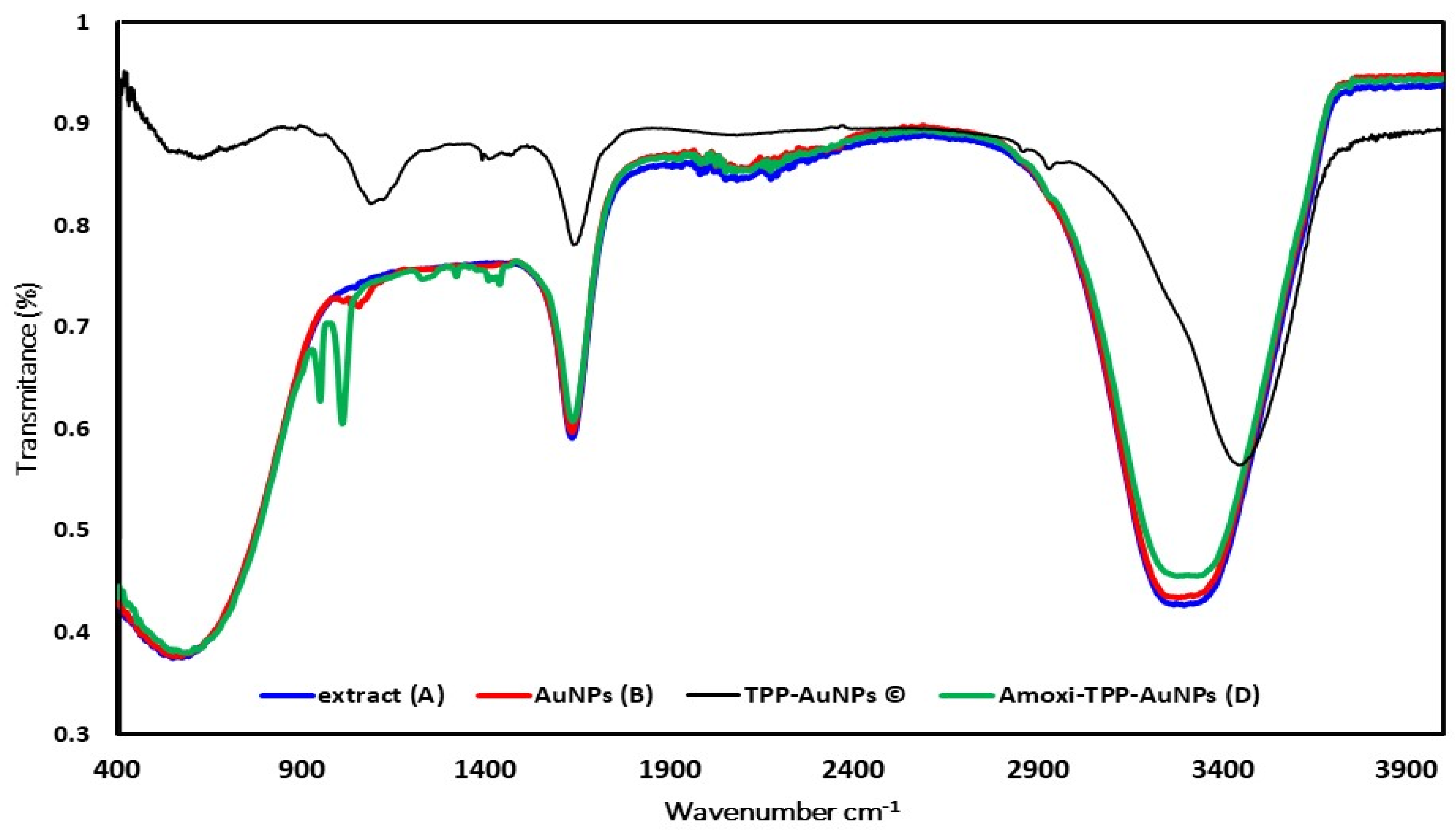
3.3.3. FTIR Analysis
3.4. Determination of Amoxi-TPP-AuNP Conjugating Efficiency
3.5. In Vitro Drug Release
3.6. Isolation and Identification of E. coli and S. aureus from Different Sources
3.7. Antimicrobial Susceptibility by Disk Diffusion Method
3.8. Molecular Characterization of E. coli and S. aureus Isolates from Different Sources 16S rRNA Analysis
3.9. Inhibition Zone, Minimum Inhibitory Concentration (MIC), and Maximum Bactericidal Concentration (MBC) of AuNPs and Amoxi-TPP-AuNPs against MDR E. coli and S. aureus Strains
3.10. Mode of Action
3.11. Killing Rate of Amoxi-TPP-AuNPs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garg, R. Tackling Antimicrobial Resistance: Optimizing Use of an Older Antibiotic-Amoxicillin. Indian J. Clin. Pract. 2014, 24, 843–845. [Google Scholar]
- Lozano, C.; Torres, C. Update on antibiotic resistance in Gram positives. Enfermedades Infecc. Y Microbiol. Clínica 2017, 35, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xiao, S.; Tang, W.; Shi, G. New insight on antimicrobial therapy adjustment strategies for Gram-negative bacterial infection. A cohort study. Medicine 2017, 96, e6439. [Google Scholar]
- Sun, Y.; Ye, J.; Hou, Y.; Chen, H.; Cao, J.; Zhou, T. Predation efficacy of Bdellovibrio bacteriovorus on multidrug-resistant clinical pathogens and their corresponding biofilms. Jpn. J. Infect. Dis. 2017, 70, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Cohn, A.C.; MacNeil, J.R.; Clark, T.A.; Ortega-Sanchez, I.R.; Briere, E.Z.; Meissner, H.C.; Baker, C.J.; Messonnier, N.E. Prevention and control of meningococcal disease: Recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. Recomm. Rep. 2013, 62, 1–28. [Google Scholar]
- Ghai, I.; Bajaj, H.; Bafna, J.A.; Hussein, H.A.E.D.; Winterhalter, M.; Wagner, R. Ampicillin permeation across OmpF, the major outer-membrane channel in Escherichia coli. J. Biol. Chem. 2018, 293, 7030–7037. [Google Scholar] [CrossRef] [PubMed]
- Tobar, K. Antimicrobial agents used in treatment of infectious disease. In Tobar’s Online Text Book of Bacteriology; Wisconsin University: Madison, WI, USA, 2011; pp. 1–10. Available online: https://textbookofbacteriology.net/antimicrobial.html (accessed on 17 July 2020).
- Ghai, I.; Pira, A.; Scorciapino, M.A.; Bodrenko, I.; Benier, L.; Ceccarelli, M.; Winterhalter, M.; Wagner, R. General method to determine the flux of charged molecules through nanopores applied to β-lactamase inhibitors and OmpF. J. Phys. Chem. Lett. 2017, 8, 1295–1301. [Google Scholar] [CrossRef]
- Lam, P.L.; Wong, W.Y.; Bian, Z.; Chui, C.H.; Gambari, R. Recent advances in green nanoparticulate systems for drug delivery: Efficient delivery and safety concern. Nanomedicine 2017, 12, 357–385. [Google Scholar] [CrossRef]
- Quan, X.Q.; Kang, L.; Yin, X.Z.; Jin, Z.H.; Gao, Z.G. Synthesis of PEGylated hyaluronic acid for loading dichloro (1,2-diaminocyclohexane) platinum (II)(DACHPt) in nanoparticles for cancer treatment. Chin. Chem. Lett. 2015, 26, 695–699. [Google Scholar] [CrossRef]
- Gupta, P.; Garcia, E.; Sarkar, A.; Kapoor, S.; Rafiq, K.; Chand, H.S.; Jayant, R.D. Nanoparticle based treatment for cardiovascular diseases. Cardiovasc. Haematol. Disord.-Drug Targets 2019, 19, 33–44. [Google Scholar] [CrossRef]
- Islan, G.A.; Mukherjee, A.; Castro, G.R. Development of biopolymer nanocomposite for silver nanoparticles and ciprof-691 loxacin controlled release. Int. J. Biol. Macromol. 2015, 72, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Srivastava, O.N. One-step green synthesis of gold nanoparticles using black cardamom and effect of pH on its synthesis. Nanoscale Res. Lett. 2015, 10, 353. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-strategies to fight multidrug resistant bacteria— “A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef] [PubMed]
- Muzammil, S.; Hayat, S.; Fakhar-E-Alam, M.; Aslam, B.; Siddique, M.H.; Nisar, M.A.; Saqalein, M.; Atif, M.; Sarwar, A.; Khurshid, A.; et al. Nanoantibiotics: Future nanotechnologies to combat antibiotic resistance. Front. Biosci. (Elite Ed.) 2018, 10, 352–374. [Google Scholar]
- Iravani, S.; Korbekandi, H.; Mirmohammadi, S.V.; Zolfaghari, B. Synthesis of silver nanoparticles: Chemical, physical and biological methods. Res. Pharm. Sci. 2014, 9, 385. [Google Scholar]
- Dewanjee, S.; Chakraborty, P.; Mukherjee, B.; De Feo, V. Plant-based antidiabetic nanoformulations: The emerging paradigm for effective therapy. Int. J. Mol. Sci. 2020, 21, 2217. [Google Scholar] [CrossRef]
- Gad El-Rab, S.M.F.; Halawani, E.M.; Alzahrani, S.S.S. Biosynthesis of silver nano-drug using Juniperus excelsa and its synergistic antibacterial activity against multidrug-resistant bacteria for wound dressing applications. 3 Biotech 2021, 11, 255. [Google Scholar] [CrossRef]
- Kalita, S.; Kandimalla, R.; Bhowal, A.C.; Kotoky, J.; Kundu, S. Functionalization of β-lactam antibiotic on lysozyme capped gold nanoclusters retrogress MRSA and its persisters following awakening. Sci. Rep. 2018, 8, 5778. [Google Scholar] [CrossRef]
- Enan, E.T.; Ashour, A.A.; Basha, S.; Felemban, N.H.; El-Rab, S.M.G. Antimicrobial activity of biosynthesized silver nanoparticles, amoxicillin, and glass-ionomer cement against Streptococcus mutans and Staphylococcus aureus. Nanotechnology 2021, 32, 215101. [Google Scholar] [CrossRef]
- Gad El-Rab, S.M.F.; Halawani, E.M.; Hassan, A.M. Formulation of ceftriaxone conjugated gold nanoparticles and their medical applications against extended-spectrum β-lactamase producing bacteria and breast cancer. J. Microbiol. Biotechnol. 2018, 28, 1563–1572. [Google Scholar] [CrossRef]
- Ashour, A.A.; Felemban, N.H.; Enan, E.T.; Basha, S.; Gad El-Rab, S.M. The Antimicrobial and Synergistic Strategies of Erythromycin Combined Synthesized Chitosan-Silver and Chitosan-Zinc Oxide Nanodrug on Oral Bacteria. J. Biobased Mater. Bioenergy 2022, 16, 408–417. [Google Scholar] [CrossRef]
- Gad El-Rab, S.M.; Ashour, A.A.; Basha, S.; Alyamani, A.A.; Felemban, N.H.; Enan, E.T. Well-Orientation Strategy Biosynthesis of Cefuroxime-Silver Nanoantibiotic for Reinforced Biodentine™ and Its Dental Application against Streptococcus mutans. Molecules 2021, 26, 6832. [Google Scholar] [CrossRef] [PubMed]
- Gad El-Rab, S.M.; Halawani, E.M.; Abo-Amer, A.E.; Mohamed, N.H.; Asiri, A.M. Biosynthesis of Silver Nano-Drug by Bacillus thuringiensis and Its Potential Application Against Extended-Spectrum β-Lactamase Producing E. coli. J. Biobased Mater. Bioenergy 2022, 16, 572–580. [Google Scholar] [CrossRef]
- Shaker, M.A.; Shaaban, M.I. Formulation of carbapenems loaded gold nanoparticles to combat multi-antibiotic bacterial resistance: In vitro antibacterial study. Int. J. Pharm. 2017, 525, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Güliz, A.K. Green Synthesis and Characterization of Magnetic Antibiotic Delivery System Using Linden Extract. Erzincan Univ. J. Sci. Technol. 2020, 13, 746–755. [Google Scholar]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. (Eds.) Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams & Wilkins: Baltimore, MD, USA, 1994; pp. 527–555. [Google Scholar]
- MaccFadin, J.K. Biochemical Test for Identification of Medical Bacteria, 3rd ed.; Lippincott Williams and Winkins; Awolter Klumer Company: Philadelphia, PA, USA; Baltimore, MD, USA; New York, NY, USA, 2000. [Google Scholar]
- Loberto, J.C.S.; Martins, C.A.; Santos, S.S.; Cortelli, J.R.; Jorge, A.O.C. Staphylococcus spp. in the oral cavity and periodontal pockets of chronic periodontitis patients. Braz. J. Microbiol. 2004, 35, 64–68. [Google Scholar] [CrossRef]
- Chaudhary, S.B.; Vives, M.J.; Basra, S.K.; Reiter, M.F. Postoperative spinal wound infections and postprocedural diskitis. J. Spinal Cord Med. 2007, 30, 441–451. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 9th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Brown, A.N.; Smith, K.; Samuels, T.A.; Lu, J.; Obare, S.O.; Scott, M.E. Nanoparticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureus. Appl. Environ. Microbiol. 2012, 78, 2768–2774. [Google Scholar] [CrossRef]
- Ashour, A.A.; Felemban, M.F.; Felemban, N.H.; Enan, E.T.; Basha, S.; Hassan, M.M.; Gad El-Rab, S.M. Comparison and Advanced Antimicrobial Strategies of Silver and Copper Nanodrug-Loaded Glass Ionomer Cement against Dental Caries Microbes. Antibiotics 2022, 11, 756. [Google Scholar] [CrossRef]
- Yu, J.; Xu, D.; Guan, H.N.; Wang, C.; Huang, L.K. Facile one-step green synthesis of gold nanoparticles using Citrus maxima aqueous extracts and its catalytic activity. Mater. Lett. 2016, 166, 110–112. [Google Scholar] [CrossRef]
- Markus, J.; Wang, D.; Kim, Y.J.; Ahn, S.; Mathiyalagan, R.; Wang, C.; Yang, D.C. Biosynthesis, characterization, and bioactivities evaluation of silver and gold nanoparticles mediated by the roots of Chinese herbal Angelica pubescens Maxim. Nanoscale Res. Lett. 2017, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Kalita, S.; Kandimalla, R.; Sharma, K.K.; Kataki, A.C.; Deka, M.; Kotoky, J. Amoxicillin functionalized gold nanoparticles reverts MRSA resistance. Mater. Sci. Eng. C 2016, 61, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Demurtas, M.; Carole, C.P. Facile one-pot synthesis of amoxicillin-coated gold nanoparticles and their antimicrobial activity. Gold Bull. 2014, 47, 103–107. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Murphy, C.J. Toxicity and cellular uptake of gold nanoparticles: What we have learned so far? J. Nanoparticle Res. 2010, 12, 2313–2333. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S. Gold nanoparticles: Their application as antimicrobial agents and vehicles of gene delivery. Adv. Biotechnol. Microbiol. 2017, 4, 555658. [Google Scholar] [CrossRef]
- Goy, R.C.; Britto, D.; Assis, O.B.G. A review of the antimicrobial activity of chitosan. Polímeros Ciência Tecnol. 2009, 19, 241–247. [Google Scholar] [CrossRef]
- Krishna, G.; Kumar, S.S.; Pranitha, V.; Alha, M.; Charaya, S. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: A study against Salmonella sp. Int. J. Pharm. Pharm. Sci. 2015, 7, 84–88. [Google Scholar]




| Characteristics | ESC Isolates | STA Isolates |
|---|---|---|
| Gram’s stain | − | + |
| Cocci | − | + |
| Rods | + | − |
| Urease | − | + |
| Nitrate Reduction | + | + |
| Motility | + | − |
| Catalase test | + | + |
| Coagulase test | − | + |
| Oxidase test | − | − |
| Indole test | + | − |
| Methyl red | + | + |
| Voges-proskaure | − | + |
| Citrate | − | + |
| H2S Production | − | − |
| Fermentation | ||
| Glucose | + | + |
| Lactose | + | + |
| Mannitol | + | + |
| Maltose | − | + |
| Probable bacteria | E. coli | S. aureus |
| Bacterial Isolates | Antibiotic Resistance Pattern | No. of Antibiotics |
|---|---|---|
| ESC1 | AMP, AMC, CLR, CIP, LVX, SXT, LEX, CLI, CFR, CRO, | 10 |
| ESC2 | AMP, AMC, CLR, CIP, LVX, SXT, LEX, CLI, CFR, AZM | 10 |
| ESC3 | AMP, AMC, CEC, FOX, FEP, CXM, CAZ, CRO, CTX, CIP, LVX, DO, SXT, LEX, CLR, AZM, CLI, FUX, CFR | 19 |
| ESC4 | AMP, AMC, CLR, DO, LEX, SXT, CLI, CFR, GEM | 9 |
| ESC5 | AMP, AMC, CEC, FOX, FEP, CXM, CAZ, CRO, CTX, CIP, LVX, DO, SXT, LEX, CLR, AZM, CLI, CFR, AK | 19 |
| ESC6 | AMP, AMC, CLR, DO, LEX, AZM, CRO, FUX | 8 |
| ESC7 | AMP, AMC, CEC, FEP, CLR, CXM, CAZ, CRO, CTX, CIP, LVX, DO, SXT, LEX, FUX | 15 |
| ESC8 | AMP, AMC, CLR, CIP, CAZ, SXT, LEX, CLI, CFR, AZM | 10 |
| # STA6 | CEC, PEN, ERY, FOX | 4 |
| ESC9 | AMP, AMC, CLR, CIP, LVX, SXT, CEC, FEP, CXM, CRO, CTX | 11 |
| ESC10 | AMP, AMC, CEC, FEP, FOX, CXM, CAZ, CRO, CTX, DO, SXT | 11 |
| ESC11 | AMP, AMC, CEC, FEP, FOX, CXM, CAZ, CRO, CTX, DO, GEM | 11 |
| # STA1 | AMP, AMC, ERY, PEN, DO | 5 |
| # STA2 | AMP, PEN, ERY, CLR, AZM | 5 |
| * STA3 | AMP, AMC, CEC, FOX, CXM, CIP, LVX, GEN, MEM, OXA, PEN, LEX, FEP, CRO, CFR | 15 |
| * STA4 | AMP, AMC, CEC, FOX, CXM, CIP, LVX, GEN, OXA, PEN, LEX, FEP, CRO, CFR | 14 |
| * STA5 | AMP, AMC, CEC, FOX, CXM, MEM, OXA, PEN, LEX, CRO, CFR | 11 |
| * STA7 | AMP, AMC, CEC, FOX, CXM, MEM, OXA, PEN, ERY | 9 |
| * STA8 | AK, AMC AZM, CEC, FOX, CXM, CIP, LVX, OXA, PEN, CLR, CLI, ERY | 13 |
| * STA9 | AMC, CEC, PEN, ERY, GEM, OXA | 6 |
| Isolate | Amoxicillin (µg/mL) | Amoxacillin-TPP-AuNP (µg/mL) | |||
|---|---|---|---|---|---|
| 1 ZIN (mm) | 2 MIC | 1 ZIN (mm) | 2 MIC | 3 MBC | |
| ESC1 | 4 ND | 108 | 32 | 4 | 16 |
| ESC2 | ND | 114 | 34 | 8 | 16 |
| ESC3 | ND | 108 | 37 | 8 | 32 |
| ESC4 | ND | 108 | 32 | 4 | 16 |
| ESC5 | ND | 114 | 34 | 8 | 32 |
| ESC6 | ND | 108 | 24 | 8 | 12 |
| ESC7 | ND | 102 | 32 | 8 | 16 |
| ESC8 | ND | 102 | 34 | 4 | 16 |
| ESC9 | ND | 108 | 34 | 4 | 16 |
| ESC10 | ND | 102 | 34 | 8 | 16 |
| ESC11 | ND | 96 | 37 | 4 | 12 |
| * STA3 | ND | 114 | 20 | 8 | 32 |
| * STA4 | ND | 108 | 22 | 8 | 32 |
| * STA5 | ND | 114 | 24 | 4 | 16 |
| * STA7 | ND | 114 | 26 | 3.6 | 16 |
| * STA8 | ND | 108 | 23 | 8 | 32 |
| * STA9 | ND | 108 | 26 | 3.6 | 12 |
| MIC (µg/mL) | |||
|---|---|---|---|
| Amoxicillin | Amoxi-TPPi-AuNPs (15.99–24.71 nm) | Amoxi-TPPi-AuNPs (50.34–100.45 nm) | |
| ESC1 | ND | 4 | 8 |
| ESC2 | ND | 8 | 16 |
| ESC3 | ND | 8 | 16 |
| ESC4 | ND | 4 | 8 |
| ESC5 | ND | 8 | 16 |
| ESC6 | ND | 8 | 16 |
| ESC7 | ND | 8 | 16 |
| ESC8 | ND | 4 | 8 |
| ESC9 | ND | 4 | 8 |
| ESC10 | ND | 8 | 16 |
| ESC11 | ND | 4 | 8 |
| * STA3 | ND | 8 | 16 |
| * STA4 | ND | 8 | 16 |
| * STA5 | ND | 4 | 8 |
| * STA7 | ND | 3.6 | 8 |
| * STA8 | ND | 8 | 16 |
| * STA9 | ND | 3.6 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halawani, E.M.S.; Alzahrani, S.S.S.; Gad El-Rab, S.M.F. Biosynthesis Strategy of Gold Nanoparticles and Biofabrication of a Novel Amoxicillin Gold Nanodrug to Overcome the Resistance of Multidrug-Resistant Bacterial Pathogens MRSA and E. coli. Biomimetics 2023, 8, 452. https://doi.org/10.3390/biomimetics8060452
Halawani EMS, Alzahrani SSS, Gad El-Rab SMF. Biosynthesis Strategy of Gold Nanoparticles and Biofabrication of a Novel Amoxicillin Gold Nanodrug to Overcome the Resistance of Multidrug-Resistant Bacterial Pathogens MRSA and E. coli. Biomimetics. 2023; 8(6):452. https://doi.org/10.3390/biomimetics8060452
Chicago/Turabian StyleHalawani, Eman M. S., Seham S. S. Alzahrani, and Sanaa M. F. Gad El-Rab. 2023. "Biosynthesis Strategy of Gold Nanoparticles and Biofabrication of a Novel Amoxicillin Gold Nanodrug to Overcome the Resistance of Multidrug-Resistant Bacterial Pathogens MRSA and E. coli" Biomimetics 8, no. 6: 452. https://doi.org/10.3390/biomimetics8060452
APA StyleHalawani, E. M. S., Alzahrani, S. S. S., & Gad El-Rab, S. M. F. (2023). Biosynthesis Strategy of Gold Nanoparticles and Biofabrication of a Novel Amoxicillin Gold Nanodrug to Overcome the Resistance of Multidrug-Resistant Bacterial Pathogens MRSA and E. coli. Biomimetics, 8(6), 452. https://doi.org/10.3390/biomimetics8060452







