A Microphysiological Model to Mimic the Placental Remodeling during Early Stage of Pregnancy under Hypoxia-Induced Trophoblast Invasion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
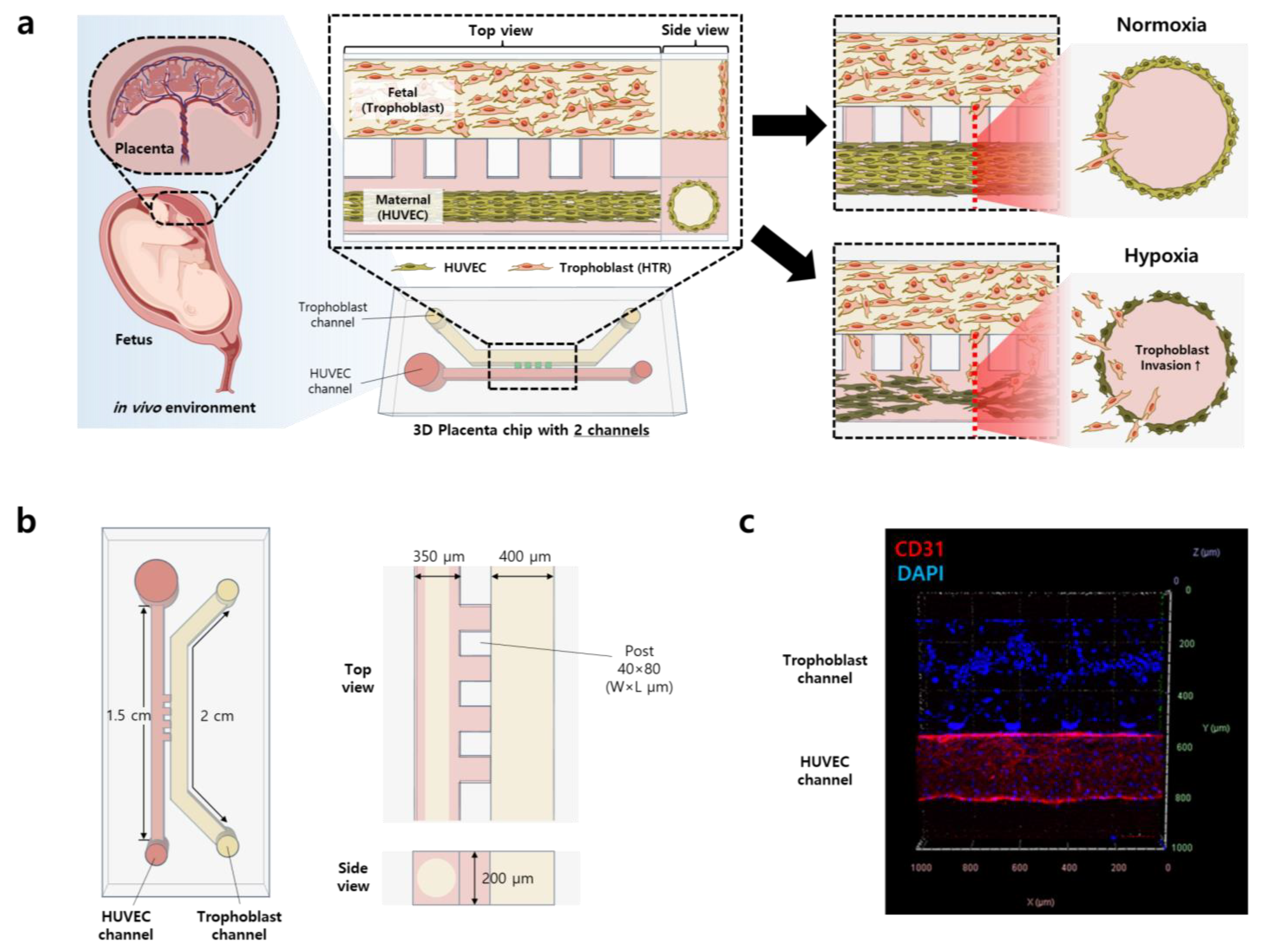
2.2. Placenta-on-a-Chip Fabrication
2.3. Vessel Formation and Cell Seeding
2.4. Hypoxia Generation on a Chip
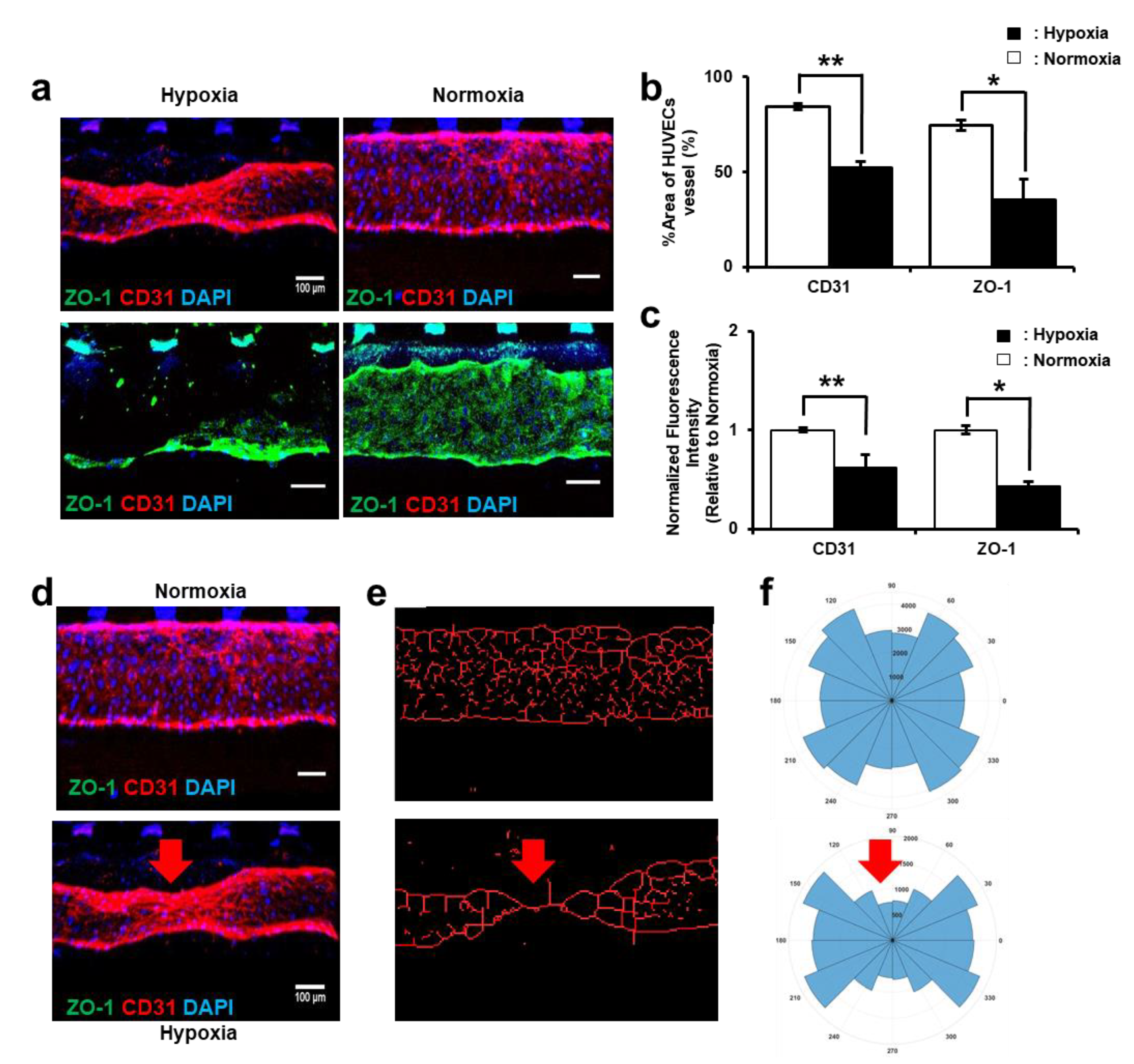
2.5. Immunofluorescence Assay
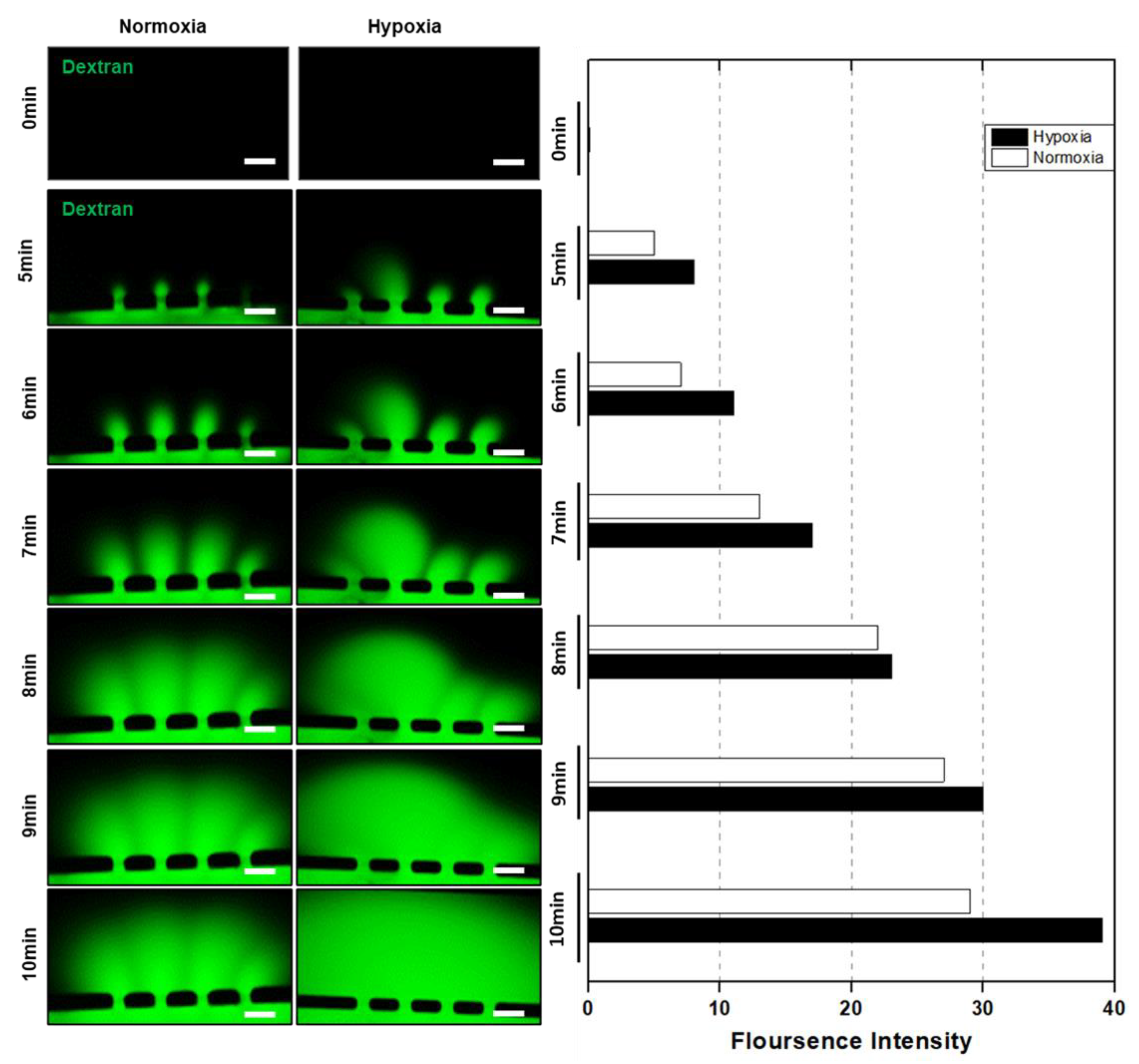
2.6. Dextran Assay for HUVEC Vessel Remodeling
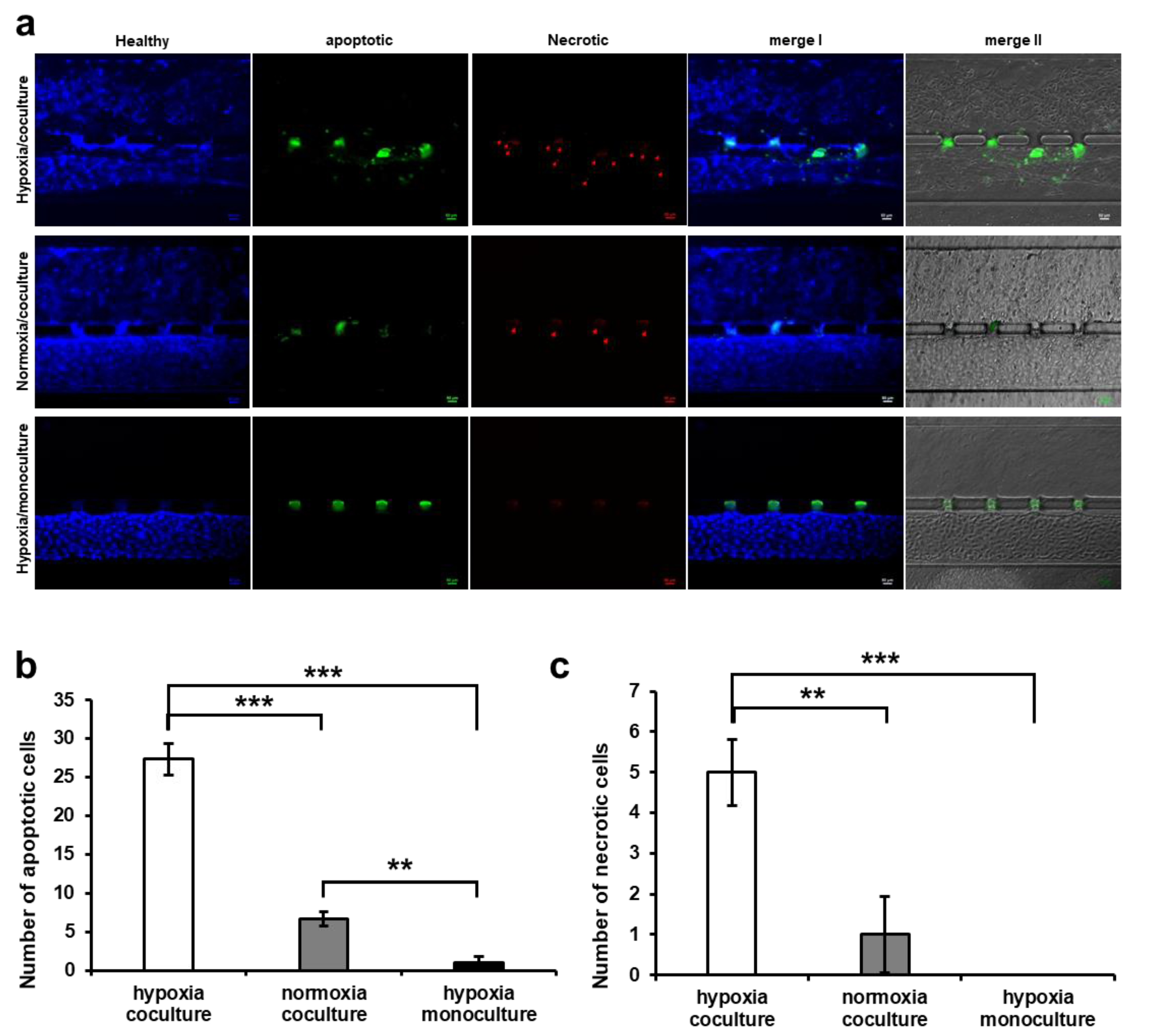
2.7. Apoptosis Assay
2.8. Statistical Analysis
3. Results and Discussion
3.1. Construction of 3D Placenta-on-a-Chip
3.2. Hypoxic Environment Promotes the Invasion Ability of Trophoblasts
3.3. HUVEC Vessel Remodeling by Trophoblast Invasion
3.4. HUVEC Cell Apoptosis Induction by Trophoblast Invasion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burton, G.J.; Fowden, A.L. Review: The Placenta and Developmental Programming: Balancing Fetal Nutrient Demands with Maternal Resource Allocation. Placenta 2012, 33, S23–S27. [Google Scholar] [CrossRef] [PubMed]
- Lager, S.; Powell, T.L. Regulation of Nutrient Transport across the Placenta. J. Pregnancy 2012, 2012, 179827. [Google Scholar] [CrossRef] [PubMed]
- Gude, N.M.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and Function of the Normal Human Placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, I.; Means, R.E.; Aldo, P.; Koga, K.; Lang, S.M.; Booth, C.; Manzur, A.; Oyarzun, E.; Romero, R.; Mor, G. Viral Infection of the Placenta Leads to Fetal Inflammation and Sensitization to Bacterial Products Predisposing to Preterm Labor. J. Immunol. 2010, 185, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Blundell, C.; Tess, E.R.; Schanzer, A.S.R.; Coutifaris, C.; Su, E.J.; Parry, S.; Huh, D. A Microphysiological Model of the Human Placental Barrier. Lab. Chip 2016, 16, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Tetro, N.; Moushaev, S.; Rubinchik-Stern, M.; Eyal, S. The Placental Barrier: The Gate and the Fate in Drug Distribution. Pharm. Res. 2018, 35, 71. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J.; Cindrova-Davies, T.; Yung, H.W.; Jauniaux, E. Hypoxia and reproductive health: Oxygen and Development of the Human Placenta. Reproduction 2021, 161, F53–F65. [Google Scholar] [CrossRef]
- Woods, L.; Perez-Garcia, V.; Hemberger, M. Regulation of Placental Development and Its Impact on Fetal Growth—New Insights From Mouse Models. Front. Endocrinol. 2018, 9, 416241. [Google Scholar] [CrossRef]
- Huang, X.; Anderle, P.; Hostettler, L.; Baumann, M.U.; Surbek, D.V.; Ontsouka, E.C.; Albrecht, C. Identification of Placental Nutrient Transporters Associated with Intrauterine Growth Restriction and Pre-Eclampsia. BMC Genom. 2018, 19, 173. [Google Scholar] [CrossRef]
- Myren, M.; Mose, T.; Mathiesen, L.; Knudsen, L.E. The Human Placenta–An Alternative for Studying Foetal Exposure. Toxicol. Vitr. 2007, 21, 1332–1340. [Google Scholar] [CrossRef]
- Pemathilaka, R.L.; Reynolds, D.E.; Hashemi, N.N. Drug Transport across the Human Placenta: Review of Placenta-on-a-Chip and Previous Approaches. Interface Focus 2019, 9, 20190031. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.P.; Pence, L.; Pinkas, G.; Song, H.; Telugu, B.P. Placental Hypoxia during Early Pregnancy Causes Maternal Hypertension and Placental Insufficiency in the Hypoxic Guinea Pig Model. Biol. Reprod. 2016, 95, 128. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.S.; Anthony, R.V. The Pregnant Sheep as a Model for Human Pregnancy. Theriogenology 2008, 69, 55–67. [Google Scholar] [CrossRef]
- Carter, A.M. Comparative Studies of Placentation and Immunology in Non-Human Primates Suggest a Scenario for the Evolution of Deep Trophoblast Invasion and an Explanation for Human Pregnancy Disorders. Reproduction 2011, 141, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, S.; Kidane, D.; Guller, S.; Luo, T.; Norwitz, N.G.; Arcuri, F.; Toti, P.; Norwitz, E.R. In Vivo and In Vitro Evidence for Placental DNA Damage in Preeclampsia. PLoS ONE 2014, 9, e86791. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.Y.; Susarla, R.; Canovas, D.; Vasilopoulou, E.; Ohizua, O.; McCabe, C.J.; Hewison, M.; Kilby, M.D. Vitamin D Promotes Human Extravillous Trophoblast Invasion In Vitro. Placenta 2015, 36, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Fuwad, A.; Ko, G.; Kim, G.J.; Jeon, T.J.; Kim, S.M. A PDMS-Based Interdigitated Platform for Trophoblast Invasion Study Under Oxygen Stress Conditions. Biochip J. 2021, 15, 362–370. [Google Scholar] [CrossRef]
- Dos Santos, E.; Moindjie, H.; Sérazin, V.; Arnould, L.; Rodriguez, Y.; Fathallah, K.; Barnea, E.R.; Vialard, F.; Dieudonné, M.N. Preimplantation Factor Modulates Trophoblastic Invasion throughout the Decidualization of Human Endometrial Stromal Cells. Reprod. Biol. Endocrinol. 2021, 19, 96. [Google Scholar] [CrossRef] [PubMed]
- Boos, J.A.; Misun, P.M.; Brunoldi, G.; Furer, L.A.; Aengenheister, L.; Modena, M.; Rousset, N.; Buerki-Thurnherr, T.; Hierlemann, A. Microfluidic Co-Culture Platform to Recapitulate the Maternal–Placental–Embryonic Axis. Adv. Biol. 2021, 5, 2100609. [Google Scholar] [CrossRef]
- Mosavati, B.; Oleinikov, A.V.; Du, E. Development of an Organ-on-a-Chip-Device for Study of Placental Pathologies. Int. J. Mol. Sci. 2020, 21, 8755. [Google Scholar] [CrossRef]
- Miura, S.; Sato, K.; Kato-Negishi, M.; Teshima, T.; Takeuchi, S. Fluid Shear Triggers Microvilli Formation via Mechanosensitive Activation of TRPV6. Nat. Commun. 2015, 6, 8871. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yin, F.; Wang, H.; Wang, L.; Yuan, J.; Qin, J. Placental Barrier-on-a-Chip: Modeling Placental Inflammatory Responses to Bacterial Infection. ACS Biomater. Sci. Eng. 2018, 4, 3356–3363. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Zhu, Y.; Zhang, M.; Yu, H.; Chen, W.; Qin, J. A 3D Human Placenta-on-a-Chip Model to Probe Nanoparticle Exposure at the Placental Barrier. Toxicol. Vitr. 2019, 54, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Pemathilaka, R.L.; Caplin, J.D.; Aykar, S.S.; Montazami, R.; Hashemi, N.N. Placenta-on-a-Chip: In Vitro Study of Caffeine Transport across Placental Barrier Using Liquid Chromatography Mass Spectrometry. Glob. Chall. 2019, 3, 1800112. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Mani, S.; Clair, G.; Olson, H.M.; Paurus, V.L.; Ansong, C.K.; Blundell, C.; Young, R.; Kanter, J.; Gordon, S.; et al. A Microphysiological Model of Human Trophoblast Invasion during Implantation. Nat. Commun. 2022, 13, 1252. [Google Scholar] [CrossRef] [PubMed]
- Blundell, C.; Yi, Y.S.; Ma, L.; Tess, E.R.; Farrell, M.J.; Georgescu, A.; Aleksunes, L.M.; Huh, D. Placental Drug Transport-on-a-Chip: A Microengineered In Vitro Model of Transporter-Mediated Drug Efflux in the Human Placental Barrier. Adv. Healthc. Mater. 2018, 7, 1700786. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.; Oefner, C.M.; Polacheck, W.J.; Gardner, L.; Farrell, L.; Sharkey, A.; Kamm, R.; Moffett, A.; Oyen, M.L. A Microfluidics Assay to Study Invasion of Human Placental Trophoblast Cells. J. R. Soc. Interface 2017, 14, 20170131. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Romero, R.; Han, Y.M.; Kim, H.C.; Kim, C.J.; Hong, J.S.; Huh, D. Placenta-on-a-Chip: A Novel Platform to Study the Biology of the Human Placenta. J. Matern.-Fetal Neonatal Med. 2016, 29, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Gingrich, J.; Veiga-Lopez, A. A 3-Dimensional Microfluidic Platform for Modeling Human Extravillous Trophoblast Invasion and Toxicological Screening. Lab. Chip 2021, 21, 546–557. [Google Scholar] [CrossRef]
- Fuwad, A.; Hossain, S.; Ryu, H.; Ansari, M.A.; Khan, M.S.I.; Kim, K.Y.; Jeon, T.J.; Kim, S.M. Numerical and Experimental Study on Mixing in Chaotic Micromixers with Crossing Structures. Chem. Eng. Technol. 2020, 43, 1866–1875. [Google Scholar] [CrossRef]
- Bischel, L.L.; Lee, S.H.; Beebe, D.J. A Practical Method for Patterning Lumens through ECM Hydrogels via Viscous Finger Patterning. SLAS Technol. 2012, 17, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Bischel, L.L.; Beebe, D.J.; Sung, K.E. Microfluidic Model of Ductal Carcinoma in Situ with 3D, Organotypic Structure. BMC Cancer 2015, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Tornavaca, O.; Chia, M.; Dufton, N.; Almagro, L.O.; Conway, D.E.; Randi, A.M.; Schwartz, M.A.; Matter, K.; Balda, M.S. ZO-1 Controls Endothelial Adherens Junctions, Cell-Cell Tension, Angiogenesis, and Barrier Formation. J. Cell Biol. 2015, 208, 821–838. [Google Scholar] [CrossRef] [PubMed]
- Privratsky, J.R.; Newman, P.J. PECAM-1: Regulator of Endothelial Junctional Integrity. Cell Tissue Res. 2014, 355, 607–619. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 334617. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesisin a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D Tumor Spheroids: An Overview on the Tools and Techniques Used for Their Analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, J.E.; Fraser, R.; Leslie, K.; Wallace, A.E.; James, J.L. Remodelling at the Maternal–Fetal Interface: Relevance to Human Pregnancy Disorders. Reproduction 2010, 140, 803–813. [Google Scholar] [CrossRef]
- Soares, M.J.; Chakraborty, D.; Kubota, K.; Renaud, S.J.; Karim Rumi, M.A. Adaptive Mechanisms Controlling Uterine Spiral Artery Remodeling during the Establishment of Pregnancy. Int. J. Dev. Biol. 2014, 58, 247–259. [Google Scholar] [CrossRef]
- Xie, B.; Wu, H.; Li, J.; Lv, X.; Zhou, Y.; Yu, Q.; Cui, S.; Zeng, L.; Li, J.; Huang, X.; et al. Mechanical Forces on Trophoblast Motility and Its Potential Role in Spiral Artery Remodeling during Pregnancy. Placenta 2022, 123, 46–53. [Google Scholar] [CrossRef]
- Aengenheister, L.; Keevend, K.; Muoth, C.; Schönenberger, R.; Diener, L.; Wick, P.; Buerki-Thurnherr, T. An Advanced Human in Vitro Co-Culture Model for Translocation Studies across the Placental Barrier. Sci. Rep. 2018, 8, 5388. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Gough, A.; Schurdak, M.E.; Vernetti, L.; Chennubhotla, C.S.; Lefever, D.; Pei, F.; Faeder, J.R.; Lezon, T.R.; Stern, A.M.; et al. Harnessing Human Microphysiology Systems as Key Experimental Models for Quantitative Systems Pharmacology. Handb. Exp. Pharmacol. 2019, 260, 327–367. [Google Scholar] [PubMed]
- Soares, C.P.; Midlej, V.; de Oliveira, M.E.W.; Benchimol, M.; Costa, M.L.; Mermelstein, C. 2D and 3D-Organized Cardiac Cells Shows Differences in Cellular Morphology, Adhesion Junctions, Presence of Myofibrils and Protein Expression. PLoS ONE 2012, 7, e38147. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Paul Solomon, F.D. 3D Cell Culture Systems: Advantages and Applications. J. Cell Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 513823. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D Cell Cultures—A Comparison of Different Types of Cancer Cell Cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Sekine, H.; Shimizu, T.; Hobo, K.; Sekiya, S.; Yang, J.; Yamato, M.; Kurosawa, H.; Kobayashi, E.; Okano, T. Endothelial Cell Coculture Within Tissue-Engineered Cardiomyocyte Sheets Enhances Neovascularization and Improves Cardiac Function of Ischemic Hearts. Circulation 2008, 118, S145–S152. [Google Scholar] [CrossRef] [PubMed]
- Soucy, P.A.; Romer, L.H. Endothelial Cell Adhesion, Signaling, and Morphogenesis in Fibroblast-Derived Matrix. Matrix Biol. 2009, 28, 273–283. [Google Scholar] [CrossRef]
- Paul, M.; Chakraborty, S.; Islam, S.; Ain, R. Trans-Differentiation of Trophoblast Stem Cells: Implications in Placental Biology. Life Sci. Alliance 2023, 6, e202201583. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.; Fuwad, A.; Yoon, S.; Jeon, T.-J.; Kim, S.M. A Microphysiological Model to Mimic the Placental Remodeling during Early Stage of Pregnancy under Hypoxia-Induced Trophoblast Invasion. Biomimetics 2024, 9, 289. https://doi.org/10.3390/biomimetics9050289
Jeong S, Fuwad A, Yoon S, Jeon T-J, Kim SM. A Microphysiological Model to Mimic the Placental Remodeling during Early Stage of Pregnancy under Hypoxia-Induced Trophoblast Invasion. Biomimetics. 2024; 9(5):289. https://doi.org/10.3390/biomimetics9050289
Chicago/Turabian StyleJeong, Seorin, Ahmed Fuwad, Sunhee Yoon, Tae-Joon Jeon, and Sun Min Kim. 2024. "A Microphysiological Model to Mimic the Placental Remodeling during Early Stage of Pregnancy under Hypoxia-Induced Trophoblast Invasion" Biomimetics 9, no. 5: 289. https://doi.org/10.3390/biomimetics9050289
APA StyleJeong, S., Fuwad, A., Yoon, S., Jeon, T.-J., & Kim, S. M. (2024). A Microphysiological Model to Mimic the Placental Remodeling during Early Stage of Pregnancy under Hypoxia-Induced Trophoblast Invasion. Biomimetics, 9(5), 289. https://doi.org/10.3390/biomimetics9050289







