EEG Motor Imagery Classification: Tangent Space with Gate-Generated Weight Classifier
Abstract
1. Introduction
- –
- Introduction of the GG-FWC classifier for categorizing covariance matrices extracted from EEG signals;
- –
- Integration of both linear and nonlinear features to enhance classification accuracy;
- –
- Comprehensive comparison of M-TSP and Cholesky decomposition methodologies;
- –
- Mitigation of noise from muscle movements, eye blinks, and electronic interference by focusing on channel relationships through covariance matrices;
- –
- Gender-based performance analysis to investigate potential differences in MI-BCI accuracy between male and female subjects.
2. Related Works
3. Methodology
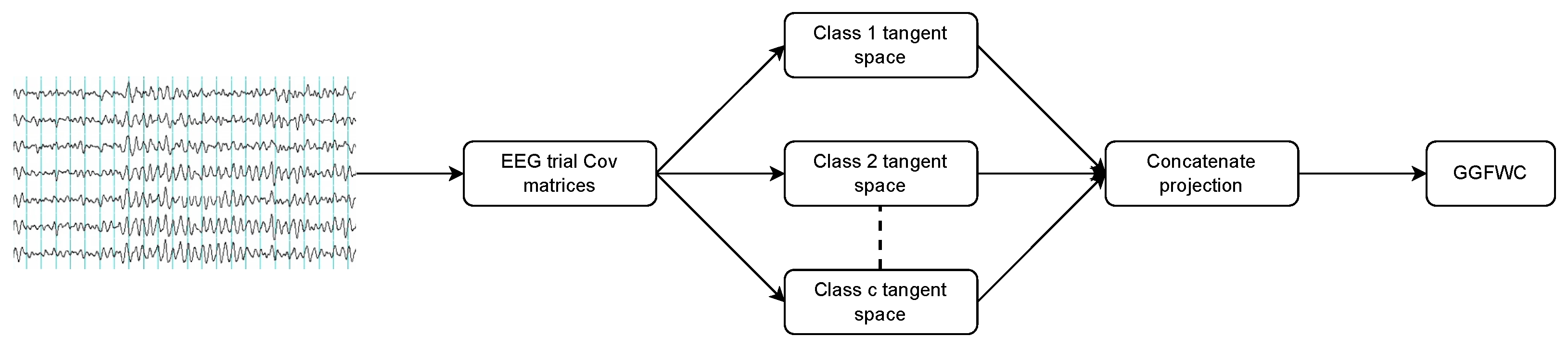
3.1. Multiple Tangent Space Projections
- –
- Computation of covariance matrices: The covariance matrices of EEG signal trials are computed. Each covariance matrix represents the statistical dependencies between different EEG channels.
- –
- Definition of tangent space: A reference point on the manifold, typically the Riemannian mean of the training set covariance matrices, is chosen. This point defines the tangent space.
- –
- Projection onto tangent space: The covariance matrices are projected onto the tangent space using the logarithm map. This projection transforms the covariance matrices from the Riemannian manifold to a Euclidean space.
- –
- Application of standard classifiers: In the Euclidean space of the tangent space, standard classifiers such as logistic regression or support vector machines are applied.
3.2. Cholesky Decomposition Projections
- –
- Computation of covariance matrices: Similar to the tangent space projection method, the covariance matrices of EEG signal trials are computed.
- –
- Cholesky decomposition: Each covariance matrix is decomposed into a lower triangular matrix such that .
- –
- Feature extraction: The elements of the lower triangular matrix are used as features for classification. These elements capture the essential characteristics of the covariance matrix in a reduced form.
- –
- Application of classifiers: The extracted features are used as inputs for classifiers such as the GG-FWC, which constructs the output through a functionally weighted linear combination of the input variables.
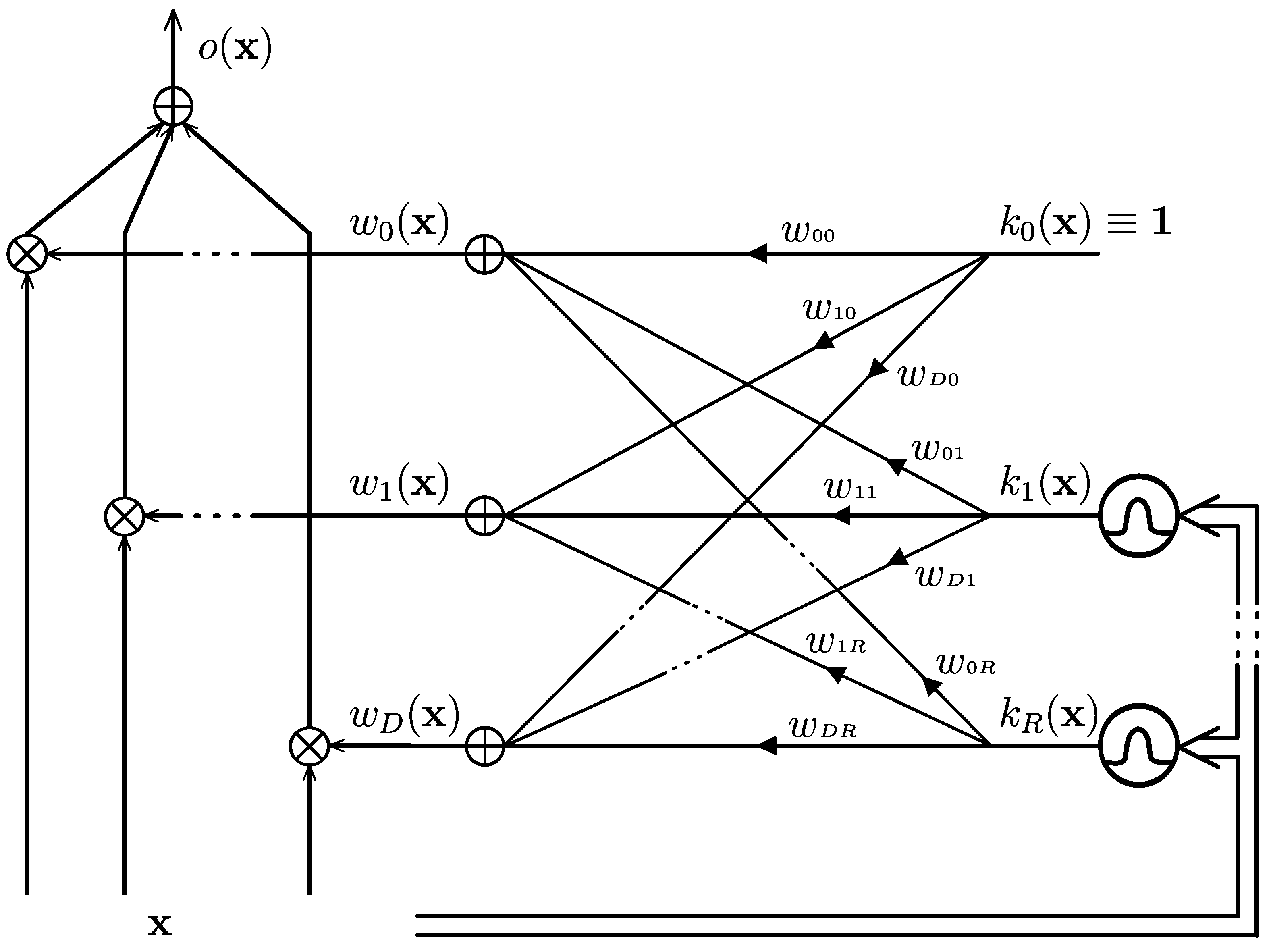
3.3. The Monolithic GG-FWC
3.4. Proposed Method
| Algorithm 1 Training the M-TSC model |
| Require: the set of training data for to M do the Riemannian mean of class m end for = empty-matrix() for to N do for to M do end for = concatenate end for model = GGFWC(model parameters) model = model.fit(, ) ▹ the vector of all Return (model) |
| Algorithm 2 Class Prediction with M-TSC |
| Require: model: trained model Require: : new sample covariance matrix Require: : the Riemannian mean of class m for to M do end for = concatenate y = model.predict Return (y) |
| Algorithm 3 Training the Cholesky model |
| Require: the set of training data = empty-matrix() for to N do = cholesky_decomposition() end for model = GGFWC(model parameters) model = model.fit(, ) ▹ the vector of all Return (model) |
| Algorithm 4 Class prediction with Cholesky model |
| Require: model: trained model Require: : new sample covariance matrix = cholesky_decomposition() y = model.predict Return (y) |
- R: the number of RBF units;
- C: the parameter for the MM algorithm;
- : the scale factor for the dispersion of RBF.
- Variation in the number of centroids, spanning from 2 to 30;
- , ranging from 1 to 400;
- Parameter C, encompassing a specified range of values: [0.0001, 0.001, 0.01, 0.1, 1, 10, 100, 1000].
4. Experiment
- LR M-TSC1: This iteration involves the multiple tangent space projection of covariance matrices, succeeded by the concatenation of the resultant data, followed by classification using LR.
- SVM M-TSC1: In this variation, the multiple tangent space projection is applied, and the ensuing data are concatenated and classified using SVMs.
- LR M-TSC2: In this version, the approach involves the multiple tangent space projection of covariance matrices. The resultant data from diverse projections undergo concatenation and subsequent normalization to achieve zero means and unity standard deviation before being employed as input for a logistic regression classifier.
- SVM M-TSC2: After applying the multiple tangent space projection of covariance matrices in this variant, the resulting data from various projections are concatenated and normalized to zero means and unity standard deviation prior to being utilized as input for a SVM.
4.1. Dataset
- DS1: This dataset [28] comprises raw labeled EEG data obtained from nine subjects and recorded through three channels (C3, C4, and Cz). Data acquisition was based on the mental reproduction of two different gestures: one corresponding to the left hand (class 1) and the other to the right hand (class 2). Each subject completed a total of five sessions, which consisted of 120 trials per session. The dataset is divided into three sessions for training and two sessions for testing, providing 360 trials for training and 240 trials for testing for each subject. In this dataset, the subject’s gender is not known.
- DS2: This dataset [29] includes raw labeled EEG data collected through a 22-electrode helmet from a cohort of nine subjects: four females and five men. Each participant actively attended two separate data capture sessions, during which they performed mental simulations that covered a variety of gestures related to left hand (class 1), right hand (class 2), both feet (class 3), and tongue (class 4) movement. For the purpose of performance comparison with the preceding work, only data corresponding to two classes, the right-hand and left-hand, were selectively retained. The dataset is divided into one session for training and one session for testing, with each session consisting of 120 trials per subject.
4.2. DS1 and DS2 Analyses
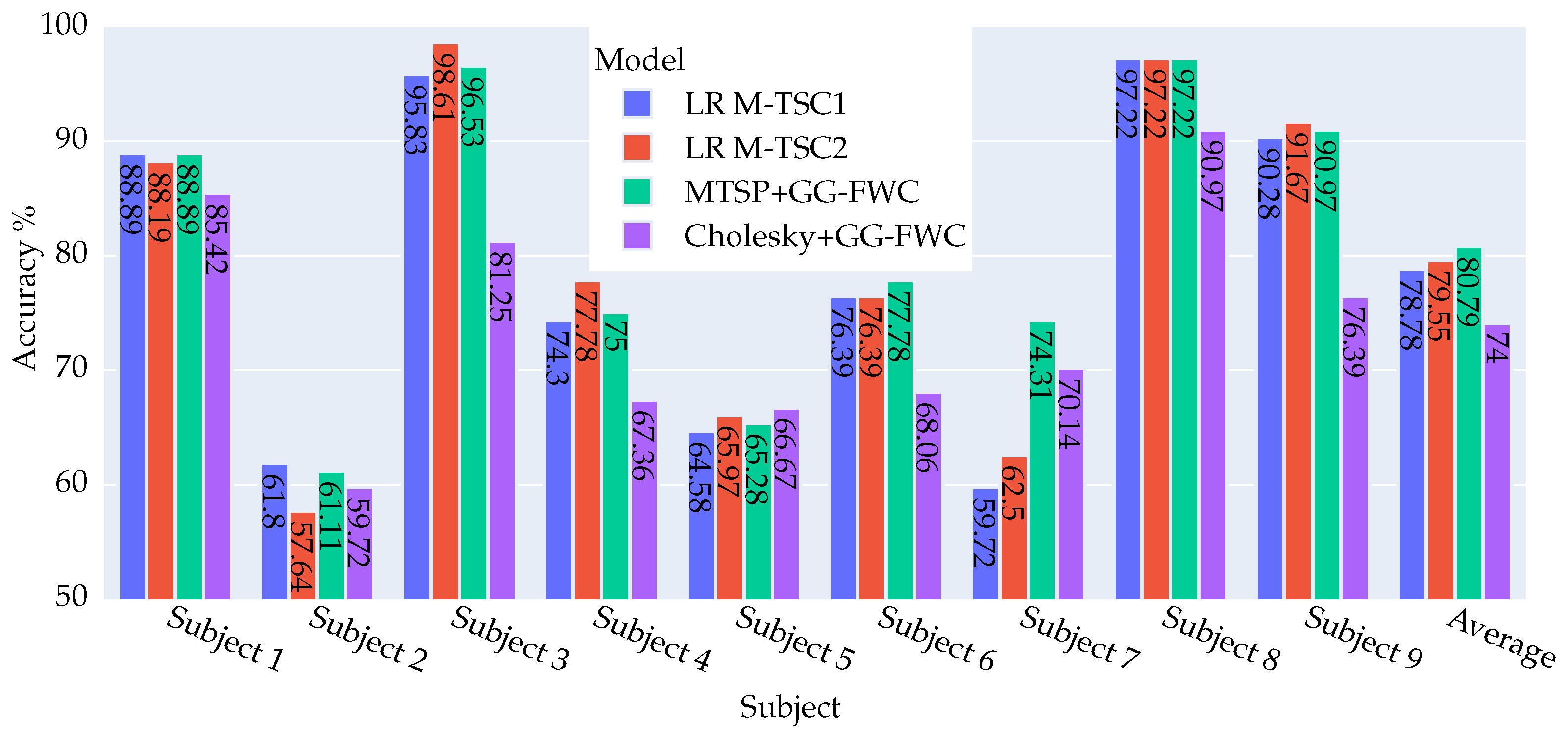
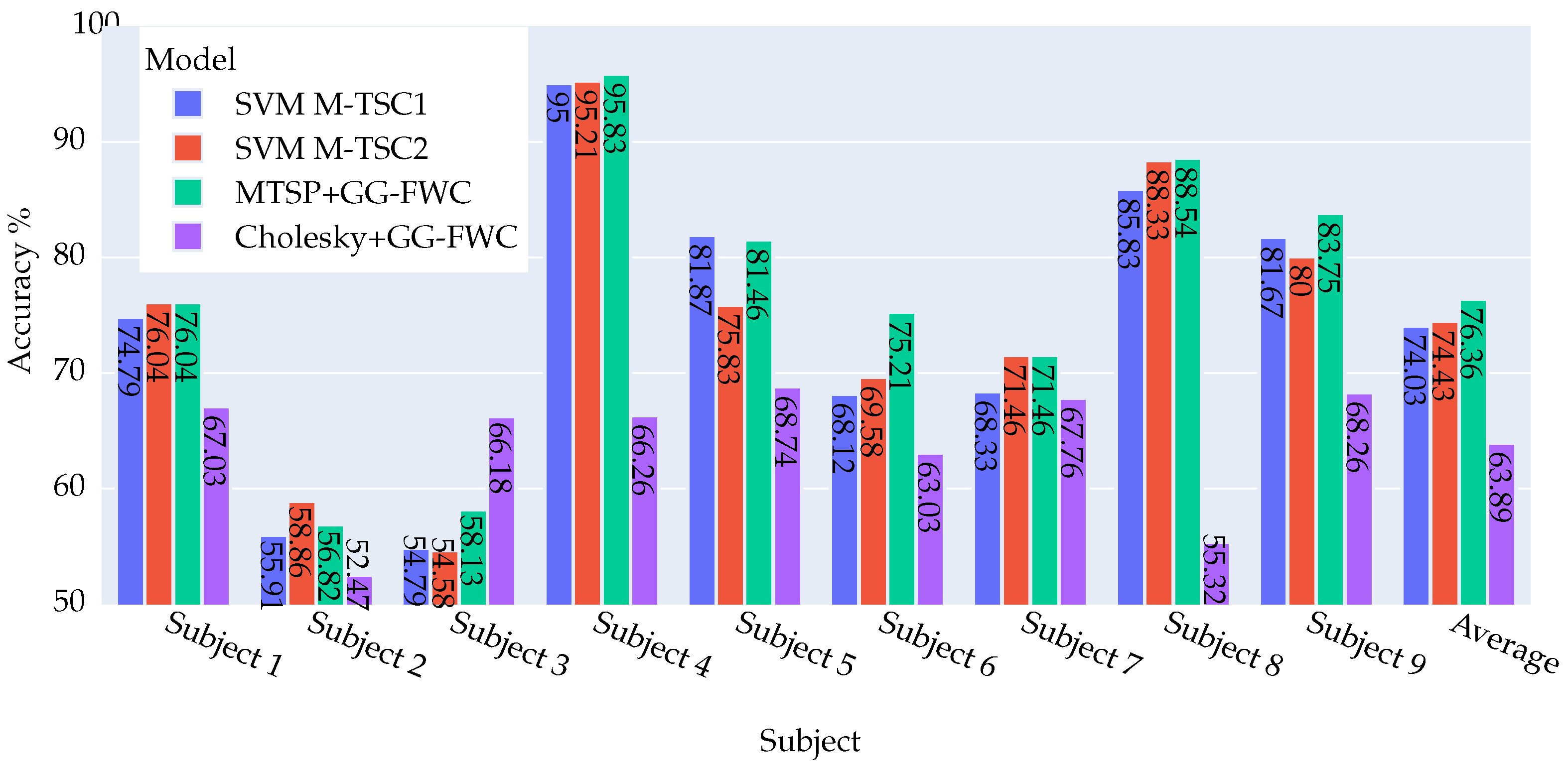
4.2.1. DS2 Dataset: Key Findings and Outcomes
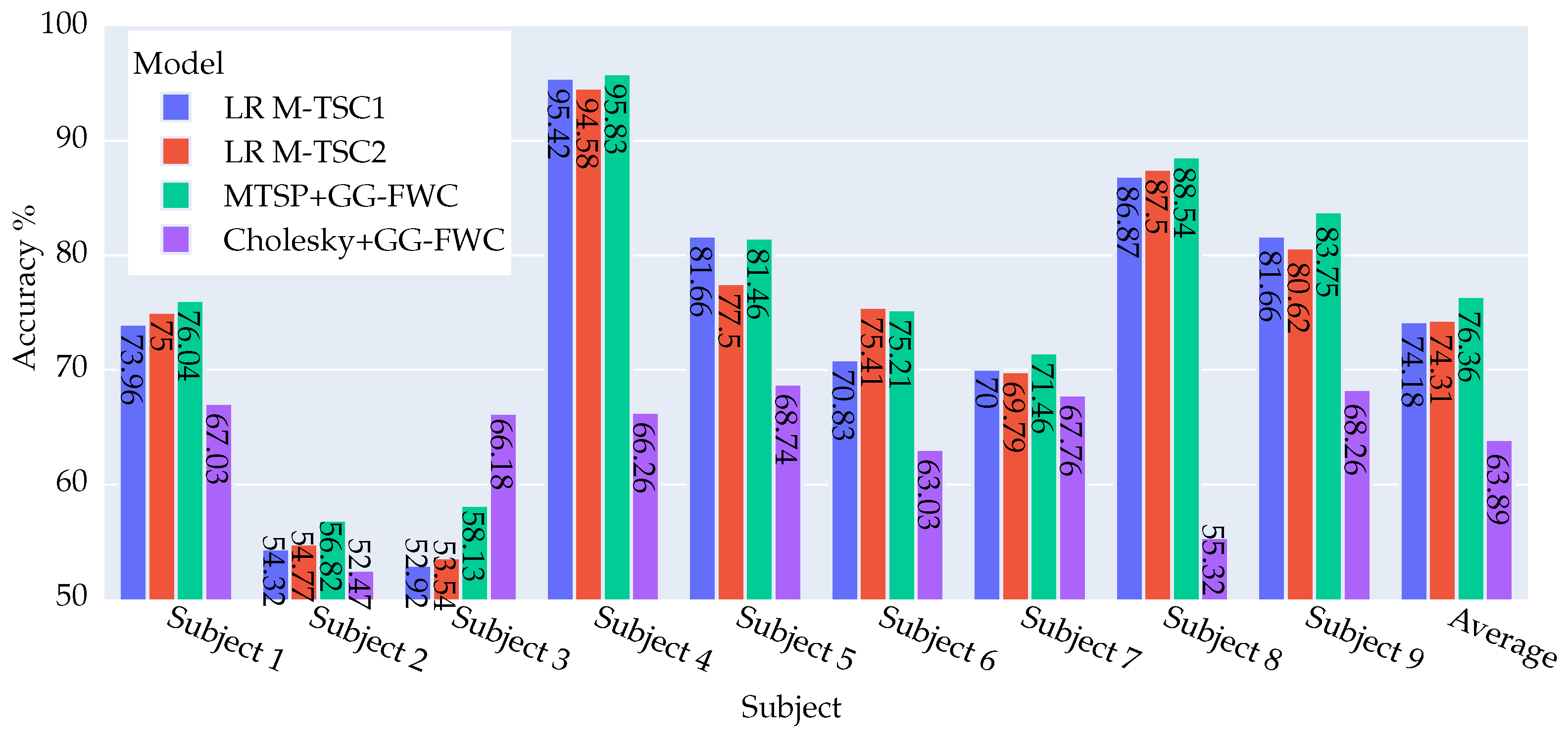
4.2.2. DS1 Dataset: Key Findings and Outcomes
4.2.3. Summary of Model Achievements
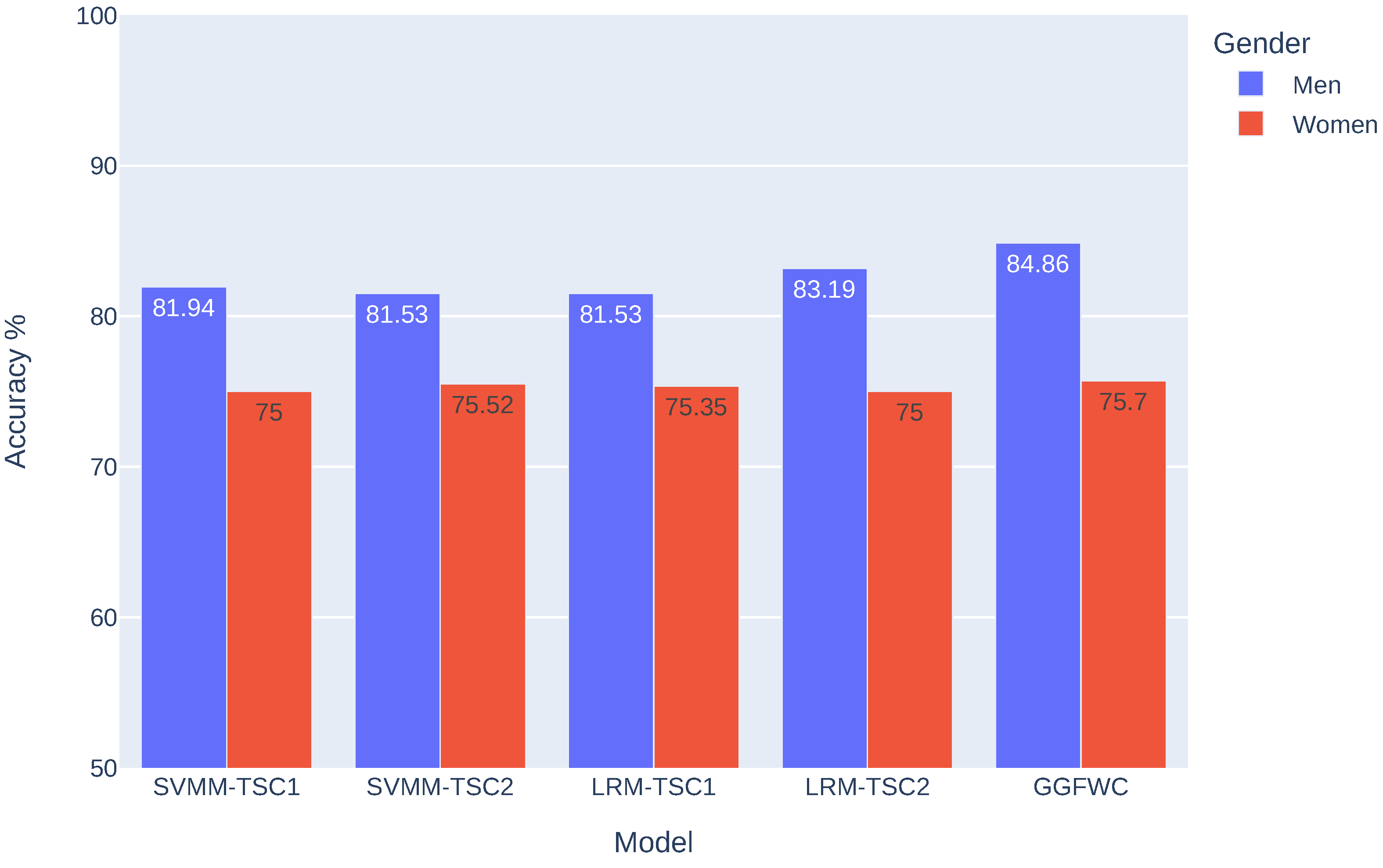
4.3. Gender-Based Analysis
5. Discussion and Future Directions
- –
- Enhanced BCI performance: The proposed methodologies significantly improve the accuracy of motor imagery classification, which can lead to more effective and reliable BCI systems for neurorehabilitation and assistive technologies.
- –
- Personalized BCI systems: The gender-based analysis reveals performance differences between male and female subjects, highlighting the need for personalized BCI systems that can adapt to individual user characteristics.
- –
- Robust feature extraction: The use of covariance matrices and the integration of linear and nonlinear features through the GG-FWC classifier demonstrate the potential for robust feature extraction methods that can capture the complex dynamics of EEG signals.
- –
- Development of a comprehensive database: Create a comprehensive database that includes demographic and psychological measures to enable more detailed user-specific studies. This will help with understanding the factors influencing individual abilities to control MI-BCIs.
- –
- Real-time application and deployment: Develop and test a real-time BCI application for deployment on robotic systems such as prosthetics, exoskeletons, and robotic arms in order to validate the practical applicability of the proposed methodologies.
- –
- Exploration of additional features: Investigate the incorporation of additional features such as EEG signal complexity measurements and feature fusion techniques to further enhance the performance and robustness of MI-BCI systems.
- –
- Gender-based adaptations: Conduct more extensive gender-based analyses using larger datasets to explore and develop gender-specific adaptations in BCI systems, ensuring optimal performance for both male and female users.
- –
- Integration with deep learning: Explore the integration of the proposed methodologies with deep learning techniques such as convolutional neural networks and recurrent neural networks to leverage their powerful feature learning capabilities.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wolpaw, J.R.; Birbaumer, N.; McFarland, D.J.; Pfurtscheller, G.; Vaughan, T.M. Brain–computer interfaces for communication and control. Clin. Neurophysiol. 2002, 113, 767–791. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Ahn, M.; Kwon, M.; Jun, S.C. A step-by-step tutorial for a motor imagery–based BCI. In Brain–Computer Interfaces Handbook; CRC Press: Boca Raton, FL, USA, 2018; pp. 445–460. [Google Scholar]
- Li, B.; Cheng, T.; Guo, Z. A review of EEG acquisition, processing and application. J. Phys. Conf. Ser. 2021, 1907, 012045. [Google Scholar] [CrossRef]
- Irimia, D.C.; Ortner, R.; Poboroniuc, M.S.; Ignat, B.E.; Guger, C. High classification accuracy of a motor imagery based brain-computer interface for stroke rehabilitation training. Front. Robot. AI 2018, 5, 130. [Google Scholar] [CrossRef] [PubMed]
- Al-Qazzaz, N.K.; Alyasseri, Z.A.A.; Abdulkareem, K.H.; Ali, N.S.; Al-Mhiqani, M.N.; Guger, C. EEG feature fusion for motor imagery: A new robust framework towards stroke patients rehabilitation. Comput. Biol. Med. 2021, 137, 104799. [Google Scholar] [CrossRef] [PubMed]
- Al-Qazzaz, N.K.; Aldoori, A.A.; Ali, S.H.B.M.; Ahmad, S.A.; Mohammed, A.K.; Mohyee, M.I. EEG Signal complexity measurements to enhance BCI-based stroke patients’ rehabilitation. Sensors 2023, 23, 3889. [Google Scholar] [CrossRef] [PubMed]
- Barachant, A. Commande Robuste d’un Effecteur par une Interface Cerveau Machine EEG Asynchrone. Ph.D. Thesis, Université de Grenoble, Grenoble, France, 2012. [Google Scholar]
- Omari, S.; Omari, A.; Abderrahim, M. Multiple tangent space projection for motor imagery EEG classification. Appl. Intell. 2023, 53, 21192–21200. [Google Scholar] [CrossRef]
- Bao, F.; Liu, W. EEG feature extraction methods in motor imagery brain computer interface. In Proceedings of the Third International Seminar on Artificial Intelligence, Networking, and Information Technology (AINIT 2022), Shanghai, China, 23–25 September 2022; SPIE: Bellingham, WA, USA, 2023; Volume 12587, pp. 375–380. [Google Scholar]
- Nandhini, A.; Sangeetha, J. A Review on Deep Learning Approaches for Motor Imagery EEG Signal Classification for Brain–Computer Interface Systems. In Computational Vision and Bio-Inspired Computing: Proceedings of ICCVBIC 2022; Springer: Singapore, 2023; pp. 353–365. [Google Scholar]
- Barachant, A.; Bonnet, S.; Congedo, M.; Jutten, C. Riemannian geometry applied to BCI classification. In Latent Variable Analysis and Signal Separation; Springer: Berlin/Heidelberg, Germany, 2010; pp. 629–636. [Google Scholar]
- Singh, A.; Lal, S.; Guesgen, H.W. Small sample motor imagery classification using regularized Riemannian features. IEEE Access 2019, 7, 46858–46869. [Google Scholar] [CrossRef]
- Barachant, A.; Bonnet, S.; Congedo, M.; Jutten, C. Multiclass brain–computer interface classification by Riemannian geometry. IEEE Trans. Biomed. Eng. 2011, 59, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Gaur, P.; Chowdhury, A.; McCreadie, K.; Pachori, R.B.; Wang, H. Logistic regression with tangent space-based cross-subject learning for enhancing motor imagery classification. IEEE Trans. Cogn. Dev. Syst. 2021, 14, 1188–1197. [Google Scholar] [CrossRef]
- Xie, X.; Yu, Z.L.; Lu, H.; Gu, Z.; Li, Y. Motor imagery classification based on bilinear sub-manifold learning of symmetric positive-definite matrices. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 25, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Miah, A.S.M.; Islam, M.R.; Molla, M.K.I. Motor imagery classification using subband tangent space mapping. In Proceedings of the 2017 20th International Conference of Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 22–24 December 2017; pp. 1–5. [Google Scholar]
- Fang, H.; Jin, J.; Daly, I.; Wang, X. Feature extraction method based on filter banks and Riemannian tangent space in motor-imagery BCI. IEEE J. Biomed. Health Inform. 2022, 26, 2504–2514. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.; Babu, D. Inter-channel Covariance Matrices Based Analyses of EEG Baselines. In Proceedings of the 2022 30th European Signal Processing Conference (EUSIPCO), Belgrade, Serbia, 29 August–2 September 2022; pp. 1303–1307. [Google Scholar]
- Abu-Dakka, F.J.; Rozo, L.; Caldwell, D.G. Force-based variable impedance learning for robotic manipulation. Robot. Auton. Syst. 2018, 109, 156–167. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, Y.; Ibrahim, J.G.; Li, Y.; Hall, C.; Lin, W. Intrinsic regression models for positive-definite matrices with applications to diffusion tensor imaging. J. Am. Stat. Assoc. 2009, 104, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Song, Y.; Liu, G.; Kompella, R.R.; Wu, X.; Sebe, N. Riemannian Multiclass Logistics Regression for SPD Neural Networks. arXiv 2023, arXiv:2305.11288. [Google Scholar]
- Omari, A.; Figueiras-Vidal, A.R. Feature combiners with gate-generated weights for classification. IEEE Trans. Neural Netw. Learn. Syst. 2012, 24, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.R.; Mika, S.; Rätsch, G.; Tsuda, K.; Schölkopf, B. An Introduction to Kernel-Based learning Algorithms. IEEE Trans. Neural Netw. 2001, 12, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Schölkopf, B.; Smola, A. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Shawe-Taylor, J.; Cristianini, N. Kernel Methods for Pattern Analysis; Cambridge University Press: New York, NY, USA, 2004. [Google Scholar]
- Omari, A.; Figueiras-Vidal, A.R. Post-aggregation of classifier ensembles. Inf. Fusion 2015, 26, 96–102. [Google Scholar] [CrossRef]
- Head, T.; Kumar, M.; Nahrstaedt, H.; Louppe, G.; Shcherbatyi, I. scikit-optimize library/scikit-optimize: v0.9.0. Zenodo 2021. [CrossRef]
- Leeb, R.; Brunner, C.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set B; Graz University of Technology: Graz, Austria, 2008; pp. 1–6. [Google Scholar]
- Brunner, C.; Leeb, R.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set A; Institute for Knowledge Discovery (Laboratory Brain-Computer Interfaces), Graz University of Technology: Graz, Austria, 2008; Volume 16, pp. 1–6. [Google Scholar]
- Alimardani, M.; Gherman, D.E. Individual differences in motor imagery bcis: A study of gender, mental states and mu suppression. In Proceedings of the 2022 10th International Winter Conference on Brain-Computer Interface (BCI), Gangwon-do, Republic of Korea, 21–23 February 2022; pp. 1–7. [Google Scholar]
- Roc, A.; Pillette, L.; N’Kaoua, B.; Lotte, F. Would motor-imagery based BCI user training benefit from more women experimenters? arXiv 2019, arXiv:1905.05587. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omari, S.; Omari, A.; Abu-Dakka, F.; Abderrahim, M. EEG Motor Imagery Classification: Tangent Space with Gate-Generated Weight Classifier. Biomimetics 2024, 9, 459. https://doi.org/10.3390/biomimetics9080459
Omari S, Omari A, Abu-Dakka F, Abderrahim M. EEG Motor Imagery Classification: Tangent Space with Gate-Generated Weight Classifier. Biomimetics. 2024; 9(8):459. https://doi.org/10.3390/biomimetics9080459
Chicago/Turabian StyleOmari, Sara, Adil Omari, Fares Abu-Dakka, and Mohamed Abderrahim. 2024. "EEG Motor Imagery Classification: Tangent Space with Gate-Generated Weight Classifier" Biomimetics 9, no. 8: 459. https://doi.org/10.3390/biomimetics9080459
APA StyleOmari, S., Omari, A., Abu-Dakka, F., & Abderrahim, M. (2024). EEG Motor Imagery Classification: Tangent Space with Gate-Generated Weight Classifier. Biomimetics, 9(8), 459. https://doi.org/10.3390/biomimetics9080459








