Current State of the Art of Newborn Screening for Lysosomal Storage Disorders
Abstract
:1. Introduction
2. Materials and Methods
2.1. Introduction
2.2. Microtiter-Plate Fluorometry
2.3. Digital Microfluidic Fluorometry
2.4. Tandem Mass Spectrometry
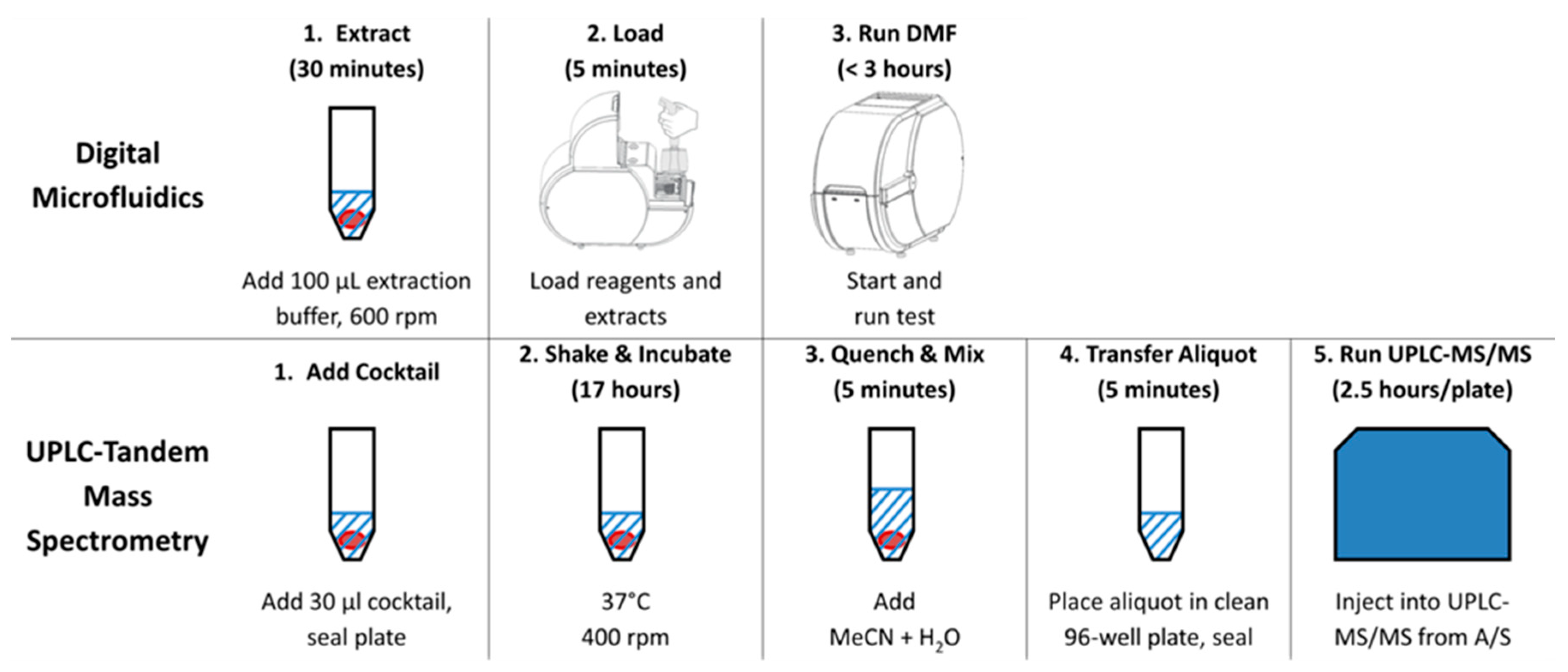
2.5. Workflows
3. Results
3.1. Results from the Taiwan Program
3.2. Results from the Missouri and Illinois State Programs
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Guthrie, R.; Susi, A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 1963, 32, 338–343. [Google Scholar] [PubMed]
- Wilson, J.; Jungner, G. Principles and Practice of Screening for Disease; Public Health Papers No. 34; WHO: Geneva, Switzerland, 1968; Available online: http://apps.who.int/iris/bitstream/handle/10665/37650/WHO_PHP_34.pdf (accessed on 22 June 2018).
- Millington, D.S.; Kodo, N.; Norwood, D.L.; Roe, C.R. Tandem mass spectrometry: A new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J. Inherit. Metab. Dis. 1990, 13, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Rashed, M.S.; Bucknell, M.P.; Little, D.; Awad, A.; Jacob, M.; Alamoudi, M.; Alwattar, M.; Ozand, P.T. Screening blood spots for inborn errors of metabolism by electrospray tandem mass spectrometry with a microplate batch process and a computer algorithm for automated flagging of abnormal profiles. Clin. Chem. 1997, 43, 1129–1141. [Google Scholar] [PubMed]
- Wilcken, B.; Wiley, V.; Hammond, J.; Carpenter, C. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N. Engl. J. Med. 2003, 348, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Lindner, M.; Kohlmuller, D.; Olgemoller, K.; Mayatepek, E.; Hoffman, G.F. Expanded newborn screening for inborn errors of metabolism by electrospray ionization-tandem mass spectrometry: Results, outcome, and implications. Pediatrics 2003, 111, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Chace, D.H.; Kalas, T.A.; Naylor, E.W. Use of tandem mass spectrometry for multianalyte screening of dried blood specimens from newborns. Clin. Chem. 2003, 49, 1797–1817. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.S.; Mann, M.Y.; Lloyd-Puryear, M.A.; Rinaldo, P.; Howell, R.R. Newborn Screening: Toward a Uniform Screening Panel and System—Executive Summary. Pediatrics 2006, 117, S296–S307. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, L.; Millington, D.S.; Therrell, B.L.; Hannon, W.H.; Popovich, B.; Watson, M.S.; Mann, M.Y.; Lloyd-Puryear, M.A.; van Dyck, P.C. Naming and Counting Disorders (Conditions) Included in Newborn Screening Panels. Pediatrics 2006, 117, S308–S314. [Google Scholar] [CrossRef] [PubMed]
- Advisory Committee on Heritable Disorders in Newborns and Children. Available online: https://www.hrsa.gov/advisory-committees/heritable-disorders/rusp/index.html (accessed on 20 June 2018).
- Bodamer, O.A.; Scott, C.R.; Giugliani, R.; On Behalf of the Pompe Disease Newborn Screening Working Group. Newborn Screening for Pompe Disease. Pediatrics 2017, 140, S4–S13. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.A.; Atherton, A.M.; Burton, B.K.; Day-Salvatore, D.L.; Kaplan, P.; Leslie, N.D.; Scott, C.R.; Stockton, D.W.; Thomas, J.A.; Muenzer, J. Mucopolysaccharidosis Type I Newborn Screening: Best Practices for Diagnosis and Management. J. Pediatr. 2017, 182, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Marsden, D.; Levy, H. Newborn screening of lysosomal storage disorders. Clin. Chem. 2010, 56, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Hattori, K.; Endo, F. Newborn screening for lysosomal storage disorders. Am. J. Med. Genet. Part C Semin. Med. Genet. 2011, 157, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Orsini, J.J.; Saavedra-Matiz, C.A.; Gelb, M.H.; Caggana, M. Newborn screening for Krabbe’s disease. J. Neurosci. Res. 2016, 94, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.V.; Campbell, C.; Klug, T.; Rogers, S.; Raburn-Miller, J.; Kiesling, J. Lysosomal storage disorder screening implementation: Findings from the first six months of full population pilot testing in Missouri. J. Pediatr. 2015, 166, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Burton, B.K.; Charrow, J.; Hoganson, G.E.; Waggoner, D.; Tinkle, B.; Braddock, S.R.; Schneider, M.; Grange, D.K.; Nash, C.; Shryock, H.; et al. Newborn Screening for Lysosomal Storage Disorders in Illinois: The Initial 15-Month Experience. J. Pediatr. 2017, 190, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Grasby, D.J.; Dean, C.J.; Lang, D.L.; Bockmann, M.; Whittle, A.M.; Fietz, M.J.; Simonsen, H.; Fuller, M.; Brooks, D.A.; et al. Newborn screening for lysosomal storage disorders. Mol. Genet. Metab. 2006, 88, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Chamoles, N.A.; Blanco, M.; Gaggioli, D. Diagnosis of α-l-iduronidase deficiency in dried blood spots on filter paper: The possibility of newborn diagnosis. Clin. Chem. 2001, 47, 780–781. [Google Scholar] [PubMed]
- Chamoles, N.A.; Blanco, M.; Gaggioli, D. Fabry disease: Enzymatic diagnosis in dried blood spots on filter paper. Clin. Chim. Acta 2001, 308, 195–196. [Google Scholar] [CrossRef]
- Chamoles, N.A.; Niizawa, G.; Blanco, M.; Gaggioli, D.; Casentini, C. Glycogen storage disease type II: Enzymatic screening in dried blood spots on filter paper. Clin. Chim. Acta 2004, 347, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.H.; Chiang, S.C.; Zhang, X.K.; Keutzer, J.; Lee, N.C.; Huang, A.C.; Chen, C.A.; Wu, M.H.; Huang, P.H.; Tsai, F.J.; et al. Early detection of Pompe disease by newborn screening is feasible: Results from the Taiwan screening program. Pediatrics 2008, 122, e39–e45. [Google Scholar] [CrossRef] [PubMed]
- Hwu, W.L.; Chien, Y.H.; Lee, N.C.; Chiang, S.-C.; Dobrovolny, R.; Huang, A.-C.; Yeh, H.Y.; Chao, M.C.; Lin, S.J.; Kitagawa, T.; et al. Newborn screening for Fabry disease in Taiwan reveals a high incidence of the later-onset GLA mutation c.936 + 919G > A (IVS4 + 919G > A). Hum. Mutat. 2009, 30, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Sista, R.S.; Eckhardt, A.E.; Wang, T.; Graham, C.; Rouse, J.L.; Norton, S.M.; Srinivasan, V.; Pollack, M.G.; Tolun, A.A.; Bali, D.; et al. Digital Microfluidic Platform for Multiplexing Enzyme Assays: Implications for Lysosomal Storage Disease Screening in Newborns. Clin. Chem. 2011, 57, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Sista, R.S.; Wang, T.; Wu, N.; Graham, C.; Eckhardt, A.; Winger, T.; Srinivasan, V.; Bali, D.; Millington, D.S.; Pamula, V.K. Multiplex Newborn Screening for Pompe, Fabry, Hunter, Gaucher, and Hurler Diseases Using a Digital Microfluidic Platform. Clin. Chim. Acta 2013, 424, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Matern, D.; Gavrilov, D.; Oglesbee, D.; Raymond, K.; Rinaldo, P.; Tortorelli, S. Newborn screening for lysosomal storage disorders. Semin. Perinatol. 2015, 39, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Burton, B.; Charrow, J.; Angle, B.; Widera, S.; Waggoner, D. A Pilot Newborn Screening Program for Lysosomal Storage Disorders (LSD) in Illinois. Mol. Genet. Metab. 2012, 105, S23–S24. [Google Scholar] [CrossRef]
- Hopkins, P.V.; Klug, T.; Vermette, L.; Raburn-Miller, J.; Kiesling, J.; Rogers, S. Incidence of 4 lysosomal storage disorders from 4 years of newborn screening. JAMA Pediatr. 2018, 172, 696–697. [Google Scholar] [CrossRef] [PubMed]
- FDA News Release. Silver Spring (MD): U.S. Food & Drug Administration. FDA Permits Marketing of First Newborn Screening System for Detection of Four, Rare Metabolic Disorders. Update on 3 February 2017. Available online: https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm539893.htm (accessed on 30 March 2018).
- Li, Y.; Brockman, K.; Turecek, F.; Scott, C.R.; Gelb, M.H. Tandem mass spectrometry for the direct assay of enzymes in dried blood spots: Application to newborn screening for Krabbe disease. Clin. Chem. 2004, 50, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Orsini, J.J.; Morrissey, M.A.; Slavin, L.N.; Wojik, M.; Biski, C.; Martin, M.; Keutzer, J.; Zhang, X.K.; Chuang, W.-L.; Elbin, C.; et al. Implementation of newborn screening for Krabbe disease: Polulation study and cut-off determination. Clin. Biochem. 2009, 42, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Scott, R.; Chamoles, N.A.; Ghavami, B.; Pinto, B.M.; Turecek, F. Direct multiplex assay of lysosomal enzymes in dried blood spots for newborn screening. Clin. Chem. 2004, 50, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Schielen, P.C.J.I.; Kemper, E.A.; Gelb, M.H. Newborn screening for lysosomal storage diseases: A concise review of the literature on screening methods, therapeutic possibilities and regional programs. Int. J. Neonatal Screen. 2017, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Spacil, Z.; Tatipaka, H.; Barcenas, M.; Scott, C.R.; Turecek, F.; Gelb, M.H. High-throughput assay of 9 lysosomal enzymes for newborn screening. Clin. Chem. 2013, 59, 502–511. [Google Scholar] [CrossRef] [PubMed]
- La Marca, G.; Casetta, B.; Malvagia, S.; Guerrini, R.; Zammarchi, E. New strategy for the screening of lysosomal storage disorders: The use of the online trapping-and-clean up liquid chromatography/mass spectrometry. Anal. Chem. 2009, 81, 6113–6121. [Google Scholar] [CrossRef] [PubMed]
- Peake, R.W.A.; Marsden, D.I.; Bodamer, O.A.; Gelb, M.H.; Millington, D.S.; Wijburg, F. Newborn screening for lysosomal storage disorders: Quo Vadis? Clin. Chem. 2016, 62, 1430–1438. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.C.; Hwu, W.L.; Lee, N.C.; Hsu, L.W.; Chien, Y.H. Algorithm for Pompe disease newborn screening: Results from the Taiwan screening program. Mol. Genet. Metab. 2012, 106, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.H.; Lee, N.C.; Huang, H.J.; Thurberg, B.L.; Tsai, F.J.; Hwu, W.L. Later-onset Pompe disease: Early detection and early treatment initiation enabled by newborn screening. J. Pediatr. 2011, 158, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.H.; Hwu, W.L.; Lee, N.C. Pompe Disease: Early Diagnosis and Early Treatment Make a Difference. Ped. Neonatol. 2013, 54, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Chong, K.W.; Hsu, J.H.; Yu, H.C.; Shih, C.C.; Huang, C.H.; Lin, S.J.; Chen, C.H.; Chiang, C.C.; Ho, H.J.; et al. High incidence of the cardiac variant of Fabry disease revealed by newborn screening in the Taiwan Chinese population. Circ. Cardiovasc. Genet. 2009, 2, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.F.; Liu, H.C.; Hsu, T.R.; Tsai, F.C.; Chiang, S.F.; Chiang, C.C.; Ho, H.C.; Lai, C.J.; Yang, T.F.; Chuang, S.Y.; et al. A large-scale nationwide newborn screening program for Pompe disease in Taiwan: Towards effective diagnosis and treatment. Am. J. Med. Genet. A 2014, 164, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.C.; Chiang, C.C.; Niu, D.M.; Wang, C.H.; Kao, S.M.; Tsai, F.J.; Huang, Y.H.; Liu, H.C.; Huang, C.K.; Gao, H.J.; et al. Detecting multiple lysosomal storage diseases by tandem mass spectrometry—A national newborn screening program in Taiwan. Clin. Chim. Acta 2014, 431, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M.H.; Scott, C.R.; Turacek, F. Newborn screening for lysosomal storage disorders. Clin. Chem. 2015, 61, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Peake, R.W.A.; Bodamer, O.A. Newborn screening for lysosomal storage disorders. J. Pediatr. Genet. 2017, 63, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.; Buroker, N.; Cournoyer, J.J.; Potier, A.M.; Trometer, J.D.; Elbin, C.; Schermer, M.J.; Kantola, J.; Boyce, A.; Turecek, F.; et al. Pilot study of newborn screening for six lysosomal storage diseases using tandem mass spectrometry. Mol. Genet. Metab. 2016, 118, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Millington, D.S.; Bali, D. Misinformation regarding tandem mass spectrometric vs fluorometric assays to screen newborns for LSDs. Mol. Genet. Metab. Rep. 2017, 12, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M.H.; Scott, C.R.; Turacek, F.; Liao, H.C. Comparison of tandem mass spectrometry to fluorimetry for newborn screening of LSDs. Mol. Genet. Metab. Rep. 2017, 12, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Millington, D.S. Response to Gelb et al.: “Comparison of tandem mass spectrometry to fluorimetry for newborn screening of LSDs”. Mol. Genet. Metab. Rep. 2017, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Minter Baerg, M.M.; Stoway, S.D.; Hart, J.; Mott, L.; Peck, D.S.; Nett, S.L.; Eckerman, J.S.; Lacey, J.M.; Turgeon, C.T.; Gavrilov, D.; et al. Precision newborn screening for lysosomal disorders. Genet. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kemper, A.R. Evidence Report: Newborn Screening for Pompe Disease. June 2013. Available online: https://www.hrsa.gov/sites/default/files/hrsa/advisory-committees/heritable-disorders/rusp/previous-nominations/pompe-external-evidence-review-report-2013.pdf (accessed on 5 June 2018).
- Kemper, A.R. Newborn Screening for MPSI: Final Report from the Condition Review Workgroup. February 2015. Available online: https://www.hrsa.gov/sites/default/files/hrsa/advisory-committees/heritable-disorders/meetings/20150212/kemper-mps1.pdf (accessed on 5 June 2018).
- Orsini, J.J.; Caggana, M. Newborn Screening for Krabbe Disease and Other Lysosomal Storage Disorders: Broad Lessons Learned. Int. J. Neonatal Screen. 2017, 3, 3. [Google Scholar] [CrossRef]
- Tortorelli, S.; Eckerman, J.S.; Orsini, J.J.; Stevens, C.; Hart, J.; Hall, P.L.; Alexander, J.J.; Gavrilov, D.; Oglesbee, D.; Raymond, K.; Matern, D.; et al. Moonlighting newborn screening markers: The incidental discovery of a second tier test for Pompe disease. Genet. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- De Ruijter, J.; de Ru, M.H.; Wagemans, T.; IJlst, L.; Lund, A.M.; Orchard, P.J.; Schaefer, G.G.; Wijburg, F.A.; van Vlies, N. Heparan sulfate and dermatan sulfate derived disaccharides are sensitive markers for newborn screening for mucopolysaccharidoses types I, II and III. Mol. Genet. Metab. 2012, 107, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M. Newborn Screening for Pompe and MPS-I: Performance Data from 11 Newborn Screening Labs Worldwide. Presented at the 2017 Newborn Screening and Genetic Testing Symposium Association of Public Health Laboratories, New Orleans, LA, USA, 10–13 September 2017. [Google Scholar]
- Singh, R.; Chopra, S.; Graham, C.; Nelson, L.; Ng, R.; Nuffer, M.; Vadlamani, P.; Norton, S.; Pamula, V. Demonstration of a digital microfluidic platform for the high throughput analysis of 12 discrete fluorimetric enzyme assays using a single newborn dried blood spot punch. Presented at the 14th Annual Meeting World Symposium on Lysosomal Disease, San Diego, CA, USA, 5–9 February 2018. [Google Scholar]
| Disorder | Published Incidence | Incidence MO [16] | Incidence IL [17] |
|---|---|---|---|
| Pompe | 1:28,000 [50] | 1:9625 | 1:22,000 |
| Hurler | 1:87,000–1:185,000 [51] | 1:154,000 | 1:220,000 |
| Fabry | 1:1500–1:13,000 [26] | 1:3277 | 1:8500 |
| Gaucher | 1:57,000 [26] | 1:61,600 | 1:44,000 |
| GAA (Pompe) | IDUA (MPS I) | GLA (Fabry) | GBA (Gaucher) | |||||
|---|---|---|---|---|---|---|---|---|
| MO (DMF) | IL (MS/MS) | MO (DMF) | IL (MS/MS) | MO (DMF) | IL (MS/MS) | MO (DMF) | IL (MS/MS) | |
| Total infants | 308,000 | 219,793 | 308,000 | 219,793 | 308,000 | 219,793 | 308,000 | 219,793 |
| Screen Positives | 161 | 139 | 133 | 151 | 179 | 107 | 37 | 117 |
| Pending | 0 | 8 | 0 | 24 | 0 | 16 | 0 | 19 |
| Refused/Lost to f.u./Died | 2 | n/a | 5 | n/a | 12 | n/a | 2 | n/a |
| Normal | 48 | 87 | 45 | 87 | 66 | 59 | 22 | 91 |
| Carrier | 39 | 15 | 8 | 5 | 0 | 0 | 5 | 0 |
| Pseudodeficient | 31 | 15 | 63 | 30 | 0 | 16 | 0 | 0 |
| Undetermined /VUS/GUS 3 | 9 | 4 | 2 | 4 | 6 | 6 | 2 | 2 |
| Confirmed Positive | 32 | 10 | 2 | 1 | 94 | 10 | 5 | 5 |
| PPV 1 | 26% | 11% | 3% | 4% | 60% | 18% | 21% | 7% |
| False Positive rate 2 | 0.04% | 0.05% | 0.04% | 0.06% | 0.02% | 0.03% | 0.01% | 0.04% |
| False positives per 100,000 | 38.3 | 53.2 | 37.7 | 55.5 | 21.4 | 34.1 | 8.8 | 41.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millington, D.S.; Bali, D.S. Current State of the Art of Newborn Screening for Lysosomal Storage Disorders. Int. J. Neonatal Screen. 2018, 4, 24. https://doi.org/10.3390/ijns4030024
Millington DS, Bali DS. Current State of the Art of Newborn Screening for Lysosomal Storage Disorders. International Journal of Neonatal Screening. 2018; 4(3):24. https://doi.org/10.3390/ijns4030024
Chicago/Turabian StyleMillington, David S., and Deeksha S. Bali. 2018. "Current State of the Art of Newborn Screening for Lysosomal Storage Disorders" International Journal of Neonatal Screening 4, no. 3: 24. https://doi.org/10.3390/ijns4030024
APA StyleMillington, D. S., & Bali, D. S. (2018). Current State of the Art of Newborn Screening for Lysosomal Storage Disorders. International Journal of Neonatal Screening, 4(3), 24. https://doi.org/10.3390/ijns4030024





