Abstract
Antimicrobial resistance (AMR) is the major issue posing a serious global health threat. Low- and middle-income countries are likely to be the most affected, both in terms of impact on public health and economic burden. Recent studies highlighted the role of resistance networks on the transmission of AMR organisms, with this network being driven by complex interactions between clinical (e.g., human health, animal husbandry and veterinary medicine) and other components, including environmental factors (e.g., persistence of AMR in wastewater). Many studies have highlighted the role of wastewater as a significant environmental reservoir of AMR as it represents an ideal environment for AMR bacteria (ARB) and antimicrobial resistant genes (ARGs) to persist. Although the treatment process can help in removing or reducing the ARB load, it has limited impact on ARGs. ARGs are not degradable; therefore, they can be spread among microbial communities in the environment through horizontal gene transfer, which is the main resistance mechanism in most Gram-negative bacteria. Here we analysed the recent literature to highlight the contribution of wastewater to the emergence, persistence and transmission of AMR under different settings, particularly those associated with mass gathering events (e.g., Hajj and Kumbh Mela).
1. Introduction
1.1. The Current Status of AMR as a Major Global Health Challenge
Antibiotics are one of the greatest tools of medicine. However, since the development of fluoroquinolones in early 1970, no new major groups of antibacterial drugs have been developed [1]. This paucity in development is accompanied by an increasing threat of antimicrobial resistant (AMR) organisms [1,2]. AMR is the major issue posing a threat to public health, with many reports warning of the significant risk of a post-antimicrobial era in which common infections can kill [1,3,4,5]. Recently, the World Health Organization (WHO) Global Antimicrobial Surveillance System (GLASS) reported increased levels of resistance in a number of serious and common bacterial infections in many regions of the world [6]. Currently, resistant infections result in 700,000 deaths every year, but the global resistance-associated mortality is estimated to top 10 million lives per year in 2050 [2]. The European Center for Disease Prevention and Control (ECDC) and the US Centers for Disease Control and Prevention (CDC) have reported that AMR infections resulted in 25,000 and 23,000 deaths every year in high-income countries in Europe and the USA, respectively [7]. In low- and middle-income countries, AMR infections have been responsible for the deaths of 58,000 children and 38,000 adults in India and Thailand, respectively [7].
1.2. WHO AMR Priority Pathogens List
Recently, the WHO identified 12 bacterial species and their accompanying AMR profiles that pose the greatest threat to human health [8]. This list mainly includes Gram-negative bacteria and the most common etiologic agents associated with hospital- and/or community-acquired infections. These AMR bacteria have been divided into three categories: critical, high and medium priority, according to their impact on human health and the urgency for the development of new antimicrobial drugs to treat resistant infections. The critical category includes Acinetobacter baumannii (carbapenem-resistant), Pseudomonas aeruginosa (carbapenem-resistant) and various Enterobacteriaceae members, including Klebsiella spp., Escherichia coli, Serratia spp., and Proteus spp. (carbapenem-resistant and extended-spectrum ß-lactamase (ESBL)-producing), which are associated with severe and, often deadly, infections, including bloodstream infections and pneumonia. The high-priority category includes Enterococcus faecium (vancomycin-resistant); Staphylococcus aureus (methicillin-resistant, vancomycin-intermediate and resistant); Helicobacter pylori (clarithromycin-resistant); Campylobacter spp. (fluoroquinolone-resistant); Salmonella spp. (fluoroquinolone-resistant) and Neisseria gonorrhoeae (cephalosporin-resistant and fluoroquinolone-resistant), which are causative agents associated with more common infections, such as general infections, gastroenteritis and gonorrhoea. The medium-priority category includes Streptococcus pneumoniae (penicillin-non-susceptible), Haemophilus influenzae (ampicillin-resistant) and Shigella spp. (fluoroquinolone-resistant).
1.3. The Main Drivers of AMR Transmission
AMR is driven by complex interacting factors that could be described as a resistance network [9]. This network forms links between clinical factors (e.g., human health, animal husbandry and veterinary medicine) and other components, including human activities (e.g., travel [10,11], human displacement and over and misuse of antimicrobial drugs [12,13,14]) and environmental factors (e.g., persistence of antimicrobial drugs and AMR organisms in soil and water). For example, the variations in AMR patterns among different regions of the world have been associated with differing rates of consumption of, and exposure to, antimicrobial drugs [2]. This is alarming, with the data available on AMR transmission suggesting increasing consumption of antibiotics in humans during the past two decades, primarily in low- and middle-income countries [15]. The selective pressure associated with the exposure to antimicrobials in healthcare, agriculture and the environment enhances the development of new AMR variants and novel resistance mechanisms [16]. Other factors, including lack of access to clean water sanitation and healthcare service, poor personal hygiene, failure of AMR detection and treatment and poor vaccination coverage [17] in the community, have been shown to also contribute to the global transmission of AMR [18].
1.4. The Environmental Reservoir of AMR from Water and Sewage
Transmission of AMR can spread between people, animals and the environment via a number of different routes [19]. The environment acts as a bridge for different compartments, between animals to compost to soil to water to sediments to sewage [20]. While the environment acts as the reservoir, it also works simultaneously to mix mobile genetic elements (MGEs) that interact and diffuse into other parts or into human and animal hosts [19,21,22].
Many studies have highlighted the impact of the diverse nature of the reservoirs of AMR genes (ARGs) on promoting the emergence and transmission of AMR organisms [23]. AMR is ancient and ubiquitous in the environment, with many lines of evidence suggesting that transfer of ARGs occurs among different environments (e.g., from environmental to pathogenic bacteria) [24,25]. Although it has been well-established that the genetic transfer of ARGs is likely to occur between closely-related species, recent studies have suggested that this transfer can also occur among phylogenetically distant species and even among organisms belonging to distinct phyla [26], adding further challenges in the continuous evolution of new variants of AMR organisms. High concentrations of antibiotic residues, ARGs and AMR organisms have been reported from environmental samples recovered from hospital and urban and treated wastewaters and soils treated with animal manure [27,28,29].
Many studies have highlighted the role of sewage as a major environmental reservoir of AMR, as it represents an ideal environment for AMR microorganisms and ARGs to persist [30,31,32]. The situation of ARGs is more complex, because they are not degradable and can be spread among microbial communities in the environment through horizontal gene transfer, which is the main resistance mechanism in Enterobacteriaceae [33,34].
In this study, we aimed to systematically review the literature to identify the role of wastewater in promoting the transmission of AMR and to characterise the key factors implicated in the persistence of ARB and ARGs in this environmental component. We extended the analysis to characterise AMR transmission in environmental samples associated with key religious mass gathering events—Kumbh Mela and Hajj in India and Saudi Arabia, respectively.
2. Materials and Methods
2.1. Search Strategy
Searches were systematically carried out in four databases: Embase, Medline, PubMed and Web of Science Core Collection to obtain all articles that reported AMR in sewage samples. The key terms “antimicrobial resistance” OR “AMR” in combination with “sewage” were used to obtain the articles available between 2009–2019, with the search conducted on 21st June 2019. EndNote X7.5 (Thomson Reuters) was used for bibliography management. The duplicates were removed, and initial screening was performed by assessing titles, abstracts and keywords with an explicit focus on the use of molecular approaches, including whole genome sequencing and metagenomics, in detecting AMR. The search was extended to include special settings, such as mass gatherings at Hajj and Kumbh Mela.
2.2. Selection Criteria
Articles were included if they were written in English and included an observational study design where sewage samples were investigated molecularly for the detection of AMR. We excluded articles if they were written in languages other than English, reviews, opinion articles and editorials. Potential articles were evaluated on the inclusion criteria by retrieving the full text and were subsequently included in the analysis (Figure 1).
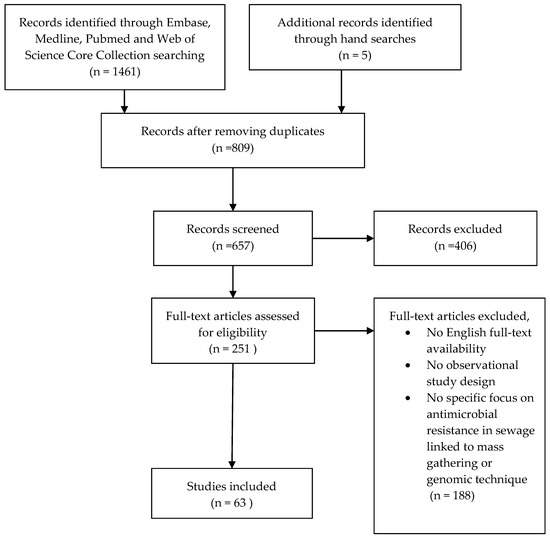
Figure 1.
Flow-chart of literature search.
2.3. Data Analysis
1461 articles were obtained in the initial literature search and five articles found by hand searches, which included 809 duplicates. After removing the duplicates, the first screening removed non-English records and irrelevant abstracts, resulting in 251 remaining articles. Full-text was retrieved to screen the articles on the selection criteria, and a total of 63 papers were eligible for inclusion in the analysis (Figure 1). All papers were dissected to summarise the key information and findings, including year of publication, country study site, source and type of wastewater, abundance of ARGs and AMR microbial communities and methods used for AMR detection.
3. Results
3.1. Dissemination of Antimicrobial Resistance in Wastewater
From the 1466 articles that were identified, 63 studies conducted on wastewater samples between 2004 and 2018 (published in the period 2009–2019) were included in the data analysis. The analysed studies documenting the detection of ARBs and/or ARGs in different types of wastewaters are listed in Table 1. The source and type of wastewater samples investigated and the key findings highlighted by these studies are summarised (Table 1). Detailed information on location and time of sample collection, structure of ARBs populations and/or ARGs detected and the technology used in AMR characterisation are provided in Table S1.

Table 1.
Overview of the studies included in this systematic review. AMR: antimicrobial resistant, ARG: antimicrobial resistant genes, ARB: antimicrobial resistant bacteria and MGE: mobile genetic elements.
These studies highlight the role of aquatic ecosystems, particularly wastewater, as a key reservoir of AMR bacteria and ARGs in the environment. High levels of both ARBs and/or ARGs were detected in samples collected from different types of wastewater, including municipal sewage [39,46,47,54,81] and influent and effluent samples from wastewater treatment plants (WWTPs) [37,42,44,55,63,64,70,71,72,78,79]. Similarly, high levels of AMR were identified in industrial [66,77,92] and agricultural wastewater and samples recovered from pharmaceutical treatment plants [49,66]. High levels of clinically relevant ARBs and/or ARGs were identified in influent and effluent samples from hospital wastewater [36,39,40,42,50,54,58,76,80,82,83,90,94,97].
A number of studies have also demonstrated elevated levels of AMR detected in samples that have been collected from downstream water [36,44]; the surface water of rivers [35,37,53,75,86,88] and tap water [56]. Few studies have detected ARBs and clinically relevant ARGs in environmental samples that have been exposed to/contaminated with sewage [62,91].
3.2. ARB Populations Associated with Wastewaters
The majority of the studies have used integrated molecular and phenotypic approaches to characterise the resistance profiles and virulence contents associated with AMR bacteria. Many studies (n = 47) have used advanced molecular approaches, including polymerase chain reaction (PCR) followed by Sanger sequencing and/or quantitative PCR, to characterise the AMR genotypes. Recent studies (n = 16) have used whole-genome sequencing and metagenomic analyses to comprehensively detect the microbial and AMR determinants in wastewater samples. The latter aimed to assess the abundance and distribution of microbes and associated AMR agents (mobile genetic elements (MGE), including plasmids, transposons, integrons and insertion sequences) and to identify the factors that determine the persistence of AMR bacteria and ARGs in wastewater.
Phenotypic characterisation demonstrated that Enterobacteriaceae members, including Escherichia coli, Klebsiella spp., Shigella spp., Salmonella spp., Vibrio spp., Acinetobacter spp. and Enterococcus spp., were among the most common AMR bacteria identified in the wastewater samples investigated in the analysed studies (Table 1). Additionally, high levels of MDR bacteria and ARGs conferring resistance to varied classes of antimicrobial drugs, including beta-lactams, carbapenems, tetracyclines, aminoglycosides, fluoroquinolones, sulphonamides, macrolides, vancomycin and erythromycin, were documented in the analysed articles (Table 1).
3.3. Selective Pressure within Wastewater Environments Promote the Emergence of Novel Variants of ARGs and ARBs
Generally, high levels of ARB, including MDR strains and diverse ARGs, have been detected in influent wastewater (untreated) collected from various sources, particularly low-income settings [41,44]; hospitals [36,39,40,42,46,47] and pharmaceutical waste [49,60]. However, many studies demonstrated that effluent samples collected from urban, hospital and pharmaceutical-treated wastewater still contain elevated levels of diverse ARGs, ARB and antimicrobial drugs [49,58,60,63]. For instance, a recent study demonstrated that the abundance of ARGs was significantly higher in effluent wastewater samples collected from low-income compared to high-income countries [41,44]. High rates of ARGs have been identified in pharmaceutical wastewater treatment plants, with the rate of those associated with clinically important antimicrobial drugs (e.g., sul1, sul2 and tet) being found to remain high throughout the different stages of the treatment process and, therefore, were subsequently discharged into the environment [49].
NDM-1 producing strains, including V. cholerae, Shigella boydii and Aeromonas caviae, which had not been previously reported to carry blaNDM-1, have been isolated for the first time from drinking water (4%; 2 out of 50) and seepage samples (17%; 12 out of 171) from New Delhi [56]. This is in addition to the previously reported NDM-1-producing species, including E. coli and K. pneumoniae [56]. The carriages of blaNDM-1-bearing plasmids by enterobacteria, aeromonads and V. cholerae have been shown to be stable, transmissible and exhibit the typical resistance pattern of NDM-1 [56]. Although the majority of strains have previously carried blaNDM-1 on plasmids, blaNDM-1-bearing chromosomes have been first identified in environmental isolates of Aeromonas caviae and V. cholerae [56]. Another study has documented the isolation of novel species of Acinetobacter cumulans from hospital wastewater [36]. These strains have been found to contain ARGs associated with resistance to clinically important drugs, including carbapenems, cephalosporines and aminoglycoside [36]. Additionally, a carbapenemase-producing K. pneumoniae strain carrying blaKPC-2, which is rarely detected in clinical settings, has been identified in WWTP effluent wastewater in Japan [38].
Additionally, many studies have detected high levels of integrons [77,78,79,81], including novel classes associated with oxacillinase gene cassette (blaOXA-109, blaOXA-368 and blaOXA-2) in varied bacterial species recovered from wastewater samples [90]. Higher prevalence of class 1 integrons was detected in bacteria recovered from sewage sludge and pig slurry (environments that contain high concentrations of antibiotic residues and detergents) compared to agricultural soils to which these waste products are amended [77]. It has been estimated that ~1019 bacteria carrying class 1 integrons enter the United Kingdom’s environment by the disposal of sewage sludge each year [77]. In another study, the investigated β–lactamase genes (blaTEM and blaCTX-M9) and mecA encoding for penicillin-binding protein were detected in all DNA phages that have been recovered from urban sewage and river water samples [75].
Collectively, the analysed studies demonstrate the potential role of wastewater (particularly untreated-, hospital- and pharmaceutical wastewaters) as an environmental reservoir that assists in the emergence and dissemination of novel variants of AMR bacteria. This is mainly promoted through the coexistence of diverse species of bacteria and high levels of ARGs in these environments, which increases the probability of the transfer of ARGs carried on mobile elements among closely related species. The untreated wastewaters also contain high levels of antimicrobial drugs, which pose an important selective pressure for the emergence and dissemination of AMR bacteria [98]. Recently, positive correlations were observed between the occurrence of heavy metals (e.g., zinc and lead and ereB, mefA&E and ermB) and antibacterial residues (e.g., triclosan with ereA, ereB, mefA&E and ermB) in urban wastewaters and the presence of erythromycin resistant genes [87]. However, the dynamic of the selective pressure and the emergence of novel variants of ARBs remain poorly documented and understood.
3.4. Hospital Wastewater and the Dissemination of Clinically Relevant ARGs and ARBs Populations
Recent studies have reported the detection of elevated levels of clinically important AMR bacteria and ARGs in hospital effluent wastewater and environmental water sources that receive untreated hospital waste [39,46,47,50,58]. Clinically important AMR bacteria, including MDR (e.g., carbapenemase-producing Enterobacteriaceae) and ESBL-producing bacteria (e.g., ESBL-producing K. pneumoniae) [39,50,58] and vancomycin and ampicillin-resistant Enterococcus spp., have been identified in hospital wastewater-associated samples [46,47].
3.5. Impact of Wastewater Treatment Processes on AMR Dissemination
Conventional and advanced WWTPs have employed different biological, physical and chemical process to clean wastewater from pollutants and contaminants so that they can be reused and/or returned back to the environment. The efficiency of removal of AMR bacteria from effluent wastewater (treated) varies according to the treatment procedure employed [99]. Therefore, it is not surprising that high levels of clinically important AMR bacteria, including MDR and ESBL-producing strains of K. pneumoniae, Enterobacter cloacae and E. coli [58]; MDR Listeria spp. [71] and MDR Vibrio spp. [72], have been detected in effluent samples. Another study demonstrated the detection of MDR E. coli strains associated with neonatal meningitis, intestinal and extraintestinal serotypes in final effluents of WWTPs [52]. High levels of resistance were identified in BFG bacteria isolated from WWTPs compared to those that have been recovered from human faeces investigated [40].
Importantly, there is growing evidence that wastewater treatment does not have a profound impact on eliminating the ARGs present in hospital wastewater, with no significant difference in ARG abundance between influent and effluent hospital wastewater samples [54]. Another study demonstrated the high abundance of ARGs and MGEs, including plasmids, transposons, integrons and insertion sequences among samples collected during different treatment processes using aerobic activation (aerobic-activated sludge (AAS)) or anaerobic digestion (anaerobically digested sludge (ADS)) [48]. However, a distinct microbial population has been identified in AAS compared to ADS samples, which suggests a role for the treatment process in promoting the dissemination of particular resistance patterns [48]. A number of recent studies have also demonstrated that novel ARG-bearing plasmids and ARGs that confer resistance to multiple clinically relevant antimicrobial drugs, including aminoglycoside and β-lactams, were highly enriched in activated sludge and effluent wastewater [57,59,61,89,92]. Additionally, activated sludge investigated in one study was found to contain varied ARBs, including ESBL-Enterobacteriaceae, MRSA and VRE, and several ARGs associated with resistance to β-lactam, vancomycin (vanA) and methicilin (mecA) [95].
Consistently, Gram-negative and -positive isolates dominated in WWTP influent and effluent samples, respectively, with the frequency of detection of tetracycline-, sulphonamide-, streptomycin- and β-lactam-resistance genes (except sulA and blaCTX-M) being higher in ARB from influent compared to effluent samples [89]. The abundance of intI1-bearing bacteria (including E. coli, Klebsiella spp. and Aeromonas veronii) were higher in effluent compared to influent wastewater, with intI1 being detected in 20.4%, 30.9% and 38.9% of bacteria recovered from influent, activated sludge and effluent wastewater, respectively [78]. In another study, MDR Enterobacteriaceae strains carrying class 1 and class 2 integrons (12.1%; 221 out of 1832) were identified in different stages of a municipal wastewater treatment plant (61.5%, 12.7% and 25.8% of ARB originated from raw sewage, aeration tank and final effluent, respectively) [81]. However, the abundance of ARGs and MDR bacteria, particularly the levels of ARG diversity and β-lactamase-producers, were higher in the final effluent samples [81].
Collectively, these studies demonstrated WWTPs as hotspots for the emergence of ARBs and highlighted the impact of the treatment technology employed and potential roles of specific stages of treatment processes, particularly those characterised by high biomass and biodiversity (e.g., activated sludge), in maintaining diverse ARGs and promoting particular populations of ARBs. Advanced treatment processes, including membrane filtration, ozonation and UV-irradiation, are highly efficient in reducing the abundance of AMR in effluent wastewater to levels observed in low-impacted surface water [99].
3.6. AMR Dissemination in Wastewater Associated with Mass Gathering Settings
Most of the studies that reported AMR in wastewater have been conducted within one or a few countries. The majority of the studies were conducted in Asia (n = 22), followed by Europe (n = 23), Africa (n = 10), South America (n = 9), North America (n = 7), Central America (n = 1) and Oceania (n = 2).
No studies have been conducted to investigate the transmission of AMR bacteria and ARGs in environmental samples associated with key religious mass gatherings (Kumbh Mela and Hajj) occurring in low-income settings. Kumbh Mela and Hajj are the largest and most diverse mass gathering events that have been associated with an increased risk of infectious disease emergence and transmission [100,101].
Kumbh Mela, the world’s largest religious gathering that attracts millions of Hindu pilgrims, is celebrated at four riverbank pilgrimage sites, including Ganges-Yamuna-mythical Saraswati rivers confluence, Ganges, Godavari and Shipra [101]. The bathing of the pilgrims in these rivers is one of the key rituals, as they believe that it cleanses them of their sins. This raises serious public health issues with regards to the dissemination of waterborne diseases in a setting known to be endemic for cholera [102,103]. Recently a number of studies using metagenomic approaches have detected high levels of ARB, ARGs and antimicrobial residues in water and sediment samples collected from the Ganges River [104]. In addition, ARGs related to different classes of clinically relevant antimicrobial drugs, including ß–lactams, aminoglycosides, fluoroquinolones, macrolides-lincosamide-streptogramins (MLS), rifampicin and sulphonamides, have been identified in samples collected from the confluence of the river Ganges with Yamuna [105].
Hajj has already been associated with an increased risk of airborne, foodborne and zoonotic infections [100]. Recent studies have demonstrated that pilgrims are at high potential risk of acquiring and transmitting AMR enteric bacteria, [106,107,108,109] including carbapenemase-producing E. coli [110] and extended-spectrum cephalosporin- and colistin-resistant non-typhoidal Salmonella [111], as well as MDR Acinetobacter spp. [110].
4. Discussion
The release of antimicrobial drugs, ARBs and ARGs originating from human and animal waste to the environment is a global problem that has serious implications on public health. Therefore, strengthening knowledge on the spread of AMR through surveillance and research was one of the key strategic objectives of the WHO global action plan that was launched in 2015 [112]. Here, we systematically analysed the recent literature to highlight the contribution of different types of wastewaters from various sources (e.g., low- and high-income countries and mass gathering settings) to the emergence, persistence and transmission of AMR in environments and their potential impacts on public health.
The analysed studies highlighted the role of wastewaters as major sources of antimicrobial agents, ARBs and ARGs in the environment. Particular types of wastewaters (e.g., untreated municipal-, hospital- and pharmaceutical wastewaters) have been characterised by high levels of clinically relevant ARBs and ARGs. These environments can provide an ideal platform allowing the transfer of ARGs among the bacterial populations either before or after being discharged into the environment. This is alarming considering that many clinically relevant bacterial species, including enterotoxigenic E. coli and typhoidal and non-typhoidal serotypes of Salmonella, have been shown to be able to persist in the environmental water for relatively long times [113,114,115].
Wastewater treatments have been shown to be effective in reducing the ARB loads in effluent samples. However, there is increasing evidence that effluent samples from wastewater treatment plants, wastewater discharges of pharmaceutical production facilities, hospitals and other healthcare facilities are hotspots (ideal platforms) for selective pressure processes that promote the emergence and dissemination of novel AMR mechanisms and new variants of ARBs and ARGs. However, it is noted that the positive selection process and the dynamics of the emergence of novel variants of ARGs and ARBs within WWTPs and their associated impacts on human health remain poorly documented and characterised. The WHO has highlighted the need for greater attention and action to develop quantitative microbial risk assessments and supporting guidance to address human health risks associated with environmental exposures to antimicrobial agents, their metabolites, ARBs and ARGs [32].
Interestingly, a recent pioneer study has proposed a culture-independent metagenomic analysis of untreated wastewater as an effective approach to track and predict the dissemination of AMR bacteria and ARGs globally [41]. The authors of this study have used a standardised metagenomic protocol to characterise the bacterial resistome content and to detect variations in the abundance and diversity of ARBs and ARGs in a global collection of untreated wastewater samples (collected from 79 sites in 60 countries). This study demonstrated that clinically relevant ARGs were more abundant in samples collected from low- and middle-income settings in Africa, Asia and South America, compared to those that have been collected from high-income settings in Europe, North America and Oceania. The variations in AMR gene abundance were found to strongly correlate with socioeconomic, health and environmental factors [41].
This approach can be applied in challenging settings (e.g., such as low-income countries and complicated mass gathering settings) to study the paradigm of AMR dynamics and epidemiology and inform on the processes leading to the emergence and the dissemination of AMR infectious agents and, therefore, help in developing management strategies.
We conducted a research study that uses the opportunity presented each year by the Hajj pilgrimage and advanced shotgun-based metagenomic approaches to characterise the global population of enteric microorganisms circulating in environmental Hajj settings. This will provide an annual snapshot of the AMR bacteria and MGEs associated with each global locality and help in identifying the dynamics of emergence and dissemination of AMR in the environment.
Hajj is a unique mass gathering event that has been associated with an increased potential for the emergence and dissemination of AMR infections, raising major public health concerns within the host country and globally. The enormously diverse population of 3 million pilgrims, originating from 190 countries all over the world come together to perform the same activities within a relatively short period of time and over a limited area of land. Importantly, the pilgrims are required to stay in tents in Mina (a nonpopulated valley covering approximately 20 km2 of land, of which only 4 km2 can be occupied by pilgrims) for at least 3–5 days. The pilgrims are distributed in campaigns across Mina according to their geographical origin (i.e., country of origin). The wastewater is disposed of through septic tanks (onsite sewage facilities) that are associated with the pilgrims’ campaign. We conducted the first study to use shotgun-based metagenomic analysis to characterise the abundance and distribution of microbial communities and resistance determinants in wastewater samples from septic tanks in Mina representing different campaigns (European, Middle East and North African (MENA) and East and Southeast Asian countries). The results indicated that high levels of ARGs, including ESBL and aminoglycoside markers, were detected in all sites tested. However, significant variations in the distribution of the bacterial species and the abundance of ARGs were identified.
Similarly, Kumbh Mela in India represents the world’s largest periodic mass gathering event that involving bathing in small-specified rivers sites. Recent studies have highlighted the striking impact of mass bathing on river ecosystems, including the AMR microbial contents and dissemination of human infectious agents [105,116,117]. A recent study found a nearly 130-fold increase in bacterial load of human origin during the event. Moreover, metagenomic analyses demonstrated an increase in virulence and ARG loads during the MGEs [118].
Many studies have highlighted the roles of surface fresh and aquatic water, rural groundwater and sewage in the dissemination of AMR pathogens. The emergence of AMRs is part of a complicated ecological and evolutionary network, with the use of antimicrobial drugs anywhere within the system potentially selecting for resistance to that drug elsewhere in the network [23]. Gram-negative bacterial resistance, in particular, is promoted through horizontal gene transfer by the acquisition of mobile elements [119,120,121]. There is also increasing evidence that ARGs found in human microbial communities are likely to have been acquired from an environmental source [122,123]. The processing of human, farm and industrial waste together has a significant impact on the emergence of AMR to a wide range of the most clinically effective antibiotics [124,125]. In addition, even treated sewage samples discharged into rivers or lakes from treatment plants may contain significant concentrations of ARGs that enhance the development of AMR bacteria and raise major public health concerns [24,126,127,128].
Supplementary Materials
The following are available online at https://www.mdpi.com/2414-6366/5/1/33/s1, Table S1: Characterisation of studies detecting disseminated AMR genes in sewage.
Author Contributions
Conceptualisation, M.A.E.G.; data extraction, K.N.A.P. and N.F.; formal analysis, K.N.A.P., N.F. and M.I.A.; writing—original draft preparation, M.A.E.G., K.N.A.P. and N.F.; writing—review and editing, G.A.H.-C. and M.A.E.G. and supervision, M.Y., A.L.A.-M., E.I.A., G.A.H.-C. and M.A.E.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014. Available online: https://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 22 June 2019).
- Review on Antimicrobial Resistance. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 25 June 2019).
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/en/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 25 June 2019).
- World Health Organization. WHO Report on Surveillance of Antibiotic Consumption 2016–2018 Early Implementation. Available online: https://www.who.int/medicines/areas/rational_use/oms-amr-amc-report-2016-2018/en/ (accessed on 22 June 2019).
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report Early Implementation 2017–2018. Available online: https://www.who.int/glass/resources/publications/early-implementation-report-2017-2018/en/ (accessed on 26 June 2019).
- Central for Disease Control and Prevention. Infographic: Antibiotic Resistance The Global Threat. Available online: https://www.cdc.gov/globalhealth/infographics/antibiotic-resistance/antibiotic_resistance_global_threat.htm (accessed on 22 June 2019).
- World Health Organization. Report on Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Dicovery, and Development of New Antibiotics. Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 26 June 2019).
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Schwartz, K.L.; Morris, S.K. Travel and the Spread of Drug-Resistant Bacteria. Curr. Infect. Dis. 2018, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Venkatesh, S.; Shibl, A.M. Impact of travel on international spread of antimicrobial resistance. Int. J. Antimicrob. Agents 2003, 21, 135–142. [Google Scholar] [CrossRef]
- Bell, B.G.; Schellevis, F.; Stobberingh, E.; Goossens, H.; Pringle, M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect. Dis. 2014, 14, 13. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef]
- Brinkac, L.; Voorhies, A.; Gomez, A.; Nelson, K.E. The Threat of Antimicrobial Resistance on the Human Microbiome. Microb. Ecol. 2017, 74, 1001–1008. [Google Scholar] [CrossRef]
- Mendelson, M.; Rottingen, J.A.; Gopinathan, U.; Hamer, D.H.; Wertheim, H.; Basnyat, B.; Butler, C.; Tomson, G.; Balasegaram, M. Maximising access to achieve appropriate human antimicrobial use in low-income and middle-income countries. Lancet 2016, 387, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial and Primary Health Care:Brief. Available online: https://apps.who.int/iris/bitstream/handle/10665/328084/WHO-HIS-SDS-2018.57-eng.pdf (accessed on 26 June 2019).
- Frost, I.; Van Boeckel, T.P.; Pires, J.; Craig, J.; Laxminarayan, R. Global Geographic Trends in Antimicrobial Resistance: The Role of International Travel. J. Travel Med. 2019. [Google Scholar] [CrossRef]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef] [PubMed]
- The Parliamentary Office of Science and Technology, H.o.P.U. Reservoir of Antimicrobial Resistance. Available online: https://researchbriefings.parliament.uk/ResearchBriefing/Summary/POST-PN-0595 (accessed on 26 June 2019).
- Food and Agricultural Organization of United Nations. Antimicrobial Resistance in the Environment: Summary Report of an FAO Meeting of Experts FAO Antimicrobial Resistance Working Group. Available online: http://www.fao.org/3/BU656en/bu656en.pdf (accessed on 26 June 2019).
- >European Medicine Agency. Reflection Paper on Antimicrobial Resistance in the Environment: Considerations for Current and Future Risk Assessment of Veterinary Medicinal Products (Draft). Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/draft-reflection-paper-antimicrobial-resistance-environment-considerations-current-future-risk_en.pdf (accessed on 26 June 2019).
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Balcazar, J.L. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014, 10, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Berglund, B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 2015, 5, 28564. [Google Scholar] [CrossRef]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotox. Environ. Safe. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- McKinney, C.W.; Dungan, R.S.; Moore, A.; Leytem, A.B. Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Rizzo, L.; Fiorentino, A.; Anselmo, A. Advanced treatment of urban wastewater by UV radiation: Effect on antibiotics and antibiotic-resistant E. coli strains. Chemosphere 2013, 92, 171–176. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial resistance: An Emerging Water, Sanitation and Hygiene Issue. Briefing Note. Available online: https://www.who.int/water_sanitation_health/emerging/AMR_briefing_note.pdf (accessed on 22 June 2019).
- Treangen, T.J.; Rocha, E.P. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011, 7, e1001284. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Denzler, R.; Maier, L.; Bernet, F.; Sanders, M.J.; Pickard, D.J.; Barthel, M.; Westendorf, A.M.; Krogfelt, K.A.; Walker, A.W.; et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2012, 109, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Nahar, A.; Islam, M.A.; Sobur, M.A.; Hossain, M.J.; Binte, S.; Zaman, M.; Rahman, B.; Kabir, S.L.; Rahman, M.T. Detection of tetracycline resistant E. coli and Salmonella spp. in sewage, river, pond and swimming pool in Mymensingh, Bangladesh. Afr. J. Microbiol. Res. 2018. [Google Scholar] [CrossRef]
- Qin, J.; Maixnerova, M.; Nemec, M.; Feng, Y.; Zhang, X.; Nemec, A.; Zong, Z. Acinetobacter cumulans sp. nov., isolated from hospital sewage and capable of acquisition of multiple antibiotic resistance genes. Syst. Appl. Microbiol. 2019, 42, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Haberecht, H.B.; Nealon, N.J.; Gilliland, J.R.; Holder, A.V.; Runyan, C.; Oppel, R.C.; Ibrahim, H.M.; Mueller, L.; Schrupp, F.; Vilchez, S.; et al. Antimicrobial-Resistant Escherichia coli from Environmental Waters in Northern Colorado. J. Environ. Public Health 2019, 2019, 3862949. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Yatsu, K.; Inamine, Y.; Segawa, T.; Nishio, M.; Kishi, N.; Kuroda, M. Complete Genome Sequence of a blaKPC-2-Positive Klebsiella pneumoniae Strain Isolated from the Effluent of an Urban Sewage Treatment Plant in Japan. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, A.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C.; et al. Hospital effluent: A reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Niestepski, S.; Harnisz, M.; Korzeniewska, E.; Aguilera-Arreola, M.G.; Contreras-Rodriguez, A.; Filipkowska, Z.; Osinska, A. The emergence of antimicrobial resistance in environmental strains of the Bacteroides fragilis group. Environ. Int. 2019, 124, 408–419. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Roder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Khan, F.A.; Soderquist, B.; Jass, J. Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front. Microbiol. 2019, 10, 688. [Google Scholar] [CrossRef]
- Tokajian, S.; Moghnieh, R.; Salloum, T.; Arabaghian, H.; Alousi, S.; Moussa, J.; Abboud, E.; Youssef, S.; Husni, R. Extended-spectrum beta-lactamase-producing Escherichia coli in wastewaters and refugee camp in Lebanon. Future Microbiol. 2018, 13, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed]
- Bougnom, B.P.; Zongo, C.; McNally, A.; Ricci, V.; Etoa, F.X.; Thiele-Bruhn, S.; Piddock, L.J.V. Wastewater used for urban agriculture in West Africa as a reservoir for antibacterial resistance dissemination. Environ. Res. 2019, 168, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Gouliouris, T.; Raven, K.E.; Moradigaravand, D.; Ludden, C.; Coll, F.; Blane, B.; Naydenova, P.; Horner, C.; Brown, N.M.; Corander, J.; et al. Detection of vancomycin-resistant Enterococcus faecium hospital-adapted lineages in municipal wastewater treatment plants indicates widespread distribution and release into the environment. Genome Res. 2019, 29, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Gaqavu, S.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in alice, eastern cape province of South Africa. Int. J. Environ. Res. Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Wang, J.; Mao, D.; Mu, Q.; Luo, Y. Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Sci. Total Environ. 2015, 526, 366–373. [Google Scholar] [CrossRef]
- Conte, D.; Palmeiro, J.K.; da Silva Nogueira, K.; de Lima, T.M.; Cardoso, M.A.; Pontarolo, R.; Degaut Pontes, F.L.; Dalla-Costa, L.M. Characterization of CTX-M enzymes, quinolone resistance determinants, and antimicrobial residues from hospital sewage, wastewater treatment plant, and river water. Ecotoxicol. Environ. Saf. 2017, 136, 62–69. [Google Scholar] [CrossRef]
- Baumlisberger, M.; Youssar, L.; Schilhabel, M.B.; Jonas, D. Influence of a non-hospital medical care facility on antimicrobial resistance in wastewater. PLoS ONE 2015, 10, e0122635. [Google Scholar] [CrossRef]
- Adefisoye, M.A.; Okoh, A.I. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiol. Open 2016, 5, 143–151. [Google Scholar] [CrossRef]
- Suzuki, S.; Ogo, M.; Koike, T.; Takada, H.; Newman, B. Sulfonamide and tetracycline resistance genes in Total- and culturable-bacterial assemblages in South African aquatic environments. Front. Microbiol. 2015, 6, 796. [Google Scholar] [CrossRef] [PubMed]
- Froes, A.M.; da Mota, F.F.; Cuadrat, R.R.; Davila, A.M. Distribution and Classification of Serine beta-Lactamases in Brazilian Hospital Sewage and Other Environmental Metagenomes Deposited in Public Databases. Front. Microbiol. 2016, 7, 1790. [Google Scholar] [CrossRef] [PubMed]
- Laht, M.; Karkman, A.; Voolaid, V.; Ritz, C.; Tenson, T.; Virta, M.; Kisand, V. Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS ONE 2014, 9, e103705. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, X.X.; Ye, L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE 2011, 6, e26041. [Google Scholar] [CrossRef]
- Chagas, T.P.; Seki, L.M.; Cury, J.C.; Oliveira, J.A.; Davila, A.M.; Silva, D.M.; Asensi, M.D. Multiresistance, beta-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2011, 111, 572–581. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.H.; Gutzkow, T.; Eichler, W.; Puhler, A.; Schluter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef]
- Li, D.; Yu, T.; Zhang, Y.; Yang, M.; Li, Z.; Liu, M.; Qi, R. Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river. Appl. Environ. Microbiol. 2010, 76, 3444–3451. [Google Scholar] [CrossRef]
- Zhang, H.; He, H.; Chen, S.; Huang, T.; Lu, K.; Zhang, Z.; Wang, R.; Zhang, X.; Li, H. Abundance of antibiotic resistance genes and their association with bacterial communities in activated sludge of wastewater treatment plants: Geographical distribution and network analysis. J. Environ. Sci. 2019, 82, 24–38. [Google Scholar] [CrossRef]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME 2015, 9, 2490–2502. [Google Scholar] [CrossRef]
- Hembach, N.; Schmid, F.; Alexander, J.; Hiller, C.; Rogall, E.T.; Schwartz, T. Occurrence of the mcr-1 Colistin Resistance Gene and other Clinically Relevant Antibiotic Resistance Genes in Microbial Populations at Different Municipal Wastewater Treatment Plants in Germany. Front. Microbiol. 2017, 8, 1282. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, I.H.; Okoh, A.I. Antibiotic susceptibility profile of Aeromonas species isolated from wastewater treatment plant. Sci. World J. 2012, 2012, 764563. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, E.O.; Obi, L.C.; Tom, M.; Okoh, A.I. Detection of potential risk of wastewater effluents for transmission of antibiotic resistance from Vibrio species as a reservoir in a peri-urban community in South Africa. Int. J. Environ. Health Res. 2011, 21, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Johnning, A.; Kristiansson, E.; Fick, J.; Weijdegard, B.; Larsson, D.G. Resistance Mutations in gyrA and parC are Common in Escherichia Communities of both Fluoroquinolone-Polluted and Uncontaminated Aquatic Environments. Front. Microbiol. 2015, 6, 1355. [Google Scholar] [CrossRef] [PubMed]
- Sahlström, L.; Rehbinder, V.; Albihn, A.; Aspan, A.; Bengtsson, B. Vancomycin resistant enterococci (VRE) in Swedish sewage sludge. Acta Vet. Scand. 2009, 51, 24. [Google Scholar] [CrossRef]
- Araújo, C.; Torres, C.; Silva, N.; Carneiro, C.; Gonçalves, A.; Radhouani, H.; Correia, S.; da Costa, P.M.; Paccheco, R.; Zarazaga, M. Vancomycin-resistant enterococci from Portuguese wastewater treatment plants. J. Basic Microbiol. 2010, 50, 605–609. [Google Scholar] [CrossRef]
- Soge, O.; Tivoli, L.; Meschke, J.; Roberts, M. A conjugative macrolide resistance gene, mef(A), in environmental Clostridium perfringens carrying multiple macrolide and/or tetracycline resistance genes. J. Appl. Microbiol. 2009, 106, 34–40. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Zhang, T.; Zhang, M.; Fang, H.H.P.; Cheng, S.-P. Characterization and quantification of class 1 integrons and associated gene cassettes in sewage treatment plants. Appl. Microbiol. Biotechnol. 2009, 82, 1169–1177. [Google Scholar] [CrossRef]
- Odjadjare, E.E.O.; Obi, L.C.; Okoh, A.I. Municipal Wastewater Effluents as a Source of Listerial Pathogens in the Aquatic Milieu of the Eastern Cape Province of South Africa: A Concern of Public Health Importance. Int. J. Environ. Res. Public Health 2010, 7, 2376–2394. [Google Scholar] [CrossRef]
- Okoh, A.I.; Igbinosa, E.O. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 2010, 10, 143. [Google Scholar] [CrossRef]
- Yang, H.; Byelashov, O.A.; Geornaras, I.; Goodridge, L.D.; Nightingale, K.K.; Belk, K.E.; Smith, G.C.; Sofos, J.N. Presence of antibiotic-resistant commensal bacteria in samples from agricultural, city, and national park environments evaluated by standard culture and real-time PCR methods. Can. J. Microbiol. 2010, 56, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Armisen, T.; Vercammen, K.; Passerat, J.; Triest, D.; Servais, P.; Cornelis, P. Antimicrobial resistance of heterotrophic bacteria in sewage-contaminated rivers. Water Res. 2011, 45, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef] [PubMed]
- Fuentefria, D.B.; Ferreira, A.E.; Corcao, G. Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: Are they genetically related? J. Environ. Manag. 2011, 92, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Zhang, L.; Abdouslam, N.A.; Hawkey, P.M.; Calvo-Bado, L.; Royle, J.; Brown, H.; Davis, S.; Kay, P.; Boxall, A.B.; et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME 2011, 5, 1253–1261. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, X.X.; Cheng, S.; Zhang, Z.; Shi, P.; Liu, B.; Wu, B.; Zhang, Y. Occurrence, abundance and elimination of class 1 integrons in one municipal sewage treatment plant. Ecotoxicology 2011, 20, 968–973. [Google Scholar] [CrossRef]
- Mokracka, J.; Koczura, R.; Jabłońska, L.; Kaznowski, A. Phylogenetic groups, virulence genes and quinolone resistance of integron-bearing Escherichia coli strains isolated from a wastewater treatment plant. Antonie Van Leeuwenhoek 2011, 99, 817–824. [Google Scholar] [CrossRef]
- Amaya, E.; Reyes, D.; Paniagua, M.; Calderón, S.; Rashid, M.U.; Colque, P.; Kühn, I.; Möllby, R.; Weintraub, A.; Nord, C.E. Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in Leon, Nicaragua. Clin. Microbiol. Infect. 2012, 18, E347–E354. [Google Scholar] [CrossRef]
- Mokracka, J.; Koczura, R.; Kaznowski, A. Multiresistant Enterobacteriaceae with class 1 and class 2 integrons in a municipal wastewater treatment plant. Water Res. 2012, 46, 3353–3363. [Google Scholar] [CrossRef]
- Spindler, A.; Otton, L.M.; Fuentefria, D.B.; Corcao, G. Beta-lactams resistance and presence of class 1 integron in Pseudomonas spp. isolated from untreated hospital effluents in Brazil. Antonie Van Leeuwenhoek 2012, 102, 73–81. [Google Scholar] [CrossRef]
- Gundogdu, A.; Jennison, A.V.; Smith, H.V.; Stratton, H.; Katouli, M. Extended-spectrum beta-lactamase producing Escherichia coli in hospital wastewaters and sewage treatment plants in Queensland, Australia. Can. J. Microbiol. 2013, 59, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Zarfel, G.; Galler, H.; Feierl, G.; Haas, D.; Kittinger, C.; Leitner, E.; Grisold, A.J.; Mascher, F.; Posch, J.; Pertschy, B.; et al. Comparison of extended-spectrum-beta-lactamase (ESBL) carrying Escherichia coli from sewage sludge and human urinary tract infection. Environ. Pollut. 2013, 173, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Mora, A.; Lopez, C.; Mamani, R.; Dahbi, G.; Marzoa, J.; Herrera, A.; Viso, S.; Blanco, J.E.; Blanco, M.; et al. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b: H4-B2-ST131 and O25b: H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J. Antimicrob. Chemother. 2013, 68, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Sadowy, E.; Luczkiewicz, A. Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol. 2014, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; He, S.; Huang, S.; Li, K.; Liu, Z.; Xue, G.; Sun, W. Impacts of coexisting antibiotics, antibacterial residues, and heavy metals on the occurrence of erythromycin resistance genes in urban wastewater. Appl. Microbiol. Biotechnol. 2015, 99, 3971–3980. [Google Scholar] [CrossRef]
- Nishiyama, M.; Iguchi, A.; Suzuki, Y. Identification of Enterococcus faecium and Enterococcus faecalis as vanC-type Vancomycin-Resistant Enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. J. Environ. Sci. Health 2015, 50, 16–25. [Google Scholar] [CrossRef]
- Zhang, S.; Han, B.; Gu, J.; Wang, C.; Wang, P.; Ma, Y.; Cao, J.; He, Z. Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere 2015, 135, 138–145. [Google Scholar] [CrossRef]
- Simo Tchuinte, P.L.; Stalder, T.; Venditti, S.; Ngandjio, A.; Dagot, C.; Ploy, M.C.; Barraud, O. Characterisation of class 3 integrons with oxacillinase gene cassettes in hospital sewage and sludge samples from France and Luxembourg. Int. J. Antimicrob. Agents 2016, 48, 431–434. [Google Scholar] [CrossRef]
- Young, S.; Nayak, B.; Sun, S.; Badgley, B.D.; Rohr, J.R.; Harwood, V.J. Vancomycin-Resistant Enterococci and Bacterial Community Structure following a Sewage Spill into an Aquatic Environment. Appl. Environ. Microbiol. 2016, 82, 5653. [Google Scholar] [CrossRef]
- Lee, J.; Shin, S.G.; Jang, H.M.; Kim, Y.B.; Lee, J.; Kim, Y.M. Characterization of antibiotic resistance genes in representative organic solid wastes: Food waste-recycling wastewater, manure, and sewage sludge. Sci. Total Environ. 2017, 579, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- An, X.L.; Chen, Q.L.; Zhu, D.; Zhu, Y.G.; Gillings, M.R.; Su, J.Q. Impact of Wastewater Treatment on the Prevalence of Integrons and the Genetic Diversity of Integron Gene Cassettes. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y. Occurrence and characteristics of extended-spectrum beta-lactamase- and carbapenemase- producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 2018, 615, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Habib, J.; Kittinger, C.; Luxner, J.; Zarfel, G. Multiresistant Bacteria Isolated from Activated Sludge in Austria. Int. J. Environ. Res. Public Health 2018, 15, 479. [Google Scholar] [CrossRef] [PubMed]
- Quach-Cu, J.; Herrera-Lynch, B.; Marciniak, C.; Adams, S.; Simmerman, A.; Reinke, R.A. The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions. Water 2018, 10, 37. [Google Scholar] [CrossRef]
- Yousfi, K.; Touati, A.; Lefebvre, B.; Garneau, P.; Brahmi, S.; Gharout-Sait, A.; Harel, J.; Bekal, S. Characterization of multidrug-resistant Gram-negative bacilli isolated from hospitals effluents: First report of a blaOXA-48-like in Klebsiella oxytoca, Algeria. Braz. J. Microbiol. 2019, 50, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Flandroy, L.; Poutahidis, T.; Berg, G.; Clarke, G.; Dao, M.C.; Decaestecker, E.; Furman, E.; Haahtela, T.; Massart, S.; Plovier, H.; et al. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci. Total Environ. 2018, 627, 1018–1038. [Google Scholar] [CrossRef]
- Hiller, C.X.; Hubner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef]
- Ahmed, Q.A.; Arabi, Y.M.; Memish, Z.A. Health risks at the Hajj. Lancet 2006, 367, 1008–1015. [Google Scholar] [CrossRef]
- Sridhar, S.; Gautret, P.; Brouqui, P. A comprehensive review of the Kumbh Mela: Identifying risks for spread of infectious diseases. Clin. Microbiol. Infect. 2015, 21, 128–133. [Google Scholar] [CrossRef]
- Mutreja, A.; Kim, D.W.; Thomson, N.R.; Connor, T.R.; Lee, J.H.; Kariuki, S.; Croucher, N.J.; Choi, S.Y.; Harris, S.R.; Lebens, M.; et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 2011, 477, 462–465. [Google Scholar] [CrossRef]
- Abd El Ghany, M.; Chander, J.; Mutreja, A.; Rashid, M.; Hill-Cawthorne, G.A.; Ali, S.; Naeem, R.; Thomson, N.R.; Dougan, G.; Pain, A. The population structure of Vibrio cholerae from the Chandigarh Region of Northern India. PLoS Negl. Trop. Dis. 2014, 8, e2981. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.; Dubey, S.K. River Ganges water as reservoir of microbes with antibiotic and metal ion resistance genes: High throughput metagenomic approach. Environ. Pollut. 2019, 246, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.; Shah, M.; Yadav, R.; Sarode, P.; Rajput, V.; Dastager, S.G.; Dharne, M.S.; Khairnar, K. Metagenomic insights to understand transient influence of Yamuna River on taxonomic and functional aspects of bacterial and archaeal communities of River Ganges. Sci. Total Environ. 2019, 674, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Abd El Ghany, M.; Alsomali, M.; Almasri, M.; Padron Regalado, E.; Naeem, R.; Tukestani, A.; Asiri, A.; Hill-Cawthorne, G.A.; Pain, A.; Memish, Z.A. Enteric Infections Circulating during Hajj Seasons, 2011–2013. Emerg. Infect. Dis. 2017, 23. [Google Scholar] [CrossRef] [PubMed]
- Abd El Ghany, M.; Al-Tawfiq, J.A.; Hill-Cawthorne, G.A.; Memish, Z.A. Hajj-Beyond traveller’s diarrhea. Travel Med. Infect. Dis. 2018, 21, 80–81. [Google Scholar] [CrossRef]
- Leangapichart, T.; Rolain, J.M.; Memish, Z.A.; Al-Tawfiq, J.A.; Gautret, P. Emergence of drug resistant bacteria at the Hajj: A systematic review. Travel Med. Infect. Dis. 2017, 18, 3–17. [Google Scholar] [CrossRef]
- Abd El Ghany, M.; Sharaf, H.; Hill-Cawthorne, G.A. Hajj vaccinations-facts, challenges, and hope. Int. J. Infect. Dis. 2016, 47, 29–37. [Google Scholar] [CrossRef][Green Version]
- Leangapichart, T.; Gautret, P.; Griffiths, K.; Belhouchat, K.; Memish, Z.; Raoult, D.; Rolain, J.M. Acquisition of a High Diversity of Bacteria during the Hajj Pilgrimage, Including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 Carbapenemase Genes. Antimicrob. Agents Chemother. 2016, 60, 5942–5948. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Dia, N.M.; Gautret, P.; Benkouiten, S.; Belhouchat, K.; Drali, T.; Parola, P.; Brouqui, P.; Memish, Z.; Raoult, D.; et al. Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj. Int. J. Antimicrob. Agents 2015, 45, 600–604. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. Available online: https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/ (accessed on 22 June 2019).
- Lothigius, A.; Sjoling, A.; Svennerholm, A.M.; Bolin, I. Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J. Appl. Microbiol. 2010, 108, 1441–1449. [Google Scholar] [CrossRef]
- Hernroth, B.; Lothigius, A.; Bolin, I. Factors influencing survival of enterotoxigenic Escherichia coli, Salmonella enterica (serovar Typhimurium) and Vibrio parahaemolyticus in marine environments. FEMS Microbiol. Ecol. 2010, 71, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, R.A.; Langridge, G.; Smith, S.E.; Makendi, C.; Fookes, M.; Wileman, T.M.; El Ghany, M.A.; Keith Turner, A.; Dyson, Z.A.; Sridhar, S.; et al. Functional analysis of Salmonella Typhi adaptation to survival in water. Environ. Microbiol. 2018, 20, 4079–4090. [Google Scholar] [CrossRef] [PubMed]
- Jani, K.; Bandal, J.; Rale, V.; Shouche, Y.; Sharma, A. Antimicrobial resistance pattern of microorganisms isolated and identified from Godavari River across the mass gathering event. J. Biosci. 2019, 44, 121. [Google Scholar] [CrossRef] [PubMed]
- Diwan, V.; Purohit, M.; Chandran, S.; Parashar, V.; Shah, H.; Mahadik, V.K.; Stalsby Lundborg, C.; Tamhankar, A.J. A Three-Year Follow-Up Study of Antibiotic and Metal Residues, Antibiotic Resistance and Resistance Genes, Focusing on Kshipra-A River Associated with Holy Religious Mass-Bathing in India: Protocol Paper. Int. J. Environ. Res. Public Health 2017, 14, 574. [Google Scholar] [CrossRef] [PubMed]
- Jani, K.; Dhotre, D.; Bandal, J.; Shouche, Y.; Suryavanshi, M.; Rale, V.; Sharma, A. World’s Largest Mass Bathing Event Influences the Bacterial Communities of Godavari, a Holy River of India. Microbial. Ecol. 2018, 76, 706–718. [Google Scholar] [CrossRef]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in Enterobacteriaceae: Mechanisms and clinical implications. BMJ 2016, 352, h6420. [Google Scholar] [CrossRef]
- Partridge, S.R. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 2011, 35, 820–855. [Google Scholar] [CrossRef]
- Partridge, S.R. Resistance mechanisms in Enterobacteriaceae. Pathology 2015, 47, 276–284. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opp. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Pruden, A. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Tech. 2014, 48, 5–14. [Google Scholar] [CrossRef]
- Amos, G.C.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 2014, 69, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Dropa, M.; Lincopan, N.; Balsalobre, L.C.; Oliveira, D.E.; Moura, R.A.; Fernandes, M.R.; da Silva, Q.M.; Matte, G.R.; Sato, M.I.; Matte, M.H. Genetic background of novel sequence types of CTX-M-8- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in Sao Paulo, Brazil. Environ. Sci. Pollut. Res. Int. 2016, 23, 4953–4958. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.; Zhang, L.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M. Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Vet. Microbiol. 2014, 171, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.X.; Huang, K.; Miao, Y.; Shi, P.; Liu, B.; Long, C.; Li, A. Metagenomic profiling of antibiotic resistance genes and mobile genetic elements in a tannery wastewater treatment plant. PLoS ONE 2013, 8, e76079. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).