Transposable Elements in the Genome of Human Parasite Schistosoma mansoni: A Review
Abstract
:1. Introduction
2. Transposable Elements: General Concepts and Their Identification in S. mansoni
3. TEs Impact on the S. mansoni Genome
3.1. Influence of Transposition Bursts in Speciation Processes
3.2. Micro-Exon Genes Coevolution Mediated by TEs in S. mansoni
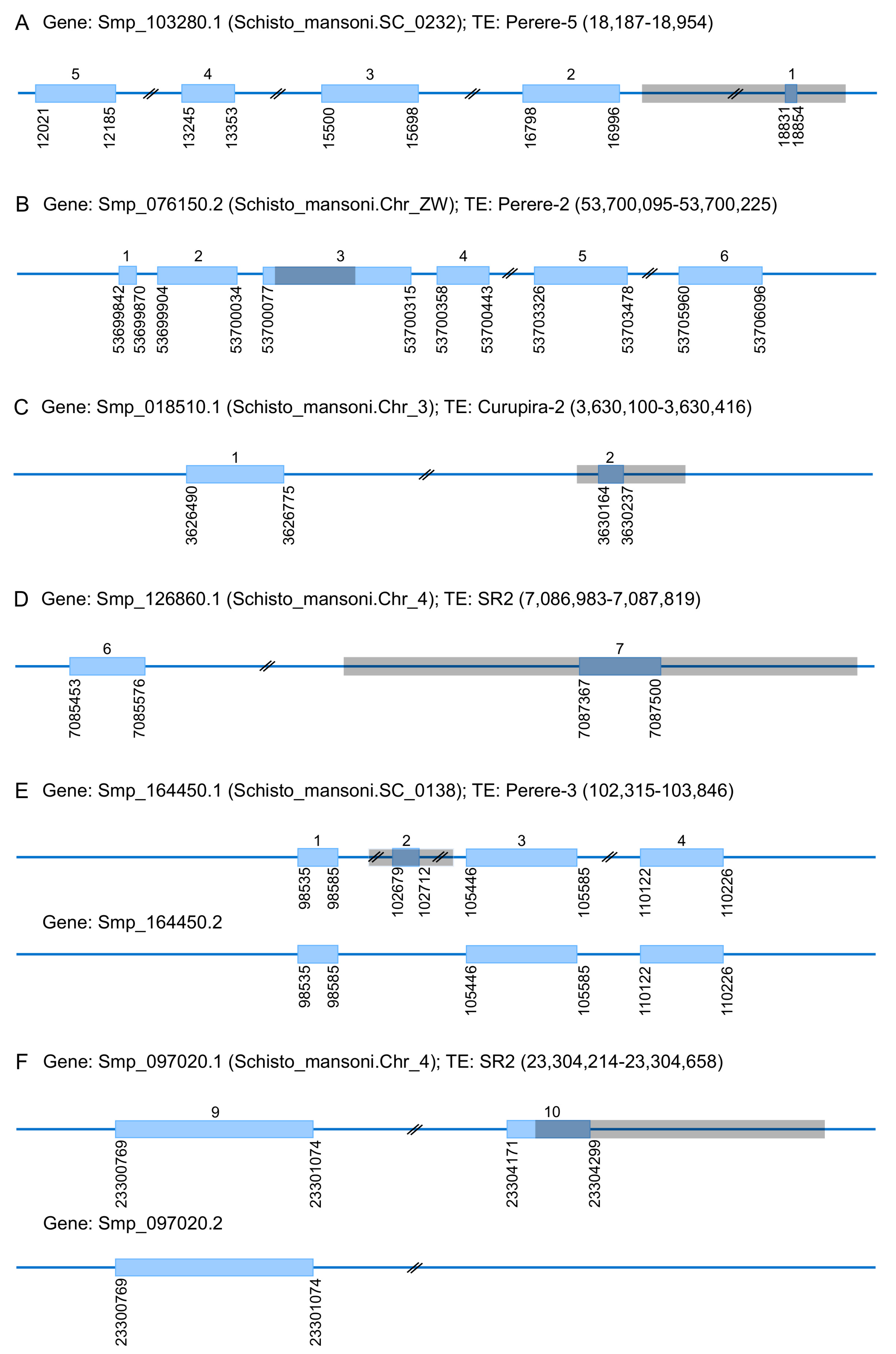
3.3. TEs Contribution to the Evolution of S. mansoni Coding Regions
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- McManus, D.P.; Dunne, D.W.; Sacko, M.; Utzinger, J.; Vennervald, B.J.; Zhou, X.-N. Schistosomiasis. Nat. Rev. Dis. Prim. 2018, 4, 13. [Google Scholar] [CrossRef]
- Molehin, A.J. Schistosomiasis vaccine development: Update on human clinical trials. J. Biomed. Sci. 2020, 27, 28. [Google Scholar] [CrossRef]
- Colley, D.G.; Bustinduy, A.L.; Secor, W.E.; King, C.H. Human schistosomiasis. Lancet 2014, 383, 2253–2264. [Google Scholar] [CrossRef]
- Verjovski-Almeida, S.; Demarco, R. Gene structure and splicing in schistosomes. J. Proteom. 2011, 74, 1515–1518. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Paz, C.; Padalino, G.; Coghlan, A.; Lu, Z.; Gradinaru, I.; Collins, J.N.R.; Berriman, M.; Hoffmann, K.F.; Collins, J.J. Large-scale RNAi screening uncovers therapeutic targets in the parasite Schistosoma mansoni. Science 2020, 369, 1649–1653. [Google Scholar] [CrossRef]
- Verjee, M.A. Schistosomiasis: Still a Cause of Significant Morbidity and Mortality. Res. Rep. Trop. Med. 2020, 10, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Zwang, J.; Olliaro, P.L. Clinical Efficacy and Tolerability of Praziquantel for Intestinal and Urinary Schistosomiasis—A Meta-analysis of Comparative and Non-comparative Clinical Trials. PLoS Negl. Trop. Dis. 2014, 8, e3286. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, R.; Verjovski-Almeida, S. Schistosomes—Proteomics studies for potential novel vaccines and drug targets. Drug Discov. Today 2009, 14, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Gobert, G.N.; You, H.; McManus, D.P. Gaining biological perspectives from schistosome genomes. Mol. Biochem. Parasitol. 2014, 196, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H., Jr. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef] [Green Version]
- Levin, H.L.; Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011, 12, 615–627. [Google Scholar] [CrossRef]
- Schrader, L.; Schmitz, J. The impact of transposable elements in adaptive evolution. Mol. Ecol. 2019, 28, 1537–1549. [Google Scholar] [CrossRef]
- Fedoroff, N.V. Transposable Elements, Epigenetics, and Genome Evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, M.; Peona, V.; Guichard, E.; Taccioli, C.; Boattini, A. Transposable Elements Activity is Positively Related to Rate of Speciation in Mammals. J. Mol. Evol. 2018, 86, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biémont, C. A Brief History of the Status of Transposable Elements: From Junk DNA to Major Players in Evolution. Genetics 2010, 186, 1085–1093. [Google Scholar] [CrossRef] [Green Version]
- Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deininger, P.L.; Batzer, M.A. Alu Repeats and Human Disease. Mol. Genet. Metab. 1999, 67, 183–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, Y.; Nishisho, I.; Horii, A.; Miyoshi, Y.; Utsunomiya, J.; Kinzler, K.W.; Vogelstein, B.; Nakamura, Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992, 52, 643–645. [Google Scholar]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Pappalardo, A.M.; Ferrito, V.; Biscotti, M.A.; Canapa, A.; Capriglione, T. Transposable Elements and Stress in Vertebrates: An Overview. Int. J. Mol. Sci. 2021, 22, 1970. [Google Scholar] [CrossRef]
- Seidl, M.F.; Thomma, B.P.H.J. Transposable Elements Direct the Coevolution between Plants and Microbes. Trends Genet. 2017, 33, 842–851. [Google Scholar] [CrossRef]
- Horváth, V.; Merenciano, M.; González, J. Revisiting the Relationship between Transposable Elements and the Eukaryotic Stress Response. Trends Genet. 2017, 33, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Han, K.; Liang, P. Role of Transposable Elements in Gene Regulation in the Human Genome. Life 2021, 11, 118. [Google Scholar] [CrossRef]
- Nekrutenko, A.; Li, W.-H. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001, 17, 619–621. [Google Scholar] [CrossRef]
- Jordan, I.K.; Rogozin, I.B.; Glazko, G.V.; Koonin, E.V. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003, 19, 68–72. [Google Scholar] [CrossRef]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar] [CrossRef]
- Joly-Lopez, Z.; Forczek, E.; Hoen, D.R.; Juretic, N.; Bureau, T.E. A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1002931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebollo, R.; Romanish, M.T.; Mager, D.L. Transposable Elements: An Abundant and Natural Source of Regulatory Sequences for Host Genes. Annu. Rev. Genet. 2012, 46, 21–42. [Google Scholar] [CrossRef]
- Janicki, M.; Rooke, R.; Yang, G. Bioinformatics and genomic analysis of transposable elements in eukaryotic genomes. Chromosome Res. 2011, 19, 787–808. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Grundy, E.E.; Diab, N.; Chiappinelli, K.B. Transposable element regulation and expression in cancer. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Deniz, Ö.; Frost, J.M.; Branco, M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef] [Green Version]
- Jurka, J.; Kapitonov, V.V.; Kohany, O.; Jurka, M.V. Repetitive Sequences in Complex Genomes: Structure and Evolution. Annu. Rev. Genom. Hum. Genet. 2007, 8, 241–259. [Google Scholar] [CrossRef] [Green Version]
- Spotila, L.D.; Hirai, H.; Rekosh, D.M.; LoVerde, P.T. A retroposon-like short repetitive DNA element in the genome of the human blood fluke, Schistosoma mansoni. Chromosoma 1989, 97, 421–428. [Google Scholar] [CrossRef]
- Drew, A.C.; Brindley, P.J. A retrotransposon of the non-long terminal repeat class from the human blood fluke Schistosoma mansoni. Similarities to the chicken-repeat-1-like elements of vertebrates. Mol. Biol. Evol. 1997, 14, 602–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drew, A.C.; Minchella, D.J.; King, L.T.; Rollinson, D.; Brindley, P.J. SR2 elements, non-long terminal repeat retrotransposons of the RTE-1 lineage from the human blood fluke Schistosoma mansoni. Mol. Biol. Evol. 1999, 16, 1256–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laha, T.; Kewgrai, N.; Loukas, A.; Brindley, P.J. Characterization of SR3 reveals abundance of non-LTR retrotransposons of the RTE clade in the genome of the human blood fluke, Schistosoma mansoni. BMC Genom. 2005, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Copeland, C.S.; Brindley, P.J.; Heyers, O.; Michael, S.F.; Johnston, D.A.; Williams, D.L.; Ivens, A.C.; Kalinna, B.H. Boudicca, a Retrovirus-Like Long Terminal Repeat Retrotransposon from the Genome of the Human Blood Fluke Schistosoma mansoni. J. Virol. 2003, 77, 6153–6166. [Google Scholar] [CrossRef] [Green Version]
- Arkhipova, I.R.; Pyatkov, K.I.; Meselson, M.; Evgen’Ev, M.B. Retroelements containing introns in diverse invertebrate taxa. Nat. Genet. 2003, 33, 123–124. [Google Scholar] [CrossRef]
- DeMarco, R.; Kowaltowski, A.T.; Machado, A.A.; Soares, M.B.; Gargioni, C.; Kawano, T.; Rodrigues, V.; Madeira, A.M.B.N.; Wilson, R.A.; Menck, C.F.M.; et al. Saci-1, -2, and -3 and Perere, Four Novel Retrotransposons with High Transcriptional Activities from the Human Parasite Schistosoma mansoni. J. Virol. 2004, 78, 2967–2978. [Google Scholar] [CrossRef] [Green Version]
- Feschotte, C. Merlin, a New Superfamily of DNA Transposons Identified in Diverse Animal Genomes and Related to Bacterial IS1016 Insertion Sequences. Mol. Biol. Evol. 2004, 21, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laha, T.; Loukas, A.; Smyth, D.J.; Copeland, C.S.; Brindley, P.J. The fugitive LTR retrotransposon from the genome of the human blood fluke, Schistosoma mansoni. Int. J. Parasitol. 2004, 34, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Copeland, C.S.; Mann, V.H.; Morales, M.E.; Kalinna, B.H.; Brindley, P.J. The Sinbad retrotransposon from the genome of the human blood fluke, Schistosoma mansoni, and the distribution of related Pao-like elements. BMC Evol. Biol. 2005, 5, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demarco, R.; Machado, A.A.; Bisson-Filho, A.W.; Verjovski-Almeida, S. Identification of 18 new transcribed retrotransposons in Schistosoma mansoni. Biochem. Biophys. Res. Commun. 2005, 333, 230–240. [Google Scholar] [CrossRef]
- Demarco, R.; Venancio, T.M.; Verjovski-Almeida, S. SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol. Biol. 2006, 6, 89. [Google Scholar] [CrossRef] [Green Version]
- Jacinto, D.S.; Muniz, H.D.S.; Venancio, T.M.; Wilson, R.A.; Verjovski-Almeida, S.; DeMarco, R. Curupira-1 and Curupira-2, two novel Mutator-like DNA transposons from the genomes of human parasites Schistosoma mansoni and Schistosoma japonicum. Parasitology 2011, 138, 1124–1133. [Google Scholar] [CrossRef]
- Berriman, M.; Haas, B.J.; LoVerde, P.T.; Wilson, R.A.; Dillon, G.P.; Cerqueira, G.C.; Mashiyama, S.T.; Al-Lazikani, B.; Andrade, L.F.; Ashton, P.D.; et al. The genome of the blood fluke Schistosoma mansoni. Nature 2009, 460, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Protasio, A.V.; Tsai, I.J.; Babbage, A.; Nichol, S.; Hunt, M.; Aslett, M.A.; De Silva, N.; Velarde, G.S.; Anderson, T.J.C.; Clark, R.C.; et al. A Systematically Improved High Quality Genome and Transcriptome of the Human Blood Fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 2012, 6, e1455. [Google Scholar] [CrossRef] [PubMed]
- Venancio, T.M.; Wilson, R.A.; Verjovski-Almeida, S.; Demarco, R. Bursts of transposition from non-long terminal repeat retrotransposon families of the RTE clade in Schistosoma mansoni. Int. J. Parasitol. 2010, 40, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.R.; Greene, W.K. Transposable elements: Powerful facilitators of evolution. BioEssays 2009, 31, 703–714. [Google Scholar] [CrossRef]
- Marques, A.C.; Dupanloup, I.; Vinckenbosch, N.; Reymond, A.; Kaessmann, H. Emergence of Young Human Genes after a Burst of Retroposition in Primates. PLoS Biol. 2005, 3, e357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belyayev, A. Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, X.-Q.; Cai, X. Gene Duplication Analysis Reveals No Ancient Whole Genome Duplication but Extensive Small-Scale Duplications during Genome Evolution and Adaptation of Schistosoma mansoni. Front. Cell. Infect. Microbiol. 2017, 7, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zheng, H.; Chen, Y.; Zhang, L.; Wang, K.; Guo, J.; Huang, Z.; Zhang, B.; Huang, W.; Jin, K.; et al. The Schistosoma Japonicum Genome Reveals Features of Host–Parasite Interplay. Nature 2009, 460, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Lockyer, A.E.; Olson, P.D.; Østergaard, P.; Rollinson, D.; Johnston, D.A.; Attwood, S.W.; Southgate, V.R.; Horák, P.; Snyder, S.D.; LE, T.H.; et al. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology 2003, 126, 203–224. [Google Scholar] [CrossRef] [Green Version]
- Snyder, S.D.; Loker, E.S. Evolutionary Relationships among the Schistosomatidae (Platyhelminthes: Digenea) and an Asian Origin for Schistosoma. J. Parasitol. 2000, 86, 283–288. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Tkacik, G.; Plotkin, J.B. The dynamics of adaptation on correlated fitness landscapes. Proc. Natl. Acad. Sci. USA 2009, 106, 18638–18643. [Google Scholar] [CrossRef] [Green Version]
- Makałowski, W. Genomic scrap yard: How genomes utilize all that junk. Gene 2000, 259, 61–67. [Google Scholar] [CrossRef]
- Crombach, A.; Hogeweg, P. Chromosome Rearrangements and the Evolution of Genome Structuring and Adaptability. Mol. Biol. Evol. 2007, 24, 1130–1139. [Google Scholar] [CrossRef]
- Croll, D.; McDonald, B.A. The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens. PLoS Pathog. 2012, 8, e1002608. [Google Scholar] [CrossRef] [Green Version]
- Tsushima, A.; Gan, P.; Kumakura, N.; Narusaka, M.; Takano, Y.; Narusaka, Y.; Shirasu, K. Genomic Plasticity Mediated by Transposable Elements in the Plant Pathogenic Fungus Colletotrichum higginsianum. Genome Biol. Evol. 2019, 11, 1487–1500. [Google Scholar] [CrossRef] [Green Version]
- DeMarco, R.; Mathieson, W.; Manuel, S.J.; Dillon, G.P.; Curwen, R.S.; Ashton, P.D.; Ivens, A.C.; Berriman, M.; Verjovski-Almeida, S.; Wilson, R.A. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 2010, 20, 1112–1121. [Google Scholar] [CrossRef] [Green Version]
- Almeida, G.T.; Amaral, M.S.; Beckedorff, F.C.F.; Kitajima, J.P.; Demarco, R.; Verjovski-Almeida, S. Exploring the Schistosoma mansoni adult male transcriptome using RNA-seq. Exp. Parasitol. 2012, 132, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Philippsen, G.S.; Wilson, R.A.; Demarco, R. Accelerated evolution of schistosome genes coding for proteins located at the host-parasite interface. Genome Biol. Evol. 2015, 7, 431–443. [Google Scholar] [CrossRef] [Green Version]
- Verjovski-Almeida, S.; DeMarco, R.; Martins, E.A.L.; Guimarães, P.E.M.; Ojopi, E.P.B.; Paquola, A.C.M.; Piazza, J.P.; Nishiyama, M.Y.; Kitajima, J.P.; Adamson, R.E.; et al. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat. Genet. 2003, 35, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Braschi, S.; Curwen, R.S.; Ashton, P.D.; Verjovski-Almeida, S.; Wilson, A. The tegument surface membranes of the human blood parasite Schistosoma mansoni: A proteomic analysis after differential extraction. Proteomics 2006, 6, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Parker-Manuel, S.J.; Ivens, A.C.; Dillon, G.P.; Wilson, R.A. Gene Expression Patterns in Larval Schistosoma mansoni Associated with Infection of the Mammalian Host. PLoS Negl. Trop. Dis. 2011, 5, e1274. [Google Scholar] [CrossRef] [Green Version]
- Endo, T.; Ikeo, K.; Gojobori, T. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 1996, 13, 685–690. [Google Scholar] [CrossRef] [Green Version]
- Copley, S.D. Evolution of new enzymes by gene duplication and divergence. FEBS J. 2020, 287, 1262–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iñiguez, L.P.; Hernández, G. The Evolutionary Relationship between Alternative Splicing and Gene Duplication. Front. Genet. 2017, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Cerbin, S.; Jiang, N. Duplication of host genes by transposable elements. Curr. Opin. Genet. Dev. 2018, 49, 63–69. [Google Scholar] [CrossRef]
- Volff, J.-N. Turning junk into gold: Domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays 2006, 28, 913–922. [Google Scholar] [CrossRef]
- Sela, N.; Kim, E.; Ast, G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010, 11, R59. [Google Scholar] [CrossRef]
- Drongitis, D.; Aniello, F.; Fucci, L.; Donizetti, A. Roles of Transposable Elements in the Different Layers of Gene Expression Regulation. Int. J. Mol. Sci. 2019, 20, 5755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapitonov, V.V.; Jurka, J. RAG1 Core and V(D)J Recombination Signal Sequences Were Derived from Transib Transposons. PLoS Biol. 2005, 3, e181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, F.R.; Carazzolle, M.F.; Pereira, G.A.G.; Colombo, C.A.; Carareto, C.M.A. Transposable elements in Coffea (Gentianales: Rubiacea) transcripts and their role in the origin of protein diversity in flowering plants. Mol. Genet. Genom. 2008, 279, 385–401. [Google Scholar] [CrossRef]
- Philippsen, G.S.; Avaca-Crusca, J.S.; Araujo, A.P.U.; DeMarco, R. Distribution patterns and impact of transposable elements in genes of green algae. Gene 2016, 594, 151–159. [Google Scholar] [CrossRef]
- Philippsen, G.S.; Demarco, R. Impact of transposable elements in the architecture of genes of the human parasite Schistosoma mansoni. Mol. Biochem. Parasitol. 2019, 228, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Sela, N.; Ast, G. TranspoGene and microTranspoGene: Transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008, 36, D47–D52. [Google Scholar] [CrossRef] [Green Version]
- Keren, H.; Lev-Maor, G.; Ast, G. Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 2010, 11, 345–355. [Google Scholar] [CrossRef]
- Gilbert, W. Why genes in pieces? Nature 1978, 271, 501. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Philippsen, G.S. Transposable Elements in the Genome of Human Parasite Schistosoma mansoni: A Review. Trop. Med. Infect. Dis. 2021, 6, 126. https://doi.org/10.3390/tropicalmed6030126
Philippsen GS. Transposable Elements in the Genome of Human Parasite Schistosoma mansoni: A Review. Tropical Medicine and Infectious Disease. 2021; 6(3):126. https://doi.org/10.3390/tropicalmed6030126
Chicago/Turabian StylePhilippsen, Gisele Strieder. 2021. "Transposable Elements in the Genome of Human Parasite Schistosoma mansoni: A Review" Tropical Medicine and Infectious Disease 6, no. 3: 126. https://doi.org/10.3390/tropicalmed6030126
APA StylePhilippsen, G. S. (2021). Transposable Elements in the Genome of Human Parasite Schistosoma mansoni: A Review. Tropical Medicine and Infectious Disease, 6(3), 126. https://doi.org/10.3390/tropicalmed6030126




