Genetic Diversity, Haplotype Relationships, and kdr Mutation of Malaria Anopheles Vectors in the Most Plasmodium knowlesi-Endemic Area of Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Sites and Sample Collection
2.3. Morphological and Molecular Species Identification
2.4. Detection of Malaria-Infected Anopheles Mosquitoes
2.5. Polymerase Chain Reaction (PCR) and Sequencing of COI and VGSC Genes
2.6. Molecular Analyses
3. Results
3.1. Anopheles Mosquitoes
3.2. Malaria Parasite Detection
3.3. Nucleotide Sequences
3.4. Phylogenetic Analysis
3.5. Genetic Diversity
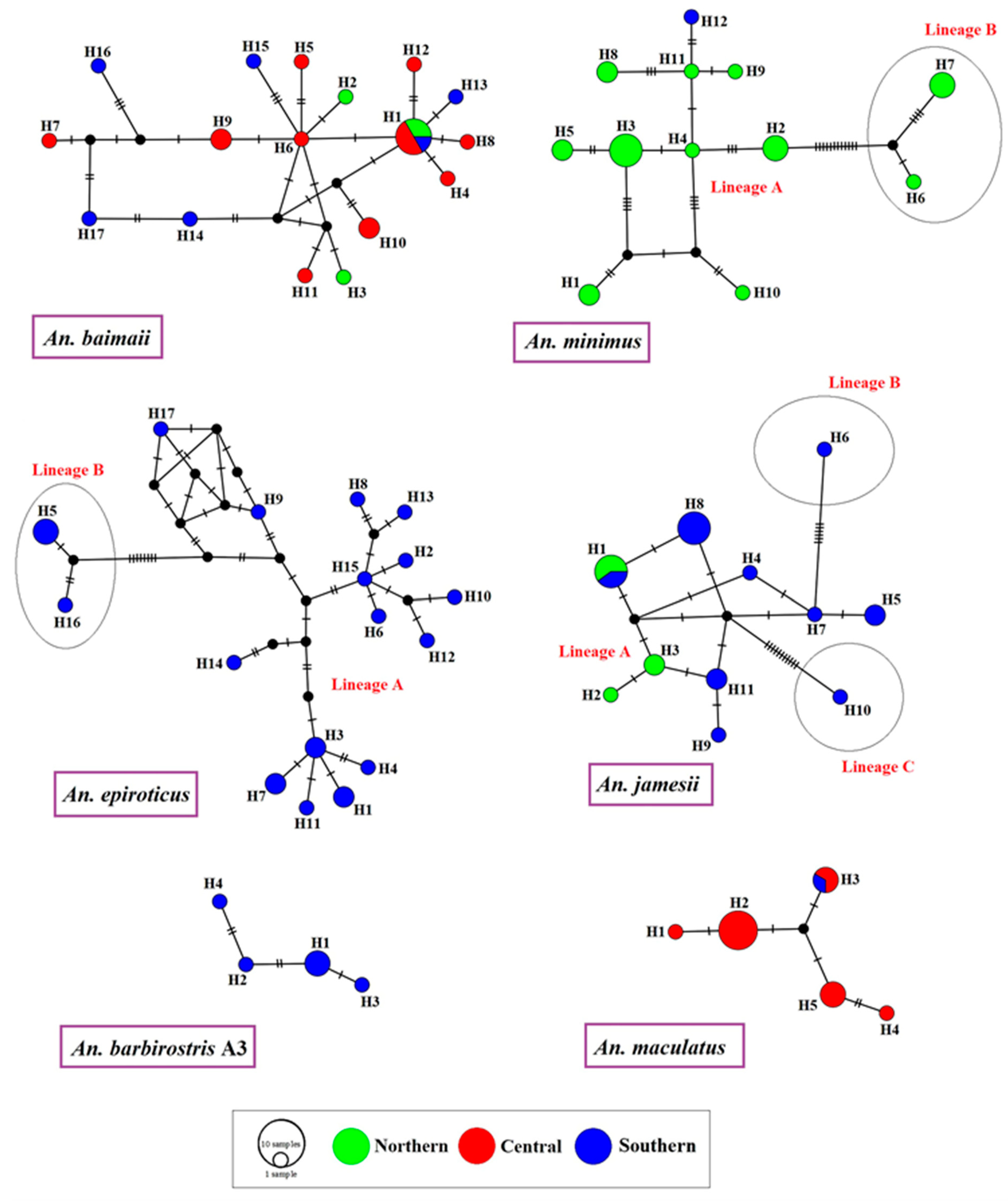
3.6. Haplotype Relationships
3.7. Screening VGSC-Mutation-Mediated Knockdown Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cox, F.E. History of the discovery of the malaria parasites and their vectors. Parasit. Vectors 2010, 3, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.I.; Mailepessov, D.; Vythilingam, I.; Lee, V.; Lam, P.; Ng, L.C.; Tan, C.H. Prevalence of simian malaria parasites in macaques of Singapore. PLoS Negl. Trop. Dis. 2021, 15, e0009110. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Schlagenhauf, P. Plasmodium Knowlesi in travellers, update 2014. Int. J. Infect. Dis. 2014, 22, 55–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, W.; Contacos, P.G.; Coatney, G.R.; Kimball, H.R. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science 1965, 149, 865. [Google Scholar] [CrossRef]
- Kantele, A.; Marti, H.; Felger, I.; Müller, D.; Jokiranta, T.S. Monkey malaria in a European traveler returning from Malaysia. Emerg. Infect. Dis. 2008, 14, 1434–1436. [Google Scholar] [CrossRef] [PubMed]
- Bronner, U.; Divis, P.C.S.; Färnert, A.; Singh, B. Swedish traveller with Plasmodium knowlesi malaria after visiting Malaysian Borneo. Malar. J. 2009, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Ehrhardt, J.; Trein, A.; Kremsner, P.G.; Frank, M. Plasmodium knowlesi and HIV co-infection in a German traveller to Thailand. Malar. J. 2013, 12, 283. [Google Scholar] [CrossRef] [Green Version]
- Froeschl, G.; Beissner, M.; Huber, K.; Bretzel, G.; Hoelscher, M.; Rothe, C. Plasmodium knowlesi infection in a returning German traveller from Thailand: A Case report on an emerging malaria pathogen in a popular low-risk travel destination. BMC Infect. Dis. 2018, 18, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Ang, J.X.D.; Kadir, K.A.; Mohamad, D.S.A.; Matusop, A.; Divis, P.C.S.; Yaman, K.; Singh, B. New vectors in northern Sarawak, Malaysian Borneo, for the zoonotic malaria parasite, Plasmodium knowlesi. Parasit. Vectors 2020, 13, 472. [Google Scholar] [CrossRef]
- Wharton, R.H.; Eyles, D.E. Anopheles hackeri, a vector of Plasmodium knowlesi in Malaya. Science 1961, 134, 279–280. [Google Scholar] [CrossRef]
- Vythilingam, I.; Tan, C.H.; Asmad, M.; Chan, S.T.; Lee, K.S.; Singh, B. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 1087–1088. [Google Scholar] [CrossRef] [PubMed]
- Marchand, R.P.; Culleton, R.; Maeno, Y.; Quang, N.T.; Nakazawa, S. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes, southern Vietnam. Emerg. Infect. Dis. 2011, 17, 1232–1239. [Google Scholar] [CrossRef] [Green Version]
- Jiram, A.I.; Vythilingam, I.; Noorazian, Y.M.; Yusof, Y.M.; Azahari, A.H.; Fong, M.Y. Entomologic investigation of Plasmodium knowlesi vectors in Kuala Lipis, Pahang, Malaysia. Malar. J. 2012, 11, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidhya, P.T.; Sunish, I.P.; Maile, A.; Zahid, A.K. Anopheles sundaicus mosquitoes as vector for Plasmodium knowlesi, andaman and Nicobar Islands, India. Emerg. Infect. Dis. 2019, 25, 817–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumruayphol, S.; Chaiphongpachara, T.; Samung, Y.; Ruangsittichai, J.; Cui, L.; Zhong, D.; Sattabongkot, J.; Sriwichai, P. Seasonal dynamics and molecular differentiation of three natural Anopheles species (Diptera: Culicidae) of the Maculatus group (Neocellia series) in malaria hotspot villages of Thailand. Parasit. Vectors 2020, 13, 574. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.M.; Carrara, V.I.; Pukrittayakamee, S.; McGready, R.; Nosten, F.H. Malaria ecology along the Thailand-Myanmar border. Malar. J. 2015, 14, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jongwutiwes, S.; Putaporntip, C.; Iwasaki, T.; Sata, T.; Kanbara, H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg. Infect. Dis. 2004, 10, 2211–2213. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Public Health, Thailand. Malaria Report. Available online: http://malaria.ddc.moph.go.th/malariaR10/index_v2.php (accessed on 1 August 2022).
- Ritthison, W.; Tainchum, K.; Manguin, S.; Bangs, M.J.; Chareonviriyaphap, T. Biting Patterns and Host Preference of Anopheles epiroticus in Chang Island, Trat Province, Eastern Thailand. J. Vector Ecol. 2014, 39, 361–371. [Google Scholar] [CrossRef]
- McCoy, K.D. The population genetic structure of vectors and our understanding of disease epidemiology. Parasite 2008, 15, 444–448. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Pedro, P.M.; Harbach, R.E.; Somboon, P.; Walton, C.; Butlin, R.K. Mitochondrial DNA variation in the malaria vector Anopheles minimus across China, Thailand and Vietnam: Evolutionary hypothesis, population structure and population history. Heredity 2011, 106, 241–252. [Google Scholar] [CrossRef]
- Weeraratne, T.C.; Surendran, S.N.; Walton, C.; Karunaratne, S.H.P.P. Genetic diversity and population structure of malaria vector mosquitoes Anopheles subpictus, Anopheles peditaeniatus, and Anopheles vagus in five districts of Sri Lanka. Malar. J. 2018, 17, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Jiang, F.; Lu, H.; Kang, X.; Wang, Y.; Zou, Z.; Wen, D.; Zheng, A.; Liu, C.; Liu, Q.; et al. Mosquito diversity and population genetic structure of six mosquito species from Hainan island. Front. Genet. 2020, 11, 602863. [Google Scholar] [CrossRef] [PubMed]
- Wangdi, K.; Furuya-Kanamori, L.; Clark, J.; Barendregt, J.J.; Gatton, M.L.; Banwell, C.; Kelly, G.C.; Doi, S.A.R.; Clements, A.C.A. Comparative effectiveness of malaria prevention measures: A systematic review and network meta-analysis. Parasit. Vectors 2018, 11, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.P.B.; Santos, J.M.M.; Martins, A.J. Mutations in the voltage-gated sodium channel gene of anophelines and their association with resistance to pyrethroids—A review. Parasit. Vectors 2014, 7, 450. [Google Scholar] [CrossRef] [Green Version]
- Syafruddin, D.; Hidayati, A.P.; Asih, P.B.; Hawley, W.A.; Sukowati, S.; Lobo, N.F. Detection of 1014F kdr mutation in four major anopheline malaria vectors in Indonesia. Malar. J. 2010, 9, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Shi, W.Q.; Wu, J.T.; Li, Y.Y.; Xue, J.B.; Zhang, Y. Resistance to pyrethroid and organophosphate insecticides, and the geographical distribution and polymorphisms of target-site mutations in voltage-gated sodium channel and acetylcholinesterase 1 genes in Anopheles sinensis populations in Shanghai, China. Parasit. Vectors 2019, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Feng, X.; Liu, N.; Li, M.; Qiu, X. Target-site mutations (AChE-G119S and kdr) in Guangxi Anopheles sinensis populations along the China-Vietnam border. Parasit. Vectors 2019, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Rattanarithikul, R.; Harrison, B.A.; Harbach, R.E.; Panthusiri, P.; Coleman, R.E. Illustrated keys to the mosquitoes of Thailand IV. Anopheles. Southeast Asian J. Trop. Med. Public Health 2006, 37 (Suppl. S2), 1–128. [Google Scholar]
- Walton, C.; Handley, J.M.; Kuvangkadilok, C.; Collins, F.H.; Harbach, R.E.; Baimai, V.; Butlin, R.K. Identification of five species of the Anopheles dirus complex from Thailand, using allele-specific polymerase chain reaction. Med. Vet. Entomol. 1999, 13, 24–32. [Google Scholar] [CrossRef]
- Walton, C.; Somboon, P.; O’Loughlin, S.M.; Zhang, S.; Harbach, R.E.; Linton, Y.M.; Chen, B.; Nolan, K.; Duong, S.; Fong, M.Y.; et al. Genetic diversity and molecular identification of mosquito species in the Anopheles Maculatus group using the ITS2 region of rDNA. Infect. Genet. Evol. 2007, 7, 93–102. [Google Scholar] [CrossRef]
- Garros, C.; Koekemoer, L.L.; Coetzee, M.; Coosemans, M.; Manguin, S. A single multiplex assay to identify major malaria vectors within the African Anopheles funestus and the Oriental An. minimus groups. Am. J. Trop. Med. Hyg. 2004, 70, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Echeverry, D.F.; Deason, N.A.; Makuru, V.; Davidson, J.; Xiao, H.; Niedbalski, J.; Yu, X.; Stevenson, J.C.; Bugoro, H.; Aparaimo, A.; et al. Fast and robust single PCR for Plasmodium sporozoite detection in mosquitoes using the cytochrome oxidase I gene. Malar. J. 2017, 16, 230. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.P.; Rajavel, A.R.; Natarajan, R.; Jambulingam, P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 2007, 44, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chaiphongpachara, T.; Changbunjong, T.; Laojun, S.; Nutepsu, T.; Suwandittakul, N.; Kuntawong, K.; Sumruayphol, S.; Ruangsittichai, J. Mitochondrial DNA barcoding of mosquito species (Diptera: Culicidae) in Thailand. PLoS ONE 2022, 17, e0275090. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Davidson, J.R.; Wahid, I.; Sudirman, R.; Small, S.T.; Hendershot, A.L.; Baskin, R.N.; Burton, T.A.; Makuru, V.; Xiao, H.; Yu, X.; et al. Molecular analysis reveals a high diversity of Anopheles species in Karama, West Sulawesi, Indonesia. Parasit. Vectors 2020, 13, 379. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Moyes, C.L.; Shearer, F.M.; Huang, Z.; Wiebe, A.; Gibson, H.S.; Nijman, V.; Mohd-Azlan, J.; Brodie, J.F.; Malaivijitnond, S.; Linkie, M.; et al. Predicting the geographical distributions of the macaque hosts and mosquito vectors of Plasmodium knowlesi malaria in forested and non-forested areas. Parasit. Vectors 2016, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Pramasivan, S.; Ngui, R.; Jeyaprakasam, N.K.; Liew, J.W.K.; Low, V.L.; Mohamed Hassan, N.; Wan Sulaiman, W.Y.; Jaraee, R.; Abdul Rahman, R.; Jelip, J.; et al. Spatial distribution of Plasmodium knowlesi cases and their vectors in Johor, Malaysia: In light of human malaria elimination. Malar. J. 2021, 20, 426. [Google Scholar] [CrossRef] [PubMed]
- Van de straat, B.; Sebayang, B.; Grigg, M.J.; Staunton, K.; Garjito, T.A.; Vythilingam, I.; Russell, T.L.; Burkot, T.R. Zoonotic malaria transmission and land use change in Southeast Asia: What is known about the vectors. Malar. J. 2022, 21, 109. [Google Scholar] [CrossRef]
- Harbach, R.E. Anopheles Classification. Available online: https://mosquito-taxonomic-inventory.myspecies.info/node/11358 (accessed on 1 August 2022).
- Tananchai, C.; Tisgratog, R.; Juntarajumnong, W.; Grieco, J.P.; Manguin, S.; Prabaripai, A.; Chareonviriyaphap, T. Species diversity and biting activity of Anopheles dirus and Anopheles baimaii (Diptera: Culicidae) in a malaria prone area of western Thailand. Parasit. Vectors 2012, 5, 211. [Google Scholar] [CrossRef] [Green Version]
- Tainchum, K.; Kongmee, M.; Manguin, S.; Bangs, M.J.; Chareonviriyaphap, T. Anopheles species diversity and distribution of the malaria vectors of Thailand. Trends Parasitol. 2015, 31, 109–119. [Google Scholar] [CrossRef]
- Chaiphongpachara, T.; Laojun, S. Variation over time in wing size and shape of the coastal malaria vector Anopheles (Cellia) epiroticus Linton and Harbach (Diptera: Culicidae) in Samut Songkhram, Thailand. J. Adv. Vet. Anim. Res. 2019, 6, 208–214. [Google Scholar] [CrossRef]
- Chaiphongpachara, T.; Laojun, S. Wing morphometric variability of the malaria vector Anopheles (Cellia) epiroticus Linton et Harbach (Diptera: Culicidae) for the duration of the rainy season in coastal areas of Samut Songkhram, Thailand. Folia Parasitol. 2020, 67, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Sumruayphol, S.; Apiwathnasorn, C.; Komalamisra, N.; Ruangsittichai, J.; Samung, Y.; Chavalitshewinkoon-Petmitr, P. Bionomic status of Anopheles epiroticus Linton & Harbach, a coastal malaria vector, in Rayong province, Thailand. Southeast Asian J. Trop. Med. Public Health 2010, 41, 541–547. [Google Scholar]
- Wilai, P.; Namgay, R.; Ali, R.S.M.; Saingamsook, J.; Saeung, A.; Junkum, A.; Walton, C.; Harbach, R.E.; Somboon, P. A multiplex PCR based on mitochondrial COI sequences for identification of members of the Anopheles barbirostris complex (Diptera: Culicidae) in Thailand and other countries in the region. Insects 2020, 11, 409. [Google Scholar] [CrossRef]
- Jeyaprakasam, N.K.; Low, V.L.; Liew, J.W.K.; Pramasivan, S.; Wan-Sulaiman, W.Y.; Saeung, A.; Vythilingam, I. Blood meal analysis of Anopheles vectors of simian malaria based on laboratory and field. Sci. Sci. Rep. 2022, 12, 354. [Google Scholar] [CrossRef] [PubMed]
- Beebe, N.W. DNA barcoding mosquitoes: Advice for potential prospectors. Parasitology 2008, 145, 622–633. [Google Scholar] [CrossRef] [Green Version]
- Chaiphongpachara, T.; Changbunjong, T.; Sumruayphol, S.; Laojun, S.; Suwandittakul, N.; Kuntawong, K. Geometric morphometrics versus DNA barcoding for the identification of malaria vectors Anopheles dirus and An. baimaii in the Thai—Cambodia border. Sci. Rep. 2022, 12, 13236. [Google Scholar] [CrossRef] [PubMed]
- Bunmee, K.; Thaenkham, U.; Saralamba, N.; Ponlawat, A.; Zhong, D.; Cui, L.; Sattabongkot, J.; Sriwichai, P. Population genetic structure of the malaria vector Anopheles minimus in Thailand based on mitochondrial DNA markers. Parasit. Vectors 2021, 14, 496. [Google Scholar] [CrossRef] [PubMed]
- Chanthran, S.S.D.; Lim, P.E.; Li, Y.; Liao, T.Y.; Poong, S.W.; Du, J.; Hussein, M.A.S.; Sade, A.; Rumpet, R.; Loh, K.H. Genetic diversity and population structure of Terapon jarbua (Forskål, 1775) (Teleostei, Terapontidae) in malaysian waters. Zookeys 2020, 911, 139–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, D.A.; Luo, H.; He, M.; Mao, C.; Kuang, Z.; Qi, H.; Xu, D.; Tan, L.; Li, Y. Genetic diversity and population differentiation of naked carp (Gymnocypris przewalskii) Revealed by cytochrome oxidase subunit I and d-loop. Front. Ecol. Evol. 2022, 10, 827654. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, J.; Wu, R.; Xue, B.; Qian, Q.; Gao, B. Genetic polymorphism study on Aedes albopictus of different geographical regions based on dna barcoding. BioMed Res. Int. 2018, 2018, 1501430. [Google Scholar] [CrossRef] [Green Version]
- Makhawi, A.M.; Liu, X.B.; Yang, S.R.; Liu, Q.Y. Genetic variations of ND5 gene of mtDNA in populations of Anopheles sinensis (Diptera: Culicidae) malaria vector in China. Parasit. Vectors 2013, 6, 290. [Google Scholar] [CrossRef] [Green Version]
- Yawson, A.E.; Weetman, D.; Wilson, M.D.; Donnelly, M.J. Ecological zones rather than molecular forms predict genetic differentiation in the malaria vector Anopheles gambiae s.s. in Ghana. Genetics 2007, 175, 751–761. [Google Scholar] [CrossRef] [Green Version]
- Syafruddinid, D.; Lestari, Y.E.; Permana, D.H.; Asih, P.B.S.; Laurent, B.S.; Zubaidah, S.; Rozi, I.E.; Kosasih, S.; Shinta; Sukowati, S.; et al. Anopheles sundaicus complex and the presence of Anopheles epiroticus in Indonesia. PLoS Negl. Trop. Dis. 2020, 14, e0008385. [Google Scholar]
- Dong, K.; Du, Y.; Rinkevich, F.; Nomura, Y.; Xu, P.; Wang, L.; Silver, K.; Zhorov, B.S. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 50, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumarnrote, A.; Overgaard, H.J.; Marasri, N.; Fustec, B.; Thanispong, K.; Chareonviriyaphap, T.; Corbel, V. Status of insecticide resistance in Anopheles mosquitoes in Ubon Ratchathani province, northeastern Thailand. Malar. J. 2017, 16, 299. [Google Scholar] [CrossRef] [PubMed]
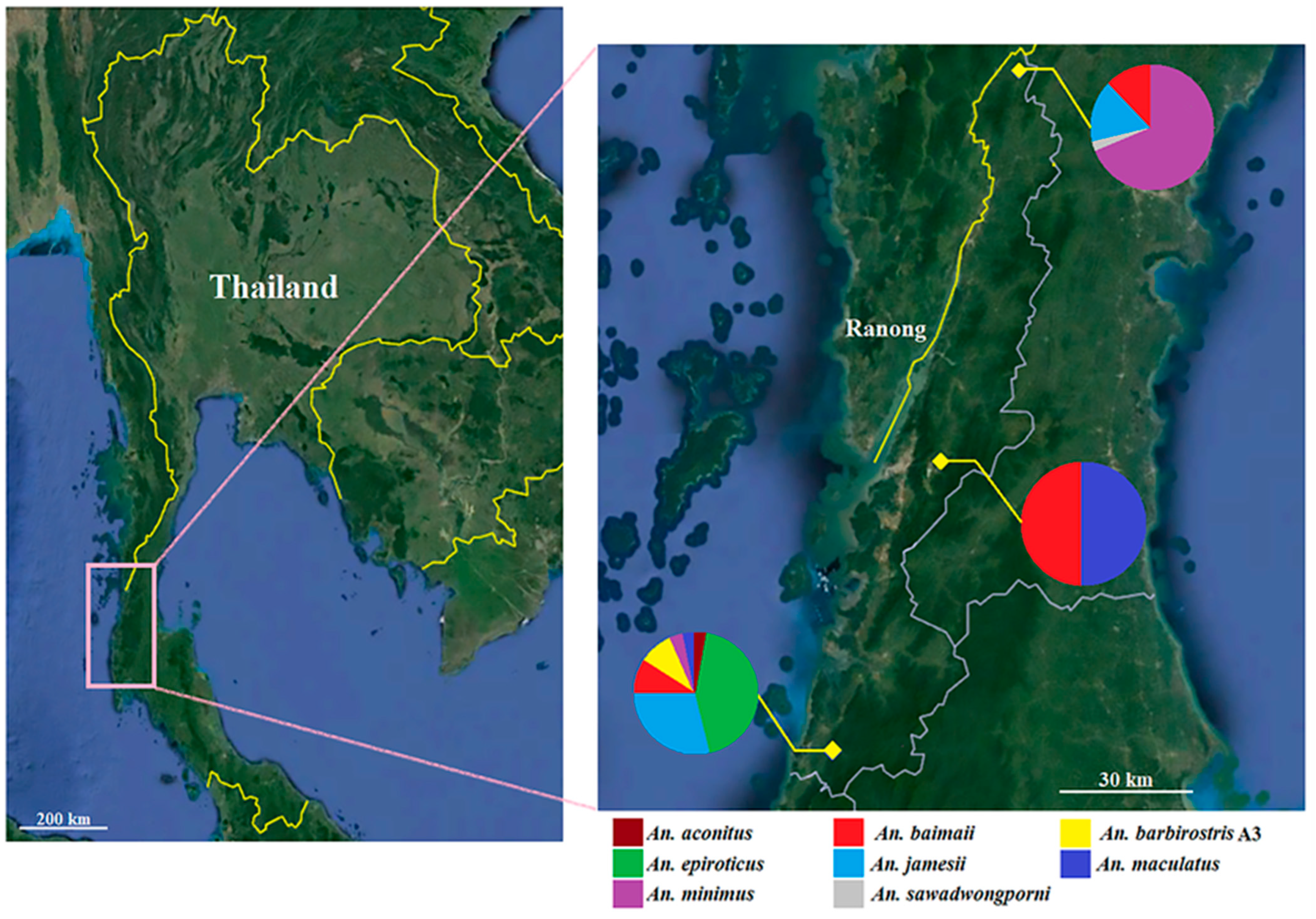


| Anopheles Species | Ranong Province | Total (%) | ||
|---|---|---|---|---|
| Northern Part | Central Part | Southern Part | ||
| An. aconitus | 0 | 0 | 1 | 1 (0.88) |
| An. baimaii | 4 | 14 | 6 | 24 (21.05) |
| An. barbirostris A3 | 0 | 0 | 6 | 6 (5.26) |
| An. epiroticus | 0 | 0 | 22 | 22 (19.30) |
| An. jamesii | 6 | 0 | 16 | 22 (19.30) |
| An. maculatus s.s. | 0 | 14 | 1 | 15 (13.16) |
| An. minimus s.s. | 22 | 0 | 1 | 23 (20.17) |
| An. sawadwongporni | 1 | 0 | 0 | 1 (0.88) |
| Total (%) | 33 (28.95) | 28 (24.56) | 53 (46.49) | 114 (100) |
| Anopheles Species | n | Location | GenBank Accession Numbers |
|---|---|---|---|
| An. Aconitus | 1 | Southern part of Ranong | OP253978 |
| An. Baimaii | 4 | Northern part of Ranong | OP253979–OP253982 |
| 14 | Central part of Ranong | OP253983–OP253996 | |
| 6 | Southern part of Ranong | OP253997–OP254002 | |
| An. barbirostris A3 | 6 | Southern part of Ranong | OP254003–OP254008 |
| An. epiroticus | 22 | Southern part of Ranong | OP254009–OP254030 |
| An. jamesii | 6 | Northern part of Ranong | OP254031–OP254036 |
| 16 | Southern part of Ranong | OP254037–OP254052 | |
| An. maculatus | 14 | Central part of Ranong | OP254053–OP254066 |
| 1 | Southern part of Ranong | OP254067 | |
| An. minimus | 22 | Northern part of Ranong | OP254068–OP254089 |
| 1 | Southern part of Ranong | OP254090 | |
| An. sawadwongporni | 1 | Northern part of Ranong | OP254091 |
| Anopheles Species | % Mean Sequence Divergence (Min–Max) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 1. An. aconitus | NA | |||||||
| 2. An. baimaii | 11.5% (11.1–11.7) | 0.5% (0.0–1.3) | ||||||
| 3. An. barbirostris A3 | 11.1% (10.9–11.2) | 9.6% (9.3–10.1) | 0.3% (0.0–0.7) | |||||
| 4. An. epiroticus | 10.7% (9.9–11.2) | 12% (10.9–12.9) | 11.6% (10.9–12.4) | 1.3% (0.0–2.7) | ||||
| 5. An. jamesii | 10% (9.8–10.4) | 11.3% (10.7–12.4) | 13.6% (13.1–14.6) | 11.8% (11.1–12.6) | 0.5% (0.0–1.9) | |||
| 6. An. maculatus | 10.2% (10.1–10.4) | 8.9% (8.3–9.8) | 10.2% (9.9–10.4) | 11.6% (10.8–12.4) | 9.5% (9.3–9.8) | 0.2% (0.0–0.7) | ||
| 7. An. minimus | 8.2% (7.9–8.4) | 10.9% (10.4–11.2) | 11.2% (10.6–11.6) | 11.5% (10.6–12.7) | 11% (10.6–12.1) | 10% (9.5–10.4) | 1.1% (0.0–2.9) | |
| 8. An. sawadwongporni | 10.6% (10.6–10.6) | 11.5% (11.2–11.9) | 10.6% (10.4–10.7) | 11.5% (10.9–11.9) | 11.7% (11.6–12.1) | 6.6% (6.5–6.6) | 9.9% (9.5–10.3) | NA |
| Anopheles Species | n | s | π (±SD) | h | Hd (±SD) | k | Neutrality Tests | |
|---|---|---|---|---|---|---|---|---|
| Fu’s Fs | Tajima’s D | |||||||
| An. aconitus | 1 | – | – | 1 | – | – | – | – |
| An. baimaii | 24 | 22 | 0.005 ± 0.001 | 17 | 0.938 ± 0.039 | 3.616 | −10.476 * | −1.621 |
| An. barbirostris A3 | 6 | 5 | 0.003 ± 0.001 | 4 | 0.800 ± 0.172 | 2.067 | −0.439 | −0.315 |
| An. epiroticus | 22 | 33 | 0.012 ± 0.001 | 17 | 0.974 ± 0.022 | 8.797 | −4.804 * | −0.220 |
| An. jamesii | 22 | 20 | 0.005 ± 0.001 | 11 | 0.900 ± 0.041 | 3.667 | −2.544 * | −1.233 |
| An. maculatus | 15 | 6 | 0.002 ± 0.000 | 5 | 0.743 ± 0.090 | 1.695 | −0.214 | −0.285 |
| An. minimus | 23 | 29 | 0.011 ± 0.002 | 12 | 0.925 ± 0.032 | 8.016 | −0.211 | 0.077 |
| An. sawadwongporni | 1 | – | – | 1 | – | – | – | – |
| Species | Allelic Frequency | |||||
|---|---|---|---|---|---|---|
| L1014 Wild | L1014C (TGT) | L1014F (TTT) | L1014S (TCA) | |||
| TTA | TTG | CTA | ||||
| An. Aconitus | 1 | 0 | 0 | 0 | 0 | 0 |
| An. baimaii | 0 | 0 | 24 | 0 | 0 | 0 |
| An. barbirostris A3 | 0 | 6 | 0 | 0 | 0 | 0 |
| An. epiroticus | 22 | 0 | 0 | 0 | 0 | 0 |
| An. jamesii | 22 | 0 | 0 | 0 | 0 | 0 |
| An. maculatus | 15 | 0 | 0 | 0 | 0 | 0 |
| An. minimus | 23 | 0 | 0 | 0 | 0 | 0 |
| An. sawadwongporni | 1 | 0 | 0 | 0 | 0 | 0 |
| Total | 84 | 6 | 24 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaiphongpachara, T.; Laojun, S.; Changbunjong, T.; Sumruayphol, S.; Suwandittakul, N.; Chookaew, S.; Atta, Y. Genetic Diversity, Haplotype Relationships, and kdr Mutation of Malaria Anopheles Vectors in the Most Plasmodium knowlesi-Endemic Area of Thailand. Trop. Med. Infect. Dis. 2022, 7, 412. https://doi.org/10.3390/tropicalmed7120412
Chaiphongpachara T, Laojun S, Changbunjong T, Sumruayphol S, Suwandittakul N, Chookaew S, Atta Y. Genetic Diversity, Haplotype Relationships, and kdr Mutation of Malaria Anopheles Vectors in the Most Plasmodium knowlesi-Endemic Area of Thailand. Tropical Medicine and Infectious Disease. 2022; 7(12):412. https://doi.org/10.3390/tropicalmed7120412
Chicago/Turabian StyleChaiphongpachara, Tanawat, Sedthapong Laojun, Tanasak Changbunjong, Suchada Sumruayphol, Nantana Suwandittakul, Sakultip Chookaew, and Yuppayong Atta. 2022. "Genetic Diversity, Haplotype Relationships, and kdr Mutation of Malaria Anopheles Vectors in the Most Plasmodium knowlesi-Endemic Area of Thailand" Tropical Medicine and Infectious Disease 7, no. 12: 412. https://doi.org/10.3390/tropicalmed7120412
APA StyleChaiphongpachara, T., Laojun, S., Changbunjong, T., Sumruayphol, S., Suwandittakul, N., Chookaew, S., & Atta, Y. (2022). Genetic Diversity, Haplotype Relationships, and kdr Mutation of Malaria Anopheles Vectors in the Most Plasmodium knowlesi-Endemic Area of Thailand. Tropical Medicine and Infectious Disease, 7(12), 412. https://doi.org/10.3390/tropicalmed7120412








