Relationship between Duffy Genotype/Phenotype and Prevalence of Plasmodium vivax Infection: A Systematic Review
Abstract
:1. Introduction
2. Methods
2.1. Registration of the Systematic Review Protocol
2.2. Guideline of Reporting Systematic Review
2.3. Research Question
2.4. Search Strategy
2.5. Eligibility Criteria and Study Selection
2.6. Quality of the Included Studies
3. Results
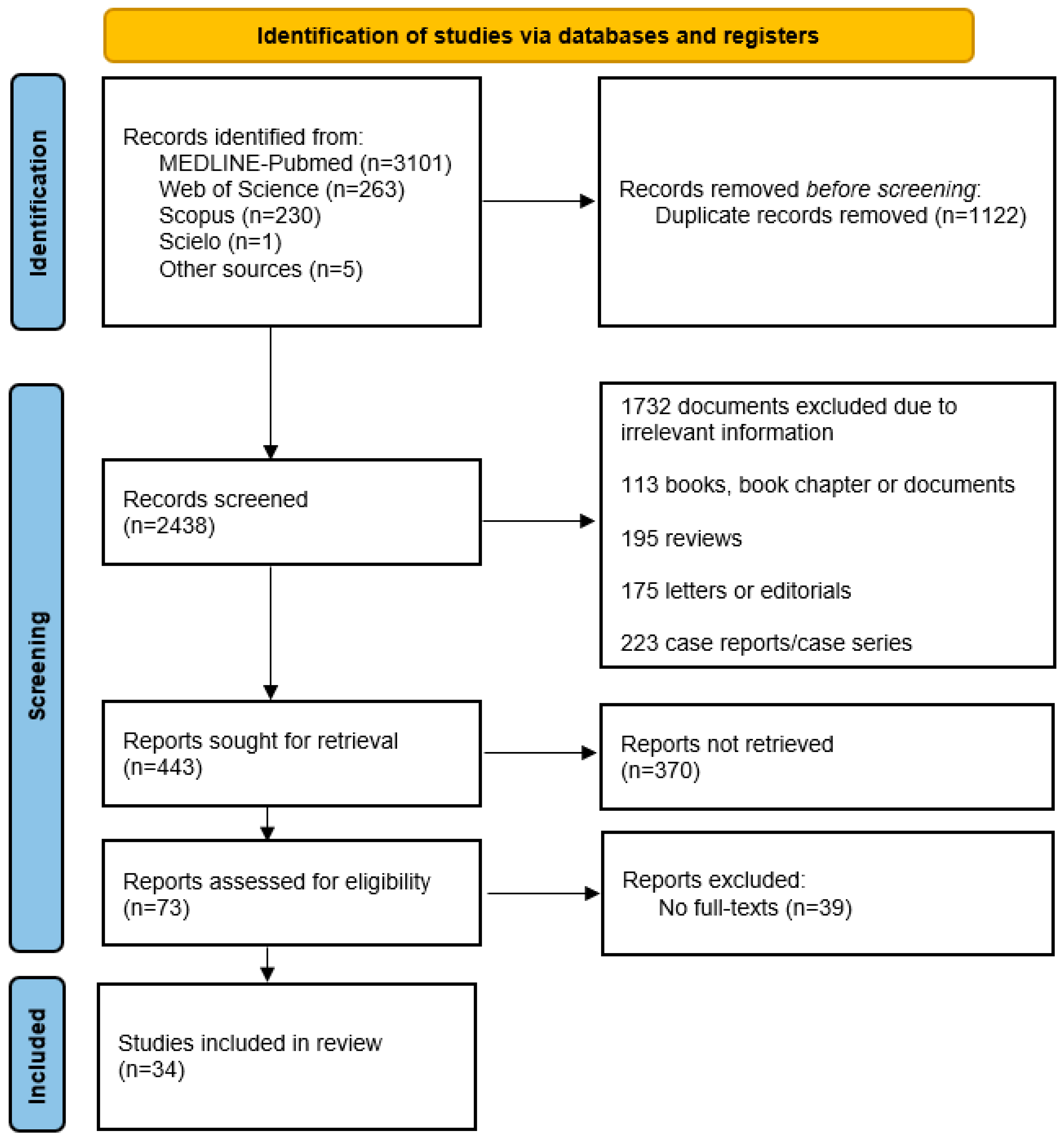
3.1. Search Results
3.2. Quality of the Included Studies
3.3. General Characteristics and Relationship between Duffy Genotype/Phenotype and Prevalence of Plasmodium vivax
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Guidelines for Malaria. Geneva: World Health Organization. 2022. Available online: https://www.who.int/publications/i/item/guidelines-for-malaria (accessed on 10 May 2023).
- World Health Organization. World Malaria Report. 2022. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022 (accessed on 26 August 2023).
- Mace, K.E.; Lucchi, N.W.; Tan, K.R. Malaria Surveillance—United States, 2018. MMWR Surveill Summ. 2022, 71, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Douglas, N.M.; Burkot, T.R.; Price, R.N. Malaria eradication revisited. Int. J. Epidemiol. 2022, 51, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Sehgal, R.; Rani, S. Duffy antigen receptor for chemokines (DARC) and susceptibility to Plasmodium vivax malaria. Parasitol. Int. 2019, 71, 73–75. [Google Scholar] [CrossRef]
- Höher, G.; Fiegenbaum, M.; Almeida, S. Molecular basis of the Duffy blood group system. Blood Transfus. 2018, 16, 93–100. [Google Scholar]
- Gai, P.P.; van Loon, W.; Siegert, K.; Wedam, J.; Kulkarni, S.S.; Rasalkar, R.; Boloor, A.; Kumar, A.; Jain, A.; Mahabala, C.; et al. Duffy antigen receptor for chemokines gene polymorphisms and malaria in Mangaluru, India. Malar. J. 2019, 18, 328. [Google Scholar] [CrossRef]
- Gunalan, K.; Niangaly, A.; Thera, M.A.; Doumbo, O.K.; Miller, L.H. Plasmodium vivax Infections of Duffy-Negative Erythrocytes: Historically Undetected or a Recent Adaptation? Trends Parasitol. 2018, 34, 420–429. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Malaria’s Impact Worldwide. Available online: https://www.cdc.gov/malaria/malaria_worldwide/impact.html (accessed on 26 August 2023).
- Brown, C.A.; Pappoe-Ashong, P.J.; Duah, N.; Ghansah, A.; Asmah, H.; Afari, E.; Koram, K.A. High frequency of the Duffy-negative genotype and absence of Plasmodium vivax infections in Ghana. Malar. J. 2021, 20, 99. [Google Scholar] [CrossRef]
- Kepple, D.; Hubbard, A.; Ali, M.M.; Abargero, B.R.; Lopez, K.; Pestana, K.; Janies, D.A.; Yan, G.; Hamid, M.M.; Yewhalaw, D.; et al. Plasmodium vivax From Duffy-Negative and Duffy-Positive Individuals Share Similar Gene Pools in East Africa. J. Infect. Dis. 2021, 224, 1422–1431. [Google Scholar] [CrossRef]
- Albsheer, M.M.A.; Pestana, K.; Ahmed, S.; Elfaki, M.; Gamil, E.; Ahmed, S.M.; Ibrahim, M.E.; Musa, A.M.; Lo, E.; Hamid, M.M.A. Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan. Genes 2019, 10, 437. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008, 61, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Schumemann, H.; Brożek, J.; Guyatt, G.; Oxman, A. GRADE Handbook. Grading of Recommendations Assessment, Development and Evaluation, Grade Working Group. 2013. Available online: https://gdt.gradepro.org/app/handbook/handbook.html (accessed on 1 September 2023).
- Gonzalez, L.; Vega, J.; Ramirez, J.L.; Bedoya, G.; Carmona-Fonseca, J.; Maestre, A. Relationship between genotypes of the Duffy blood groups and malarial infection in different ethnic groups of Choco, Colombia. Colomb. Med. 2012, 43, 189–195. [Google Scholar] [CrossRef]
- De Silva, J.R.; Lau, Y.L.; Fong, M.Y. Genotyping of the Duffy blood group among Plasmodium knowlesi-infected patients in Malaysia. PLoS ONE 2014, 9, e10895. [Google Scholar] [CrossRef]
- Abdelraheem, M.H.; Albsheer, M.M.A.; Mohamed, H.S.; Amin, M.; Mahdi Abdel Hamid, M. Transmission of Plasmodium vivax in Duffy-negative individuals in central Sudan. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Fru-Cho, J.; Bumah, V.V.; Safeukui, I.; Nkuo-Akenji, T.; Titanji, V.P.; Haldar, K. Molecular typing reveals substantial Plasmodium vivax infection in asymptomatic adults in a rural area of Cameroon. Malar. J. 2014, 13, 170. [Google Scholar] [CrossRef] [PubMed]
- Ngassa Mbenda, H.G.; Das, A. Molecular evidence of Plasmodium vivax mono and mixed malaria parasite infections in Duffy-negative native Cameroonians. PLoS ONE 2014, 9, e103262. [Google Scholar] [CrossRef]
- Woldearegai, T.G.; Kremsner, P.G.; Kun, J.F.J.; Mordmüller, B. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 328–331. [Google Scholar] [CrossRef]
- Carvalho, T.A.; Queiroz, M.G.; Cardoso, G.L.; Diniz, I.G.; Silva, A.N.; Pinto, A.Y.; Guerreiro, J.F. Plasmodium vivax infection in Anajás, State of Pará: No differential resistance profile among Duffy-negative and Duffy-positive individuals. Malar. J. 2012, 11, 430. [Google Scholar] [CrossRef]
- Lo, E.; Yewhalaw, D.; Zhong, D.; Zemene, E.; Degefa, T.; Tushune, K.; Ha, M.; Lee, M.C.; James, A.A.; Yan, G. Molecular epidemiology of Plasmodium vivax and Plasmodium falciparum malaria among Duffy-positive and Duffy-negative populations in Ethiopia. Malar. J. 2015, 14, 84. [Google Scholar] [CrossRef]
- Weppelmann, T.A.; Carter, T.E.; Chen, Z.; von Fricken, M.E.; Victor, Y.S.; Existe, A.; Okech, B.A. High frequency of the erythroid silent Duffy antigen genotype and lack of Plasmodium vivax infections in Haiti. Malar. J. 2013, 12, 30. [Google Scholar] [CrossRef]
- Miri-Moghaddam, E.; Bameri, Z.; Mohamadi, M. Duffy blood group genotypes among malaria Plasmodium vivax patients of Baoulch population in southeastern Iran. Asian Pac. J. Trop. Med. 2014, 7, 206–207. [Google Scholar] [CrossRef] [PubMed]
- Lo, E.; Hostetler, J.B.; Yewhalaw, D.; Pearson, R.D.; Hamid, M.M.A.; Gunalan, K.; Kepple, D.; Ford, A.; Janies, D.A.; Rayner, J.C.; et al. Frequent expansion of Plasmodium vivax Duffy Binding Protein in Ethiopia and its epidemiological significance. PLoS Negl. Trop. Dis. 2019, 13, e0007222. [Google Scholar] [CrossRef] [PubMed]
- Kano, F.S.; de Souza, A.M.; de Menezes Torres, L.; Costa, M.A.; Souza-Silva, F.A.; Sanchez, B.A.M.; Fontes, C.J.F.; Soares, I.S.; de Brito, C.F.A.; Carvalho, L.H.; et al. Susceptibility to Plasmodium vivax malaria associated with DARC (Duffy antigen) polymorphisms is influenced by the time of exposure to malaria. Sci. Rep. 2018, 8, 13851. [Google Scholar] [CrossRef] [PubMed]
- Abou-Ali, R.K.; Dhyani, A.; Terço, A.L.; Toro, D.M.; Gomes, K.S.; Tezza, L.C.; Negreiros, M.A.; Batista, C.S.; Souza, M.K.S.; Sanguino, E.C.B.; et al. Impact of Duffy polymorphisms on parasite density in Brazilian Amazonian patients infected by Plasmodium vivax. Malar. J. 2019, 18, 289. [Google Scholar] [CrossRef]
- De Silva, J.R.; Amir, A.; Lau, Y.L.; Ooi, C.H.; Fong, M.Y. Distribution of the Duffy genotypes in Malaysian Borneo and its relation to Plasmodium knowlesi malaria susceptibility. PLoS ONE 2019, 14, e0222681. [Google Scholar] [CrossRef]
- Popovici, J.; Roesch, C.; Carias, L.L.; Khim, N.; Kim, S.; Vantaux, A.; Mueller, I.; Chitnis, C.E.; King, C.L.; Witkowski, B. Amplification of Duffy binding protein-encoding gene allows Plasmodium vivax to evade host anti-DBP humoral immunity. Nat. Commun. 2020, 11, 953. [Google Scholar] [CrossRef]
- Haiyambo, D.H.; Aleksenko, L.; Mumbengegwi, D.; Bock, R.; Uusiku, P.; Malleret, B.; Rénia, L.; Quaye, I.K. Children with Plasmodium vivax infection previously observed in Namibia were Duffy negative and carried a c.136G > A mutation. BMC Infect. Dis. 2021, 21, 856. [Google Scholar] [CrossRef]
- Hongfongfa, P.; Kuesap, J. Genotyping of ABO and Duffy blood groups among malaria patients in Thailand. J. Parasit. Dis. 2022, 46, 178–185. [Google Scholar] [CrossRef]
- Russo, G.; Faggioni, G.; Paganotti, G.M.; Djeunang Dongho, G.B.; Pomponi, A.; De Santis, R.; Tebano, G.; Mbida, M.; Sobze, M.S.; Vullo, V.; et al. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar. J. 2017, 16, 74. [Google Scholar] [CrossRef]
- Oboh, M.A.; Badiane, A.S.; Ntadom, G.; Ndiaye, Y.D.; Diongue, K.; Diallo, M.A.; Ndiaye, D. Molecular identification of Plasmodium species responsible for malaria reveals Plasmodium vivax isolates in Duffy negative individuals from southwestern Nigeria. Malar. J. 2018, 17, 439. [Google Scholar] [CrossRef]
- Lo, E.; Russo, G.; Pestana, K.; Kepple, D.; Abargero, B.R.; Dongho, G.B.D.; Gunalan, K.; Miller, L.H.; Hamid, M.M.A.; Yewhalaw, D.; et al. Contrasting Epidemiology and Genetic Variation of Plasmodium vivax Infecting Duffy Negatives across Africa. Int. J. Infect. Dis. 2021, 108, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Howes, R.E.; Franchard, T.; Rakotomanga, T.A.; Ramiranirina, B.; Zikursh, M.; Cramer, E.Y.; Tisch, D.J.; Kang, S.Y.; Ramboarina, S.; Ratsimbasoa, A.; et al. Risk Factors for Malaria Infection in Central Madagascar: Insights from a Cross-Sectional Population Survey. Am. J. Trop. Med. Hyg. 2018, 99, 995. [Google Scholar] [CrossRef] [PubMed]
- Brazeau, N.F.; Whitesell, A.N.; Doctor, S.M.; Keeler, C.; Mwandagalirwa, M.K.; Tshefu, A.K.; Likwela, J.L.; Juliano, J.J.; Meshnick, S.R. Plasmodium vivax Infections in Duffy-Negative Individuals in the Democratic Republic of the Congo. Am. J. Trop. Med. Hyg. 2018, 99, 1128–1133. [Google Scholar] [CrossRef]
- Niangaly, A.; Gunalan, K.; Ouattara, A.; Coulibaly, D.; Sá, J.M.; Adams, M.; Travassos, M.A.; Ferrero, F.; Laurens, M.B.; Kone, A.K.; et al. Plasmodium vivax Infections over 3 Years in Duffy Blood Group Negative Malians in Bandiagara, Mali. Am. J. Trop. Med. Hyg. 2017, 97, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Roesch, C.; Popovici, J.; Bin, S.; Run, V.; Kim, S.; Ramboarina, S.; Rakotomalala, E.; Rakotoarison, R.L.; Rasoloharimanana, T.; Andriamanantena, Z.; et al. Genetic diversity in two Plasmodium vivax protein ligands for reticulocyte invasion. PLoS Negl. Trop. Dis. 2018, 12, e0006555. [Google Scholar] [CrossRef] [PubMed]
- Brazeau, N.F.; Mitchell, C.L.; Morgan, A.P.; Deutsch-Feldman, M.; Watson, O.J.; Thwai, K.L.; Gelabert, P.; van Dorp, L.; Keeler, C.Y.; Waltmann, A.; et al. The epidemiology of Plasmodium vivax among adults in the Democratic Republic of the Congo. Nat. Commun. 2021, 12, 4169. [Google Scholar] [CrossRef]
- Niang, M.; Sane, R.; Sow, A.; Sadio, B.D.; Chy, S.; Legrand, E.; Diallo, M.; Sall, A.A.; Menard, D.; Toure-Balde, A. Asymptomatic Plasmodium vivax infections among Duffy-negative population in Kedougou, Senegal. Trop. Med. Health 2018, 46, 45. [Google Scholar] [CrossRef]
- Hoque, M.R.; Elfaki, M.M.A.; Ahmed, M.A.; Lee, S.K.; Muh, F.; Ali Albsheer, M.M.; Hamid, M.M.A.; Han, E.-T. Diversity pattern of Duffy binding protein sequence among Duffy-negatives and Duffy-positives in Sudan. Malar. J. 2018, 17, 297. [Google Scholar] [CrossRef]
- Djeunang Dongho, G.B.; Gunalan, K.; L’Episcopia, M.; Paganotti, G.M.; Menegon, M.; Efeutmecheh Sangong, R.; Mayaka, G.B.; Fondop, J.; Severini, C.; Sobze, M.S.; et al. Plasmodium vivax Infections Detected in a Large Number of Febrile Duffy-Negative Africans in Dschang, Cameroon. Am. J. Trop. Med. Hyg. 2021, 104, 987–992. [Google Scholar] [CrossRef]
- Ferreira, N.S.; Mathias, J.L.S.; Albuquerque, S.R.L.; Almeida, A.C.G.; Dantas, A.C.; Anselmo, F.C.; Lima, E.S.; Lacerda, M.V.G.; Nogueira, P.A.; Ramasawmy, R.; et al. Duffy blood system and G6PD genetic variants in vivax malaria patients from Manaus, Amazonas, Brazil. Malar. J. 2022, 21, 144. [Google Scholar] [CrossRef]
- Oboh, M.A.; Singh, U.S.; Ndiaye, D.; Badiane, A.S.; Ali, N.A.; Bharti, P.K.; Das, A. Presence of additional Plasmodium vivax malaria in Duffy negative individuals from Southwestern Nigeria. Malar. J. 2020, 19, 229. [Google Scholar] [CrossRef] [PubMed]
- Onyiah, A.P.; Ajayi, I.O.; Dada-Adegbola, H.O.; Adedokun, B.O.; Balogun, M.S.; Nguku, P.M.; Ajumobi, O.O. Long-lasting insecticidal net use and asymptomatic malaria parasitaemia among household members of laboratory-confirmed malaria patients attending selected health facilities in Abuja, Nigeria, 2016: A cross-sectional survey. PLoS ONE 2018, 13, e0203686. [Google Scholar] [CrossRef] [PubMed]
- Umunnakwe, F.A.; Idowu, E.T.; Ajibaye, O.; Etoketim, B.; Akindele, S.; Shokunbi, A.O.; Otubanjo, O.A.; Awandare, G.A.; Amambua-Ngwa, A.; Oyebola, K.M. High cases of submicroscopic Plasmodium falciparum infections in a suburban population of Lagos, Nigeria. Malar. J. 2019, 18, 433. [Google Scholar] [CrossRef] [PubMed]
- Swana, E.K.; Yav, T.I.; Ngwej, L.M.; Mupemba, B.N.; Suprianto; Mukeng, C.K.; Hattingh, I.; Luboya, O.N.; Kakoma, J.B.S.; Bangs, M.J. School-based malaria prevalence: Informative systematic surveillance measure to assess epidemiological impact of malaria control interventions in the Democratic Republic of the Congo. Malar. J. 2018, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Paton, R.S.; Kamau, A.; Akech, S.; Agweyu, A.; Ogero, M.; Mwandawiro, C.; Mturi, N.; Mohammed, S.; Mpimbaza, A.; Kariuki, S.; et al. Malaria infection and severe disease risks in Africa. Science 2021, 373, 926–931. [Google Scholar] [CrossRef]
- Tsegaye, A.T.; Ayele, A.; Birhanu, S. Prevalence and associated factors of malaria in children under the age of five years in Wogera district, northwest Ethiopia: A cross-sectional study. PLoS ONE 2021, 16, e0257944. [Google Scholar] [CrossRef]
- Bawuah, A.; Ampaw, S. Ownership and use of insecticide-treated nets under Ghana’s National Malaria Control Program: What are the correlates? Trop. Med. Int. Health 2021, 26, 1593–1608. [Google Scholar] [CrossRef]
- Kanmiki, E.W.; Awoonor-Williams, J.K.; Phillips, J.F.; Kachur, S.P.; Achana, S.F.; Akazili, J.; Bawah, A.A. Socio-economic and demographic disparities in ownership and use of insecticide-treated bed nets for preventing malaria among rural reproductive-aged women in northern Ghana. PLoS ONE 2019, 14, e0211365. [Google Scholar] [CrossRef]
- Wilairatana, P.; Masangkay, F.R.; Kotepui, K.U.; De Jesus Milanez, G.; Kotepui, M. Prevalence and risk of Plasmodium vivax infection among Duffy-negative individuals: A systematic review and meta-analysis. Sci. Rep. 2022, 12, 3998. [Google Scholar] [CrossRef]
- Cardona-Arias, J.A.; Carmona-Fonseca, J. Meta-analysis of the prevalence of malaria associated with pregnancy in Colombia 2000–2020. PLoS ONE 2021, 16, e0255028. [Google Scholar] [CrossRef]
- Bilal, J.A.; Malik, E.E.; Al-Nafeesah, A.; Adam, I. Global prevalence of congenital malaria: A systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 252, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Halawani, A.J.; Saboor, M.; Abu-Tawil, H.I.; Mahzari, A.A.; Mansor, A.S.; Bantun, F. Prevalence of Duffy Blood Group Antigens and Phenotypes among Saudi Blood Donors in Southwestern Saudi Arabia. Clin. Lab. 2021, 67, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Santacruz, M.; Flórez Elvira, L.; Mejía Hurtado, A.F.; Macia Mejía, C. Estimated prevalence of the Duffy null phenotype Fy (a-b-) among black blood donors in Southwestern Colombia. Transfus. Apher. Sci. Off. J. World Apher. Assoc. Off. J. Eur. Soc. Haemapheresis. 2020, 59, 102884. [Google Scholar] [CrossRef] [PubMed]
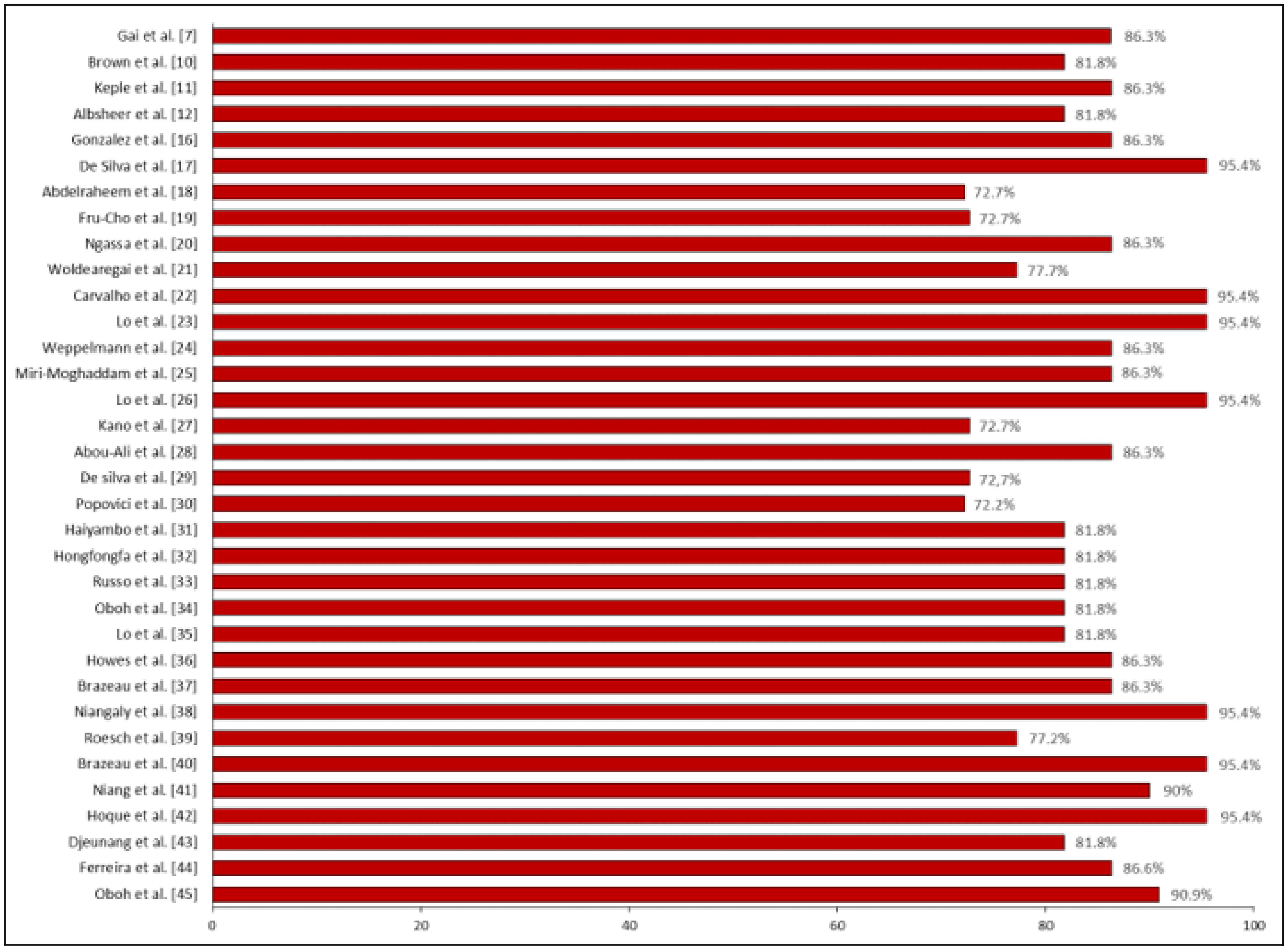
| Authors | Country | Results | P. vivax Prevalence | Risk of Bias | Certainty | Significance |
|---|---|---|---|---|---|---|
| Brown et al. [10] | Ghana | -952 adults -Absence of FY*BES allele in 90.5% of the population -No cases of P. vivax | 0% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Oboh et al. [45] | Nigeria | -242 malaria cases -All were Duffy negative genotype | 2.7% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Ferreira et al. [44] | Brazil | -225 malaria cases -Fy(a+b−): 31.1% -Fy(a+b+): 42.7% -Fy(a−b+): 24.8% -Fy(a−b−): 0.44% | 0.4% for Fy(a−b−) | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Djeunang et al. [43] | Cameroon | -1001 malaria cases -181 caused by P. vivax with Duffynegative genotype | 18% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Hoque et al. [42] | Sudan | -42 malaria cases -83.3% Duffy-positive (10 homozygous/25 heterozygous) | 16.7% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Niang et al. [41] | Senegal | -74 malaria cases -Pure infection by P. falciparum: 79.7% | 20.3% for negative Duffy | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Brazeau et al. [40] | Democratic Republic of Congo | -172 infections by P. vivax -14 infections in Duffy-negative individuals | 8.3% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Roesch et al. [39] | Cambodia and Madagascar | -174 malaria cases -T/T substitution in 100% in Cambodia/44% T/T—56% T/C in Madagascar | 100% for positive Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Niangaly et at. [38] | Mali | -Screening of 300 children -1 to 3 cases per 25 Duffy-negative children | - | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Albsheer et al. [12] | Sudan | -992 samples -190 infections by P. vivax (Fy(a−b+): 67.9%/Fy(a+b−): 14.2%/Fy(a−b−): 17.9% | 67.9% Fy(a−b+)/17.9% Fy(a−b−) | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Brazeau et al. [37] | Democratic Republic of Congo | -17,972 samples -579 infections by P. vivax and 467 sequencings (n = 464/467 for Duffy-negative) | 99.3% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Howes et al. [36] | Madagascar | -1878 adults -48.7% Duffy-negative -86 and 44 infections by P. vivax with Duffy-positive and Duffy-negative, respectively | 8.9% for negative Duffy/4.8% for positive Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Kepple et al. [11] | Sudan and Ethiopia | -107 and 305 individuals infected with P. vivax for Duffy-negative and Duffy-positive | 14.95% for negative Duffy/13.77% for positive Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Lo et al. [35] | Sudan and Ethiopia | -1963 samples -332 infections by P. vivax (49 for Duffy-negative) | 9.2%–86% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Oboh et al. [34] | Nigeria | -436 samples and 256 cases -5 infections by P. vivax (all Duffy-negative homozygotes) | 1.95% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Russo et al. [33] | Cameroon | -484 samples -27 infections by P. vivax (all Duffy-negative) | 5.6% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Hongfongfa et al. [32] | Thailand and Myanmar | -900 cases of P. vivax -FY*A/*A: 83.5% of cases | 0% for Fy(a−b−) | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Haiyambo et al. [31] | Namibia | -33 cases and 47 controls -3 infections by P. vivax (all Duffy-negative) | 9% for negative Duffy | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Popovici et al. [30] | Cambodia | -22 Duffy-positive cases (16 FY*A/*A homozygotes) | - | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Gai et al. [7] | India | -909 malaria cases (43.9% FY*A/A vs. 44.1% FYA/*B) -633 infections by P. vivax (44.2% FY*A/A vs. 43.7% FYA/*B) | 0.3% for negative Duffy | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| De Silva et al. [29] | Malaysia | -79 infections by P. knowlesi -Equal distribution of FY*A/A and FYA/*B genotypes | - | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Abou-Ali et al. [28] | Brazil | -287 infected by P. vivax -23.7% FYA/FYA; 42.8% FYA/FYB; 3% FYB/FYB | - | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Kano et al. [27] | Brazil | -Reduction in risk of clinical P. vivax malaria by 19% and 91% for FYA/BES and FYBES/BES genotypes, compared to FYA/*B | - | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Lo et al. [26] | Ethiopia | -145 symptomatic individuals infected by P. vivax -69.7% FY*A/BES or FYB/*BES -1.4% FY*BES/*BES (Duffy negative homozygotes) | - | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Miri- Moghaddam et al. [25] | Iran | -160 infections by Plasmodium -FY*A/*B: 51.9% -FY*A/*A: 16.3% -FY*B/*B: 13.8% -FY*A/*BES: 10% | 0.6% for negative Duffy | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Weppelmann et al. [24] | Haiti | -164 cases -99.4% FYES allele | - | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Lo et al. [23] | Ethiopia | -416 samples and 94 cases for Duffy-negative -3 cases of P. vivax in Duffy negative | 3.1% for negative Duffy | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Carvalho et al. [22] | Brazil | -678 cases and 94 infections by P. vivax -29 Duffy-negative individuals (2 cases of P. vivax) | 6.9% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Woldearegai et al. [21] | Ethiopia | -1931 adults -111 cases of P. vivax | 20% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Ngassa et al. [20] | Cameroon | -201 symptomatic cases -8 cases of P. vivax infection | 3.9% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Fru-Cho et al. [19] | Cameroon | -87 malaria cases -12 infections by P. vivax (6 in Duffy-negative individuals) | 6.8% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| Abdelraheem et al. [18] | Sudan | -126 suspected cases -48 confirmed cases of P. vivax (4 in Duffy-negative individuals) | 8.3% for negative Duffy | Serious | ⨁⨁⨁◯ (Moderate) | Important |
| De Silva et al. [17] | Malaysia | -111 samples -Fy(a+b−): 89.2% -FY*A/*A: 48 cases | 0% for negative Duffy | Non-serious | ⨁⨁⨁⨁ (High) | Critical |
| Gonzalez et al. [16] | Colombia | -52 individuals infected by Plasmodium (14 with P. vivax) -Amerindians and mestizos: T-46 allele in 90%–100%/Afro-Colombians 50% | - | Serious | ⨁⨁⨁◯ (Moderate) | Important |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picón-Jaimes, Y.A.; Lozada-Martinez, I.D.; Orozco-Chinome, J.E.; Molina-Franky, J.; Acevedo-Lopez, D.; Acevedo-Lopez, N.; Bolaño-Romero, M.P.; Visconti-Lopez, F.J.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Relationship between Duffy Genotype/Phenotype and Prevalence of Plasmodium vivax Infection: A Systematic Review. Trop. Med. Infect. Dis. 2023, 8, 463. https://doi.org/10.3390/tropicalmed8100463
Picón-Jaimes YA, Lozada-Martinez ID, Orozco-Chinome JE, Molina-Franky J, Acevedo-Lopez D, Acevedo-Lopez N, Bolaño-Romero MP, Visconti-Lopez FJ, Bonilla-Aldana DK, Rodriguez-Morales AJ. Relationship between Duffy Genotype/Phenotype and Prevalence of Plasmodium vivax Infection: A Systematic Review. Tropical Medicine and Infectious Disease. 2023; 8(10):463. https://doi.org/10.3390/tropicalmed8100463
Chicago/Turabian StylePicón-Jaimes, Yelson Alejandro, Ivan David Lozada-Martinez, Javier Esteban Orozco-Chinome, Jessica Molina-Franky, Domenica Acevedo-Lopez, Nicole Acevedo-Lopez, Maria Paz Bolaño-Romero, Fabriccio J. Visconti-Lopez, D. Katterine Bonilla-Aldana, and Alfonso J. Rodriguez-Morales. 2023. "Relationship between Duffy Genotype/Phenotype and Prevalence of Plasmodium vivax Infection: A Systematic Review" Tropical Medicine and Infectious Disease 8, no. 10: 463. https://doi.org/10.3390/tropicalmed8100463
APA StylePicón-Jaimes, Y. A., Lozada-Martinez, I. D., Orozco-Chinome, J. E., Molina-Franky, J., Acevedo-Lopez, D., Acevedo-Lopez, N., Bolaño-Romero, M. P., Visconti-Lopez, F. J., Bonilla-Aldana, D. K., & Rodriguez-Morales, A. J. (2023). Relationship between Duffy Genotype/Phenotype and Prevalence of Plasmodium vivax Infection: A Systematic Review. Tropical Medicine and Infectious Disease, 8(10), 463. https://doi.org/10.3390/tropicalmed8100463









