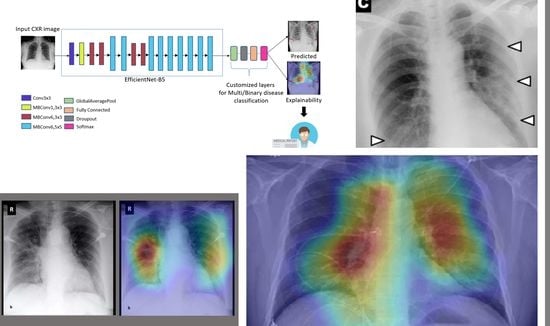
Explainable COVID-19 Detection on Chest X-rays Using an End-to-End Deep Convolutional Neural Network Architecture
Abstract
:1. Introduction
2. Proposed Approach
Deep Learning Model
3. Datasets
3.1. COVID-19 Image Data Collection (CIDC)
3.2. COVID-19 Radiography
3.3. BIMCV COVID19+
3.4. RSNA
3.5. Chest X-ray Images Pneumonia (CXRIP)
3.6. Montgomery County X-ray
3.7. Shenzhen Hospital X-ray
3.8. National Institute of Health (NIH)
3.9. Montfort Dataset
4. Experimental Results
4.1. Data Distribution for Multi-Class and Binary Models
4.2. Training Parameters
4.3. Metrics
4.4. Results
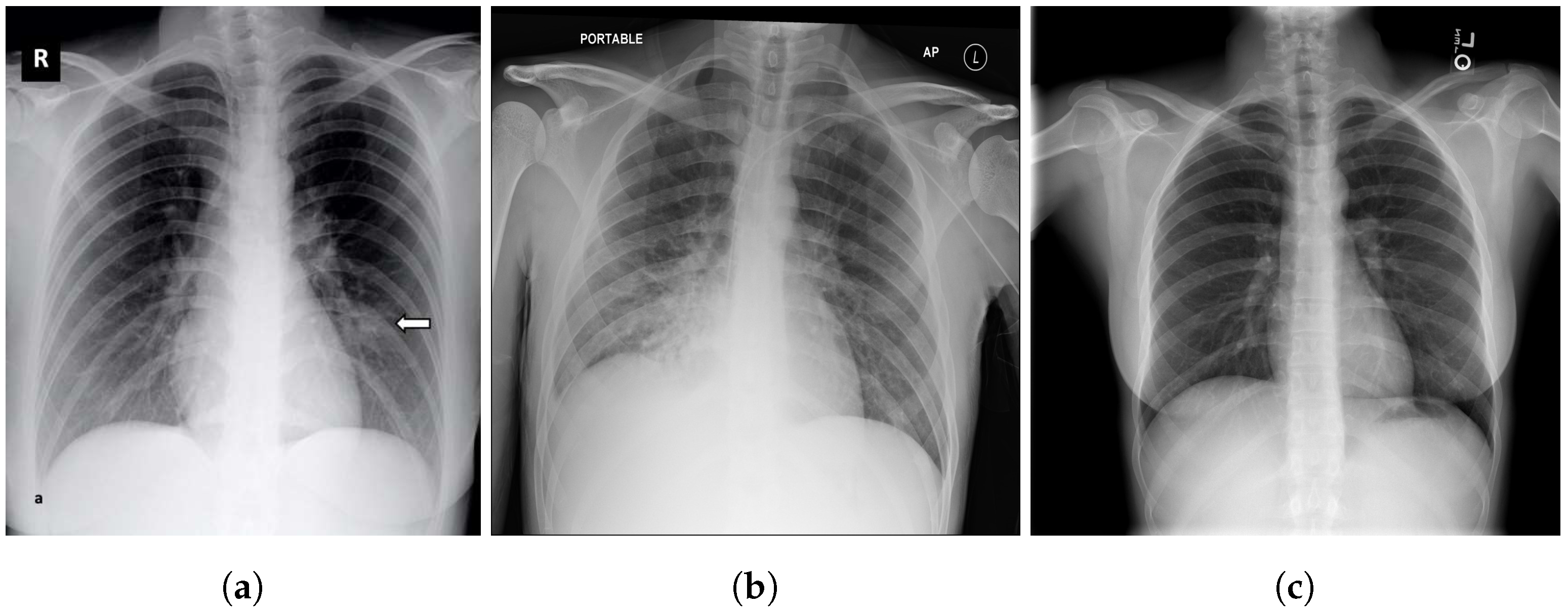
4.5. Explainability
4.6. Performance Comparison
5. Individual Tests
5.1. DeepCCXR-Bin for Individual Datasets
- CIDC dataset: The CIDC dataset does not contain the ’normal’ class. We added 1128 CXR normal images from RSNA dataset (not in the training set), and we kept all the CXR images of COVID-19 (654 CXR images).
- CXRIP dataset: The original CXRIP dataset contains pneumonia and normal classes. We added 1228 COVID-19 CXR images in place of the pneumonia class, and we kept the 1435 CXR images for normal class.
- MONTGOMERY, SHENZHEN, NIH and RSNA datasets: As with the CXRIP dataset, the original MONTGOMERY, SHENZHEN, NIH and RSNA datasets contain two categories, pneumonia and normal. We kept their normal classes and we replaced the pneumonia class with COVID-19 CXR images.
- COVID-19 RADIOGRAPHY dataset: The COVID-19 RADIOGRAPHY dataset contains only 218 CXR images of COVID-19. We added 1,128 normal CXR images from the RSNA dataset.
- The Montfort dataset contain three categories (COVID-19, normal and pneumonia). We removed the pneumonia class and we kept the normal and COVID-19 classes.
5.2. DeepCCXR-Multi for Individual Datasets
- CIDC dataset: This dataset provides the pneumonia class with 369 CXR images and 654 of COVID-19, to build a dataset with three categories. We added 1128 CXR normal images from RSNA dataset.
- The COVID-19 RADIOGRAPHY dataset contains the COVID-19 CXR class. We added normal and pneumonia classes of 1,128 CXR images and 1498 from RSNA dataset, respectively.
- The remaining datasets come with normal and pneumonia classes, and we added COVID-19 CXR images to build the datasets with three categories. The Montfort contains the three classes (normal, pneumonia and COVID-19).
6. Model Limitations
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Coronavirus Disease 2020. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 16 October 2021).
- WHO. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV). 2020. Available online: https://www.who.int/ (accessed on 16 October 2021).
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. 2020. Available online: https://www.who.int (accessed on 16 October 2021).
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Fan, K.S.; Ghani, S.A.; Machairas, N.; Lenti, L.; Fan, K.H.; Richardson, D.; Scott, A.; Raptis, D.A. COVID-19 prevention and treatment information on the internet: A systematic analysis and quality assessment. BMJ Open 2020, 10, e040487. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al Emadi, N.; et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Domingues, I.; Pereira, G.; Martins, P.; Duarte, H.; Santos, J.; Abreu, P.H. Using deep learning techniques in medical imaging: A systematic review of applications on CT and PET. Artif. Intell. Rev. 2020, 53, 4093–4160. [Google Scholar] [CrossRef]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2021, 14, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Chetoui, M.; Akhloufi, M.A. Explainable end-to-end deep learning for diabetic retinopathy detection across multiple datasets. J. Med. Imaging 2020, 7, 044503. [Google Scholar] [CrossRef]
- Chetoui, M.; Akhloufi, M.A. Explainable Diabetic Retinopathy using EfficientNET. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1966–1969. [Google Scholar]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Ghaderzadeh, M.; Asadi, F. Deep Learning in the Detection and Diagnosis of COVID-19 Using Radiology Modalities: A Systematic Review. J. Healthc. Eng. 2021, 2021, 6677314. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Hafeez, A. Covid-resnet: A deep learning framework for screening of covid19 from radiographs. arXiv 2020, arXiv:2003.14395. [Google Scholar]
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Wehbe, R.M.; Sheng, J.; Dutta, S.; Chai, S.; Dravid, A.; Barutcu, S.; Wu, Y.; Cantrell, D.R.; Xiao, N.; Allen, B.D.; et al. DeepCOVID-XR: An Artificial Intelligence Algorithm to Detect COVID-19 on Chest Radiographs Trained and Tested on a Large U.S. Clinical Data Set. Radiology 2021, 299, E167–E176. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Chetoui, M.; Akhloufi, M.A. Deep Efficient Neural Networks for Explainable COVID-19 Detection on CXR Images. In Advances and Trends in Artificial Intelligence, Artificial Intelligence Practices; IEA/AIE: Kuala Lumpur, Malaysia, 2021; Chapter 29. [Google Scholar] [CrossRef]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- imageNet. Large Scale Visual Recognition Challenge (ILSVRC). Available online: https://image-net.org/ (accessed on 16 October 2021).
- Pan, I.; Cadrin-Chênevert, A.; Cheng, P.M. Tackling the radiological society of north america pneumonia detection challenge. Am. J. Roentgenol. 2019, 213, 568–574. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L.; Roth, K.; Duong, T.Q.; Ghassemi, M. COVID-19 Image Data Collection: Prospective Predictions Are the Future. arXiv 2020, arXiv:2006.11988. [Google Scholar]
- de la Iglesia Vayá, M.; Saborit, J.M.; Montell, J.A.; Pertusa, A.; Bustos, A.; Cazorla, M.; Galant, J.; Barber, X.; Orozco-Beltrán, D.; García-García, F.; et al. BIMCV COVID-19+: A large annotated dataset of RX and CT images from COVID-19 patients. arXiv 2020, arXiv:2006.01174. [Google Scholar]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine. Tuberculosis Chest X-ray Image Data Sets. Available online: https://lhncbc.nlm.nih.gov/publication/pub9931 (accessed on 16 October 2021).
- Malhotra, A.; Mittal, S.; Majumdar, P.; Chhabra, S.; Thakral, K.; Vatsa, M.; Singh, R.; Chaudhury, S.; Pudrod, A.; Agrawal, A. Multi-Task Driven Explainable Diagnosis of COVID-19 using Chest X-ray Images. arXiv 2020, arXiv:2008.03205. [Google Scholar] [CrossRef] [PubMed]
- Montfort, H. Hopital Montfort. 2020. Available online: https://hopitalmontfort.com/ (accessed on 16 October 2021).
- Heaven, W.D. Google’s Medical AI Was Super Accurate in a Lab. Real Life Was a Different Story. 2020. Available online: https://www.technologyreview.com/2020/04/27/1000658/google-medical-ai-accurate-lab-real-life-clinic-covid-diabetes-retina-disease/ (accessed on 2 July 2020).
- Sandler, M.; Howard, A.G.; Zhu, M.; Zhmoginov, A.; Chen, L. Inverted Residuals and Linear Bottlenecks: Mobile Networks for Classification, Detection and Segmentation. arXiv 2018, arXiv:1801.04381. [Google Scholar]
- Kızrak, A. Comparison of activation functions for deep neural networks. Available online: https://towardsdatascience.com/comparison-of-activation-functions-for-deep-neural-networks-706ac4284c8a (accessed on 16 October 2021).
- Tamaki, Y.; Akiyama, F.; Iwase, T.; Kaneko, T.; Tsuda, H.; Sato, K.; Ueda, S.; Mano, M.; Masuda, N.; Takeda, M.; et al. Molecular detection of lymph node metastases in breast cancer patients: Results of a multicenter trial using the one-step nucleic acid amplification assay. Clin. Cancer Res. 2009, 15, 2879–2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, G.; Agarwal, D.; de la Torre Díez, I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020, 96, 106691. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.; Aslam, N.; Anwar, T.; Aljameel, S.S.; Ullah, M.; Khan, R.; Rehman, A.; Akhtar, N. Remote Diagnosis and Triaging Model for Skin Cancer Using EfficientNet and Extreme Gradient Boosting. Complexity 2021, 2021, 5591614. [Google Scholar] [CrossRef]
- Rahman, T. COVID-19 Radiography Database. 2020. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 16 October 2021).
- Shih, G.; Wu, C.C.; Halabi, S.S.; Kohli, M.D.; Prevedello, L.M.; Cook, T.S.; Sharma, A.; Amorosa, J.K.; Arteaga, V.; Galperin-Aizenberg, M.; et al. Augmenting the National Institutes of Health chest radiograph dataset with expert annotations of possible pneumonia. Radiol. Artif. Intell. 2019, 1, e180041. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 16 October 2021).
- Microsoft. Microsoft Azure. Available online: https://azure.microsoft.com/ (accessed on 16 October 2021).
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2016, arXiv:1609.04747. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Huang, W.; Song, G.; Li, M.; Hu, W.; Xie, K. Adaptive weight optimization for classification of imbalanced data. In Proceedings of the International Conference on Intelligent Science and Big Data Engineering, Beijing, China, 31 July–2 August 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 546–553. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Panwar, H.; Gupta, P.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Janssens, O.; Park, H.m.; Zuallaert, J.; Van Hoecke, S.; De Neve, W. Web applicable computer-aided diagnosis of glaucoma using deep learning. arXiv 2018, arXiv:1812.02405. [Google Scholar]
- Kowsari, K.; Sali, R.; Ehsan, L.; Adorno, W.; Ali, A.; Moore, S.; Amadi, B.; Kelly, P.; Syed, S.; Brown, D. Hmic: Hierarchical medical image classification, a deep learning approach. Information 2020, 11, 318. [Google Scholar] [CrossRef] [PubMed]
- Stephanie, S.; Shum, T.; Cleveland, H.; Challa, S.R.; Herring, A.; Jacobson, F.L.; Hatabu, H.; Byrne, S.C.; Shashi, K.; Araki, T.; et al. Determinants of chest x-ray sensitivity for covid-19: A multi-institutional study in the united states. Radiol. Cardiothorac. Imaging 2020, 2, e200337. [Google Scholar] [CrossRef] [PubMed]
- Chetoui, M.; Traoré, A.; Akhloufi, M.A. Deep Learning for COVID-19 Detection on Chest X-Ray and CT Scan. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar]
- Couturier, A.; Chetoui, M.; Akhloufi, M.A. COVID-19 Detection Using Deep Learning. 2020. Available online: https://covid19.primeai.ca/ (accessed on 16 October 2021).
























| Stage | Operator | Resolution H × W | #Channels | Layers |
|---|---|---|---|---|
| 1 | Conv3 × 3 | 244 × 224 | 32 | 1 |
| 2 | MBConv1, K3 × 3 | 112 × 112 | 16 | 1 |
| 3 | MBConv6, K3 × 3 | 112 × 112 | 24 | 2 |
| 4 | MBConv6, K5 × 5 | 56 × 56 | 40 | 2 |
| 5 | MBConv6, K3 × 3 | 28 × 28 | 80 | 3 |
| 6 | MBConv6, K5 × 5 | 14 × 14 | 112 | 3 |
| 7 | MBConv6, K5 × 5 | 14 × 14 | 192 | 4 |
| 8 | MBConv6, K3 × 3 | 7 × 7 | 320 | 1 |
| 9 | Conv1 × 1 and pooling and FC | 7 × 7 | 1280 | 1 |
| Configuration | Value |
|---|---|
| Optimizer | SGD |
| Epoch | 200 complete training |
| Batch size | 16 |
| Learning rate | 0.003 |
| Batch normalization | True |
| Dropout | 50% after dense layers |
| Model checkpoint | Monitor = ‘val_acc’, save_best_only = True, mode = ‘auto’ |
| Metrics | COVID-19 | Normal | Pneumonia |
|---|---|---|---|
| Sensitivity | 0.99 | 0.88 | 0.92 |
| Specificity | 0.99 | 0.96 | 0.94 |
| Ref. | Dataset | #COVID-19 Images | ACC | AUC | SN | SP | EXP |
|---|---|---|---|---|---|---|---|
| Apostolopoulos et al. [11] | CIDC | 224 | 0.93 | - | 0.98 | 0.96 | No |
| Sethy et al. [14] | CIDC | 25 | 0.95 | - | 0.97 | 0.93 | No |
| Chetoui et al. [54] | CIDC | 192 | - | 0.97 | 0.95 | 0.96 | Yes |
| Chetoui and Akhloufi [25] | Multiple datasets | 2385 | 0.95 | 0.95 | 0.97 | 0.90 | Yes |
| Wang et al. [16] | ActualMed, CIDC | 226 | 0.93 | - | 0.91 | - | Yes |
| Hemdan et al. [18] | CIDC | 25 | 0.90 | - | 1.00 | 0.83 | No |
| Wehbe et al. [20] | Multiple institutions | 4253 | 0.83 | 0.90 | 0.71 | 0.92 | Yes |
| Minaee et al. [26] | CIDC | 203 | - | 0.98 | 0.98 | 0.90 | Yes |
| DeepCCXR-Multi | Multiple datasets | 3288 | 0.93 | 0.97 | 0.97 | 0.94 | Yes |
| DeepCCXR-Bin | Multiple datasets | 3288 | 0.96 | 0.98 | 0.94 | 0.98 | Yes |
| Dataset | ACC | AUC | SP | SN |
|---|---|---|---|---|
| CIDC | 0.91 | 0.94 | 0.90 | 0.91 |
| CXRIP | 0.98 | 0.99 | 0.99 | 0.99 |
| RSNA | 0.98 | 0.99 | 0.99 | 0.99 |
| NIH | 0.96 | 0.98 | 0.96 | 0.97 |
| MONTGOMERY | 0.98 | 0.99 | 0.86 | 0.99 |
| SHENZHEN | 0.98 | 0.97 | 0.93 | 0.95 |
| Montfort | 0.95 | 0.98 | 0.91 | 0.97 |
| BIMCV COVID19+ | 0.95 | 0.96 | 0.94 | 0.93 |
| COVID-19 RADIOGRAPHY | 0.93 | 0.96 | 0.86 | 0.95 |
| Dataset | ACC | AUC | SP | SN |
|---|---|---|---|---|
| CIDC | 0.85 | 0.98 | 0.83 | 0.87 |
| CXRIP | 0.93 | 0.99 | 0.95 | 0.94 |
| RSNA | 0.92 | 0.95 | 0.92 | 0.93 |
| NIH | 0.90 | 0.90 | 0.91 | 0.89 |
| MONTGOMERY | 0.88 | 0.98 | 0.54 | 0.75 |
| SHENZHEN | 0.90 | 0.98 | 0.78 | 0.92 |
| Montfort | 0.70 | 0.80 | 0.50 | 0.51 |
| BIMCV COVID19+ | 0.87 | 0.86 | 0.88 | 0.86 |
| COVID-19 RADIOGRAPHY | 0.90 | 0.84 | 0.92 | 0.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chetoui, M.; Akhloufi, M.A.; Yousefi, B.; Bouattane, E.M. Explainable COVID-19 Detection on Chest X-rays Using an End-to-End Deep Convolutional Neural Network Architecture. Big Data Cogn. Comput. 2021, 5, 73. https://doi.org/10.3390/bdcc5040073
Chetoui M, Akhloufi MA, Yousefi B, Bouattane EM. Explainable COVID-19 Detection on Chest X-rays Using an End-to-End Deep Convolutional Neural Network Architecture. Big Data and Cognitive Computing. 2021; 5(4):73. https://doi.org/10.3390/bdcc5040073
Chicago/Turabian StyleChetoui, Mohamed, Moulay A. Akhloufi, Bardia Yousefi, and El Mostafa Bouattane. 2021. "Explainable COVID-19 Detection on Chest X-rays Using an End-to-End Deep Convolutional Neural Network Architecture" Big Data and Cognitive Computing 5, no. 4: 73. https://doi.org/10.3390/bdcc5040073
APA StyleChetoui, M., Akhloufi, M. A., Yousefi, B., & Bouattane, E. M. (2021). Explainable COVID-19 Detection on Chest X-rays Using an End-to-End Deep Convolutional Neural Network Architecture. Big Data and Cognitive Computing, 5(4), 73. https://doi.org/10.3390/bdcc5040073