Prediction of Preeclampsia Using Machine Learning and Deep Learning Models: A Review
Abstract
:1. Introduction
2. Related Study
2.1. Predicting Preeclampsia Using General Approaches
2.2. General Approaches for Predicting Preeclampsia
Machine Learning Based Model for Predicting Preeclampsia
2.3. Deep Learning Based Model for Predicting Preeclampsia
3. Discussion
4. Challenges and Opportunities
4.1. Identifying the Disease
4.2. Patients’ Data Security and Privacy
4.3. Reliability of the Models
4.4. Issues Related to the Datasets
4.5. Model Interpretation
4.6. Human Barriers with AI Adoption in Healthcare
4.7. Model Bias
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fabjan-Vodusek, V.; Kumer, K.; Osredkar, J.; Verdenik, I.; Gersak, K.; Premru-Srsen, T. Correlation between Uterine Artery Doppler and the SFlt-1/PlGF Ratio in Different Phenotypes of Placental Dysfunction. Hypertens. Pregnancy 2019, 38, 32–40. [Google Scholar] [CrossRef]
- Alrowaili, M.M.; Zakari, N.M.A.; Hamadi, H.Y.; Moawed, S. Management of Gestational Hypertension Disorders in Saudi Arabia by Primary Care Nurses. Saudi Crit. Care J. 2020, 4, 103. [Google Scholar] [CrossRef]
- Roberts, J.M.; Gammill, H.S. Preeclampsia. Hypertension 2005, 46, 1243–1249. [Google Scholar] [CrossRef]
- Pelícia, S.M.D.C.; Fekete, S.M.W.; Corrente, J.E.; Rugolo, L.M.S.D.S. Impact of Early-Onset Preeclampsia on Feeding Tolerance and Growth of Very Low Birth Weight Infants during Hospitalization. Rev. Paul. Pediatr. 2023, 41, e2021203. [Google Scholar] [CrossRef]
- Govender, S.; Naicker, T. The Contribution of Complement Protein C1q in COVID-19 and HIV Infection Comorbid with Preeclampsia: A Review. Int. Arch. Allergy Immunol. 2022, 183, 1114–1126. [Google Scholar] [CrossRef]
- Rokotyanskaya, E.A.; Panova, I.A.; Malyshkina, A.I.; Fetisova, I.N.; Fetisov, N.S.; Kharlamova, N.V.; Kuligina, M.V. Technologies for Prediction of Preeclampsia. Sovrem. Tehnol. V Med. 2020, 12, 78–86. [Google Scholar] [CrossRef]
- Soomro, T.A.; Zheng, L.; Afifi, A.J.; Ali, A.; Yin, M.; Gao, J. Artificial Intelligence (AI) for Medical Imaging to Combat Coronavirus Disease (COVID-19): A Detailed Review with Direction for Future Research. Artif. Intell. Rev. 2022, 55, 1409–1439. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial Intelligence in Medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef]
- Kumar, N.; Aggarwal, D. LEARNING-Based Focused WEB Crawler. IETE J. Res. 2021, 67, 1–9. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, S.; Qin, J.; Liu, Y.; Huang, B.; Chen, H. Application of Deep Learning in Automated Analysis of Molecular Images in Cancer: A Survey. Contrast Media Mol. Imaging 2017, 2017, 9512370. [Google Scholar] [CrossRef]
- Bakator, M.; Radosav, D. Deep Learning and Medical Diagnosis: A Review of Literature. Multimodal Technol. Interact. 2018, 2, 47. [Google Scholar] [CrossRef]
- Tahir, M.; Badriyah, T.; Syarif, I. Classification Algorithms of Maternal Risk Detection For Preeclampsia With Hypertension During Pregnancy Using Particle Swarm Optimization. EMITTER Int. J. Eng. Technol. 2018, 6, 236–253. [Google Scholar] [CrossRef]
- Kodepogu, K.R.; Annam, J.R.; Vipparla, A.; Krishna, B.V.N.V.S.; Kumar, N.; Viswanathan, R.; Gaddala, L.K.; Chandanapalli, S.K. A Novel Deep Convolutional Neural Network for Diagnosis of Skin Disease. Traitement Du Signal 2022, 39, 1873–1877. [Google Scholar] [CrossRef]
- Soongsatitanon, A.; Phupong, V. Prediction of Preeclampsia Using First Trimester Placental Protein 13 and Uterine Artery Doppler. J. Matern. Fetal Neonatal Med. 2022, 35, 4412–4417. [Google Scholar] [CrossRef]
- Serra, B.; Mendoza, M.; Scazzocchio, E.; Meler, E.; Nolla, M.; Sabrià, E.; Rodríguez, I.; Carreras, E. A New Model for Screening for Early-Onset Preeclampsia. Am. J. Obstet. Gynecol. 2020, 222, e1–e608. [Google Scholar] [CrossRef]
- Byonanuwe, S.; Fajardo, Y.; Nápoles, D.; Alvarez, A.; Cèspedes, Y.; Ssebuufu, R. Predicting Risk of Chronic Hypertension in Women with Preeclampsia Based on Placenta Histology. A Prospective Cohort Study in Cuba. 2020. Available online: https://www.researchsquare.com/article/rs-44764/v1 (accessed on 12 December 2022).
- Modak, R.; Pal, A.; Pal, A.; Ghosh, M.K. Prediction of Preeclampsia by a Combination of Maternal Spot Urinary Protein-Creatinine Ratio and Uterine Artery Doppler. Int. J. Reprod. Contracept. Obstet. Gynecol. 2020, 9, 635. [Google Scholar] [CrossRef]
- Marić, I.; Tsur, A.; Aghaeepour, N.; Montanari, A.; Stevenson, D.K.; Shaw, G.M.; Winn, V.D. Early Prediction of Preeclampsia via Machine Learning. Am. J. Obstet. Gynecol. MFM 2020, 2, 100100. [Google Scholar] [CrossRef]
- Jhee, J.H.; Lee, S.; Park, Y.; Lee, S.E.; Kim, Y.A.; Kang, S.-W.; Kwon, J.-Y.; Park, J.T. Prediction Model Development of Late-Onset Preeclampsia Using Machine Learning-Based Methods. PLoS ONE 2019, 14, e0221202. [Google Scholar] [CrossRef]
- Marin, I.; Pavaloiu, B.-I.; Marian, C.-V.; Racovita, V.; Goga, N. Early Detection of Preeclampsia Based on a Machine Learning Approach. In Proceedings of the 2019 E-Health and Bioengineering Conference (EHB), Iasi, Romania, 21–23 November 2019; pp. 1–4. [Google Scholar]
- Liu, M.; Yang, X.; Chen, G.; Ding, Y.; Shi, M.; Sun, L.; Huang, Z.; Liu, J.; Liu, T.; Yan, R.; et al. Development of a Prediction Model on Preeclampsia Using Machine Learning-Based Method: A Retrospective Cohort Study in China. Front. Physiol. 2022, 13, 896969. [Google Scholar] [CrossRef]
- Li, Y.; Shen, X.; Yang, C.; Cao, Z.; Du, R.; Yu, M.; Wang, J.; Wang, M. Novel Electronic Health Records Applied for Prediction of Pre-Eclampsia: Machine-Learning Algorithms. Pregnancy Hypertens. 2021, 26, 102–109. [Google Scholar] [CrossRef]
- Carreno, J.F.; Qiu, P. Feature Selection Algorithms for Predicting Preeclampsia: A Comparative Approach. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 2626–2631. [Google Scholar]
- Martínez-Velasco, A.; Martínez-Villaseñor, L.; Miralles-Pechuán, L. Machine Learning Approach for Pre-Eclampsia Risk Factors Association. In Proceedings of the 4th EAI International Conference on Smart Objects and Technologies for Social Good—Goodtechs ’18, Bologna, Italy, 28–30 November 2018; ACM Press: New York, NY, USA, 2018; pp. 232–237. [Google Scholar]
- Bosschieter, T.M.; Xu, Z.; Lan, H.; Lengerich, B.J.; Nori, H.; Sitcov, K.; Souter, V.; Caruana, R. Using Interpretable Machine Learning to Predict Maternal and Fetal Outcomes. arXiv 2022, arXiv:2207.05322. [Google Scholar]
- Schmidt, L.J.; Rieger, O.; Neznansky, M.; Hackelöer, M.; Dröge, L.A.; Henrich, W.; Higgins, D.; Verlohren, S. A Machine-Learning–Based Algorithm Improves Prediction of Preeclampsia-Associated Adverse Outcomes. Am. J. Obstet. Gynecol. 2022, 227, e1–e77. [Google Scholar] [CrossRef]
- Sufriyana, H.; Wu, Y.-W.; Su, E.C.-Y. Prediction of Preeclampsia and Intrauterine Growth Restriction: Development of Machine Learning Models on a Prospective Cohort. JMIR Med. Inform. 2020, 8, e15411. [Google Scholar] [CrossRef]
- Sufriyana, H.; Wu, Y.-W.; Su, E.C.-Y. Artificial Intelligence-Assisted Prediction of Preeclampsia: Development and External Validation of a Nationwide Health Insurance Dataset of the BPJS Kesehatan in Indonesia. EBioMedicine 2020, 54, 102710. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Salerno, S.; Li, Y.; Zhou, L.; Zeng, X.; Li, H. Prediction of Severe Preeclampsia in Machine Learning. Med. Nov. Technol. Devices 2022, 15, 100158. [Google Scholar] [CrossRef]
- Lin, Y.C.; Mallia, D.; Clark-sevilla, A.O.; Catto, A.; Leshchenko, A.; Haas, D.M.; Raja, A.; Salleb-aouissi, A. Preeclampsia Predictor with Machine Learning: A Comprehensive and Bias-Free Machine Learning Pipeline. medRxiv 2022. [Google Scholar] [CrossRef]
- Villalaín, C.; Herraiz, I.; Domínguez-Del Olmo, P.; Angulo, P.; Ayala, J.L.; Galindo, A. Prediction of Delivery Within 7 Days After Diagnosis of Early Onset Preeclampsia Using Machine-Learning Models. Front. Cardiovasc. Med. 2022, 9, 910701. [Google Scholar] [CrossRef]
- Tahir, M.; Badriyah, T.; Syarif, I. Neural Networks Algorithm to Inquire Previous Preeclampsia Factors in Women with Chronic Hypertension During Pregnancy in Childbirth Process. In Proceedings of the 2018 International Electronics Symposium on Knowledge Creation and Intelligent Computing (IES-KCIC), Bali, Indonesia, 29–30 October 2018; pp. 51–55. [Google Scholar]
- Sakinah, N.; Tahir, M.; Badriyah, T.; Syarif, I. LSTM With Adam Optimization-Powered High Accuracy Preeclampsia Classification. In Proceedings of the 2019 International Electronics Symposium (IES), Surabaya, Indonesia, 27–28 September 2019; pp. 314–319. [Google Scholar]
- Manoochehri, Z.; Manoochehri, S.; Soltani, F.; Tapak, L.; Sadeghifar, M. Predicting Preeclampsia and Related Risk Factors Using Data Mining Approaches: A Cross-Sectional Study. Int. J. Reprod. Biomed. 2021, 19, 959–968. [Google Scholar] [CrossRef]
- Han, Q.; Zheng, W.; Guo, X.D.; Zhang, D.; Liu, H.F.; Yu, L.; Yan, J.Y. A New Predicting Model of Preeclampsia Based on Peripheral Blood Test Value. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7222–7229. [Google Scholar]
- Bennett, R.; Mulla, Z.D.; Parikh, P.; Hauspurg, A.; Razzaghi, T. An Imbalance-Aware Deep Neural Network for Early Prediction of Preeclampsia. PLoS ONE 2022, 17, e0266042. [Google Scholar] [CrossRef]
- Dugoff, L.; Hobbins, J.C.; Malone, F.D.; Porter, T.F.; Luthy, D.; Comstock, C.H.; Hankins, G.; Berkowitz, R.L.; Merkatz, I.; Craigo, S.D.; et al. First-Trimester Maternal Serum PAPP-A and Free-Beta Subunit Human Chorionic Gonadotropin Concentrations and Nuchal Translucency Are Associated with Obstetric Complications: A Population-Based Screening Study (The FASTER Trial). Am. J. Obstet. Gynecol. 2004, 191, 1446–1451. [Google Scholar] [CrossRef]
- Seastedt, K.P.; Schwab, P.; O’Brien, Z.; Wakida, E.; Herrera, K.; Marcelo, P.G.F.; Agha-Mir-Salim, L.; Frigola, X.B.; Ndulue, E.B.; Marcelo, A.; et al. Global Healthcare Fairness: We Should Be Sharing More, Not Less, Data. PLOS Digit. Health 2022, 1, e0000102. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.; Cheng, Y.; Kang, Y.; Chen, T.; Yu, H. Federated Learning; Synthesis Lectures on Artificial Intelligence and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2019; Volume 13, pp. 1–207. [Google Scholar] [CrossRef]
- Jiang, L.; Wu, Z.; Xu, X.; Zhan, Y.; Jin, X.; Wang, L.; Qiu, Y. Opportunities and Challenges of Artificial Intelligence in the Medical Field: Current Application, Emerging Problems, and Problem-Solving Strategies. J. Int. Med. Res. 2021, 49, 030006052110001. [Google Scholar] [CrossRef]
- Hicks, S.A.; Strümke, I.; Thambawita, V.; Hammou, M.; Riegler, M.A.; Halvorsen, P.; Parasa, S. On Evaluation Metrics for Medical Applications of Artificial Intelligence. Sci. Rep. 2022, 12, 5979. [Google Scholar] [CrossRef]
- Balagurunathan, Y.; Mitchell, R.; El Naqa, I. Requirements and Reliability of AI in the Medical Context. Phys. Med. 2021, 83, 72–78. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Jiang, Y. Overfitting and Underfitting Analysis for Deep Learning Based End-to-End Communication Systems. In Proceedings of the 2019 11th International Conference on Wireless Communications and Signal Processing (WCSP), Xi’an, China, 23–25 October 2019; pp. 1–6. [Google Scholar]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New Data Preprocessing Trends Based on Ensemble of Multiple Preprocessing Techniques. TrAC Trends Anal. Chem. 2020, 132, 116045. [Google Scholar] [CrossRef]
- Feng, S.; Keung, J.; Yu, X.; Xiao, Y.; Zhang, M. Investigation on the Stability of SMOTE-Based Oversampling Techniques in Software Defect Prediction. Inf. Softw. Technol. 2021, 139, 106662. [Google Scholar] [CrossRef]
- Petch, J.; Di, S.; Nelson, W. Opening the Black Box: The Promise and Limitations of Explainable Machine Learning in Cardiology. Can. J. Cardiol. 2022, 38, 204–213. [Google Scholar] [CrossRef]
- Speith, T. A Review of Taxonomies of Explainable Artificial Intelligence (XAI) Methods. In Proceedings of the 2022 ACM Conference on Fairness, Accountability, and Transparency, Seoul, Republic of Korea, 21–24 June 2022; ACM: New York, NY, USA; pp. 2239–2250. [Google Scholar]
- Grant, K.; McParland, A.; Mehta, S.; Ackery, A.D. Artificial Intelligence in Emergency Medicine: Surmountable Barriers with Revolutionary Potential. Ann. Emerg. Med. 2020, 75, 721–726. [Google Scholar] [CrossRef]
- Atallah, R.; Al-Mousa, A. Heart Disease Detection Using Machine Learning Majority Voting Ensemble Method. In Proceedings of the 2019 2nd International Conference on new Trends in Computing Sciences (ICTCS), Amman, Jordan, 9–11 October 2019; pp. 1–6. [Google Scholar]
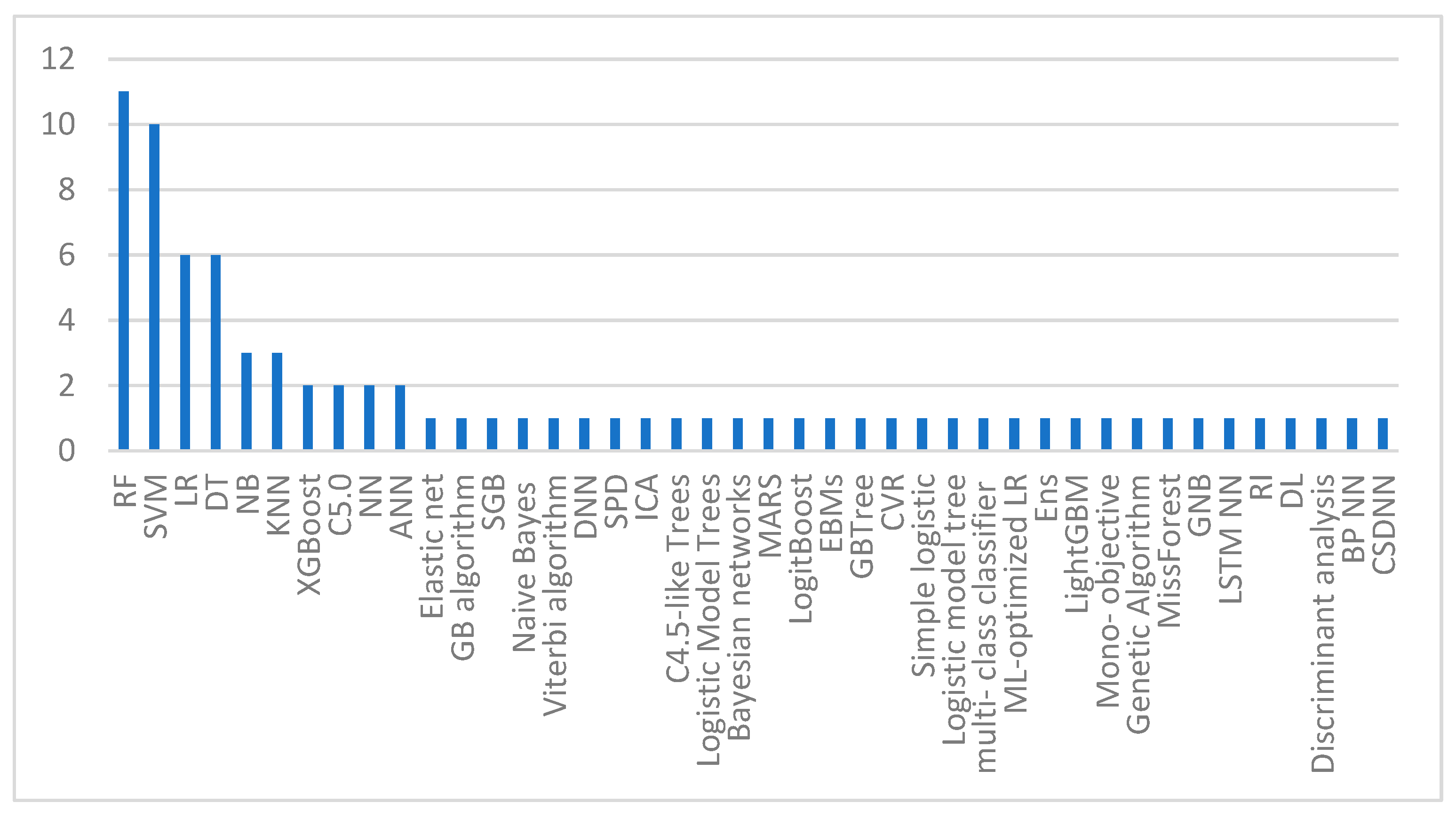

| Ref | Year | Domain | Method | Dataset | Result |
|---|---|---|---|---|---|
| [6] | 2020 | General approaches | LR method with Open Epi system | 12 features and 457 samples taken between 22 and 36 weeks of gestation | AUC of 0.733 |
| [14] | 2020 | General approaches | The predictive values of serum PP13 and UA Doppler tests were calculated using SPSS software package | 15 features and a sample of 353 from the Faculty of Medicine at Chulalongkorn University | Serum PP13 levels and UA PI together resulted in PPV of 12.4%, NPV of 94.4%, specificity of 62.9%, and sensitivity of 58.6% |
| [15] | 2020 | General approaches | Multivariate Gaussian distribution model for preeclampsia screening | 13 features from 6893 general population singleton deliveries at the university hospitals of Vall d’Hebron and Dexeus in Spain | 94% for 10% FPR and 59% for a 5% FPR with AUC of 0.96 (95% CI: 0.94 to 0.98). The detection ratio raised from 59% to 94% by including the placental growth factor in biophysical indicators |
| [16] | 2020 | General approaches | After delivery, the placenta was histologically examined and analyzed | 178 samples of placenta tissues and ultrasound scans, and 10 features in total at the Cuban teaching hospital Carlos Manuel de Cespedes | Villositary infarcts (0.048 p, 1.657 HR, and 95% CI of 1.264–2.848), chorioamnionitis (0.038 p, 1.697 HR, and 95% CI of 1.443–3.416), endarteritis (0.025 p, 1.242 HR, and 95% CI of 1.115–1.804), intervillositay thrombus (0.020 p, 1.529 HR, and 95% CI of 1.231–3.197) |
| [17] | 2019 | General approaches | Midstream urine sample, modified Jaffe’s method, and immunoturbidimetric micro albumin method | 116 pregnant women, with 7 features, in two tertiary teaching hospitals in eastern India | In ROC curve, the AUC for the spot UPCR was 0.949 (95% CI: 0.891–1.000) |
| Ref | Year | Domain | Method | Dataset | Specificity | Sensitivity | Accuracy | AUC |
|---|---|---|---|---|---|---|---|---|
| [18] | 2020 | Machine learning-based | The elastic net, the gradient boosting algorithm | 16,370 deliveries collected from April 2014 to January 2018 at Lucile Packard Children’s Hospital at Stanford, CA | - | 45.2% | - | 0.79 |
| [19] | 2019 | Machine learning-based | RF algorithm, SVM, LR, DT model, SGB, and naïve Bayes classification method | 11,006 records collected from Yonsei University Hospital | 0.991 | 0.603 | 0.973 | - |
| [20] | 2019 | Machine learning-based | The Viterbi algorithm of the i-bracelet system | 105 pregnant women | 72% | 92.5% | 80% | - |
| [21] | 2022 | Machine learning-based | LR, SVM, DNN, DT, and RF | 11,152 records collected from Hospital of Jinan University between December 2015 and September 2019; among them, 143 had preeclampsia, 95 had GH, and there were 10,914 normal pregnancies | - | 0.42 | 0.74 | 0.86 |
| [22] | 2021 | Machine learning-based | RF, XGBoost, LR, and SVM. | 3759 pregnant women who received prenatal care at Xinhua hospital during July 2016 and December 2019 | - | 0.789 | 0.92 | 0.955 |
| [23] | 2020 | Machine learning-based | RF, SVM, SPD, and ICA | Public dataset containing 202 patient records | - | - | - | 0.93 |
| [24] | 2018 | Machine learning-based | RF, SVM, C4.5-like Trees, C5.0, logistic model trees, Bayesian networks, NN, NB, multivariate adaptive regression spline, and boosted logistic regression | Collected from a public study; the dataset included 1634 records | 0.8614 | 0.6846 | 0.8530 | - |
| [25] | 2022 | Machine learning-based | RF and XGBoost | The OBCOAP of the Foundation for Healthcare Quality provided the dataset | - | - | - | 0.770 ± 0.006 |
| [26] | 2022 | Machine learning-based | RF classifier and GBTree | During July 2010 until March 2019, 114 features from pregnancies at the Charité Universitätsmedizin in Germany were used | 97% ± 2% | 66% ± 5% | 89% ± 3% | - |
| [27] | 2020 | Machine learning-based | RF, NB, LR | A sample of 95 women and considering 13 features from a public dataset from a study conducted at Ljubljana University Medical Center | - | - | 90.6%. | - |
| [28] | 2020 | Machine learning-based | SVM, ensemble, ANN, ML-optimized LR, DT, and RF | The BPJS Kesehatan dataset consisting of 95 features was preprocessed to separate 3318 cases of preeclampsia/eclampsia and 19,883 cases of normotensive pregnant women | - | - | - | 95% |
| [29] | 2022 | Machine learning-based | RF, LightGBM, and DT | 248 records, with 10 features from West China Second University Hospital, Sichuan University | 77.27% | 88.37% | - | 89.74% |
| [30] | 2022 | Machine learning-based | LR, RF, SVM, and XGBoost | At eight clinical sites dispersed throughout the US, information was acquired from four visits. The dataset used was created using only 37 training cases | - | - | - | 0.84 ± 0.09 |
| [31] | 2022 | Machine learning-based | Mono-objective, genetic algorithm, MissForest, SVM, KNN, GNB, and DT | 215 samples of the National Institute for Health and Care Excellence (NICE), with 15 features | 80.1% | 77.3% | - | - |
| Ref | Year | Domain | Method | Dataset | Specificity | Sensitivity | Accuracy | AUC |
|---|---|---|---|---|---|---|---|---|
| [32] | 2018 | Deep learning-based | ANN | 239 sample of 2016–2017 medical records from Surabaya Hajj Hospital | - | - | 96.66% | - |
| [33] | 2019 | Deep learning-based | LSTM NN with ADAM optimization | Samples taken from General Hospital of Surabaya Hajj with 16 features | - | - | 90.22% | - |
| [12] | 2018 | Deep learning-based | PSO approach for feature selection. NB, K-NN, DT, NN, SVM, RI, and DL used for classification | 9 features and 1077 patient records collected between 12 December 2017 and 12 February 2018, at two hospitals in Makassar and the Haji General Hospital in Surabaya | - | 90.51% | 95.68% | - |
| [34] | 2021 | Deep learning-based | RF, LR, discriminant analysis, KNN, SVM, and C5.0 DT | Medical records of 1452 with nine features were submitted to Fatemieh Hospital in Hamadan City, located in Iran from April 2005 until March 2015 | 0.780 | 0.800 | 0.791 | - |
| [35] | 2020 | Deep learning-based | Three-layer BP NN: input layer, hidden layer, output layer | 25 features from 568 pregnant women (216 with preeclampsia, 216 with normal pregnancies, and 36 with GH) from the Fujian Maternal and Child Health Hospital for 4 years starting from September 2014 | - | - | 79.8% | - |
| [36] | 2022 | Deep learning-based | CSDNN | 20 features from the Oklahoma and Texas PUDF, and MOMI databases to represent several different minority populations in the US | 0.739 | 0.591 | 0.722 | 0.724 |
| Ref | Demographic | Clinical and Laboratory |
|---|---|---|
| [18] | Maternal age, age group, height, weight, blood type, race, ethnicity, gravida, preeclampsia, diabetes, gestational diabetes, ART, autoimmune conditions, renal disease, anemia, hypertension, obesity, medical history | SBP, DBP, platelet, WBC, red blood cell, UA, hemoglobin, hematocrit, creatinine, glucose, protein, chlamydia, rubella, hepatitis B, varicella |
| [19] | Maternal age, parity number, height and maternal weight, medical history, hypertension, diabetes, preeclampsia | SBP, DBP, UPCR, hemoglobin, platelet count, WBC, creatinine, BUN, AST, ALT, potassium, calcium, magnesium, total bilirubin, TCO2 |
| [20] | Maternal age, BMI, hypertension, preeclampsia, diabetes | SBP, DBP, platelet, EEG, proteinuria, PLGF test for PIGF, UtA doppler, calcium |
| [21] | Age, weight, height, BMI, gestational age, MAP, parity, parous, nulliparity, smoking, hypertension, diabetes, preeclampsia, medical history | SBP, DBP, FGR, PAPP-A, β-HCG, UtA-PI, CRL, proteinuria, creatinine, SLE, APS |
| [23] | Age, BMI, race, gravidity, parity | TLR4, BCL6 protein, IL-2, IL-24, adiponectin, SHBG, CA4, TK1, AFM, PLXNB2 |
| [24] | Age, birth order, birth weight, gestational age, Hispanic origin, race, country of birth, marital status, education, one and five-minute APGAR scores, number of prenatal visits, month of pregnancy when prenatal care began, weight gained during pregnancy, obstetric procedures performed, medical risk, delivery complications, congenital anomalies, parity, gravidity, diabetes, asthma, hypertension, depression, anxiety | - |
| [26] | Age, BMI, height, weight, race, gestational age, antiphospholipid syndrome, diabetes, elevated liver enzymes, epigastric pain, parity, hypertension, headache, preeclampsia, visual disturbances, renal disease | Creatinine, proteinuria, SBP, DBP, PIGF, sFlt, prothrombin time, sodium, thrombocyte count, urea, APTT, LDH, Kalium level, hemoglobin, HCT, ALT, AST, umbilical uterine, UA, PI for middle cerebral artery |
| [29] | Maternal age, parity, BMI, number of fetuses, thyroid disease, diabetes | WBC, RBC, hemoglobin, hematocrit, globulin, GGT, LDH, urea, creatinine, FPG, fibrinogen, TT, PT, INR, APTT, ALT, AST, total bilirubin, protein, albumin, platelet count |
| [30] | Weight, BMI, diet intake, diabetes, hypertension, waist circumference, MAP | Blood pressure, UA, serum biomarkers, PIGF, sFlt-1, PAPP-A, inhibin A, ADAM12, VEGF, sFlt-1 to PIGF ratio, placental analytes, cholesterol, endoglin |
| [31] | Gestational age, height, pre-pregnancy weight, pre-pregnancy BMI, smoking status, race, Risk factors for preeclampsia, previous preeclampsia, chronic hypertension, pre-pregnancy diabetes, nulliparity, family history of preeclampsia, method of conception | Platelets, blood pressure, creatinine, Transaminases, sFlt-1, PIGF, ultrasound data, low-dose aspirin intake, heparin prophylaxis, chronic kidney disease, thrombophilia, systemic lupus erythematosus |
| [32] | Age, MAP, BMI, first pregnancy, childbirth process, medical risk, preeclampsia, hypertension, diabetes | Glucose, proteinuria, SBP, DBP |
| [34] | Age, gravidity, number of children, job, fetus gender, pregnancy season, kidney and heart diseases, diabetes, hypertension, blood group | - |
| [36] | Ethnicity, race, age, border country, insurance, month of delivery, marital status, weight, previous pregnancies, number of abortions and deliveries, infant number, prenatal weight, hypertension, obesity, pre-existing diabetes mellitus, multiple gestations, gestational diabetes mellitus, UTI, infections of genitourinary tract in pregnancy, chronic kidney, Obstructive sleep apnea, and hypertensive heart disease. Primigravida, anemia NOS, iron deficiency anemia, asthma, anxiety, pure hypercholesterolemia, tobacco use disorder, inadequate prenatal care, history of premature delivery, amphetamine dependence, unspecified vitamin D deficiency | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljameel, S.S.; Alzahrani, M.; Almusharraf, R.; Altukhais, M.; Alshaia, S.; Sahlouli, H.; Aslam, N.; Khan, I.U.; Alabbad, D.A.; Alsumayt, A. Prediction of Preeclampsia Using Machine Learning and Deep Learning Models: A Review. Big Data Cogn. Comput. 2023, 7, 32. https://doi.org/10.3390/bdcc7010032
Aljameel SS, Alzahrani M, Almusharraf R, Altukhais M, Alshaia S, Sahlouli H, Aslam N, Khan IU, Alabbad DA, Alsumayt A. Prediction of Preeclampsia Using Machine Learning and Deep Learning Models: A Review. Big Data and Cognitive Computing. 2023; 7(1):32. https://doi.org/10.3390/bdcc7010032
Chicago/Turabian StyleAljameel, Sumayh S., Manar Alzahrani, Reem Almusharraf, Majd Altukhais, Sadeem Alshaia, Hanan Sahlouli, Nida Aslam, Irfan Ullah Khan, Dina A. Alabbad, and Albandari Alsumayt. 2023. "Prediction of Preeclampsia Using Machine Learning and Deep Learning Models: A Review" Big Data and Cognitive Computing 7, no. 1: 32. https://doi.org/10.3390/bdcc7010032
APA StyleAljameel, S. S., Alzahrani, M., Almusharraf, R., Altukhais, M., Alshaia, S., Sahlouli, H., Aslam, N., Khan, I. U., Alabbad, D. A., & Alsumayt, A. (2023). Prediction of Preeclampsia Using Machine Learning and Deep Learning Models: A Review. Big Data and Cognitive Computing, 7(1), 32. https://doi.org/10.3390/bdcc7010032






