An Efficient Algorithm for Sorting and Duplicate Elimination by Using Logarithmic Prime Numbers
Abstract
1. Introduction
2. Current Related Methods
2.1. Pairwise Comparison Test
2.2. Sequential Sort Method
3. The Proposed Method
3.1. Logarithmic Prime Number and Algorithm Description
- It assigns a unique LPN to each distinct set, ensuring no two different sets share the same number. This is essential for accurately distinguishing between sets and identifying duplicates.
- The LPN, L(s), increases consistently as elements are added to the set s. This property helps maintain the relative order of sets when they are sorted by their LPNs.
- The LPN, L(s), offers a compact representation of the set s, encoding the entire set with just one value. This reduces memory usage and enhances the efficiency of sorting and comparison processes.
3.2. Pseudo-Code
| Algorithm 1: Logarithmic Prime Number Approach |
| Input: S = { s1, s2, …, sm} and P = { p1, p2, …, pr}, which means there are a total of r different elements in all the sets. Output: a new S containing the sorted and duplicate-free sets. Step 0:. Let L be an empty map (dictionary) to store the LPNs of elements. Calculate LPNs as l(ei) = log(pi), for i = 1, 2, …, r, and let L(s) = 0 for each set s in S. Step 1: Calculate the LPN for each set s in S using L(s) = L(s) + l(e). Step 2: Sort the sets based on their LPNs. Step 3: Remove duplicate sets in S if their LPNs are equal. |
3.3. Time Complexity Analysis
3.4. Advantages of the Proposed Algorithm
- Efficiency: The LPNA enhances computational efficiency and improves time complexity in processing large datasets. By calculating the LPN of each element only once and directly using it for the related sets, the method reduces redundant computations, which is especially beneficial when the number of sets significantly exceeds the number of elements. Based on the complexity results, Table 4 below provides all the time complexities together to have a more convenient comparison of the algorithms.
- 2.
- Simplicity: The proposed algorithm streamlines the sorting process and simplifies the overall approach in the following ways. Firstly, by representing sets as LPNs, it reduces the complexity of sorting from handling intricate set structures to merely organizing a list of numbers. This simplification not only makes the sorting operation more efficient but also easier to implement. Secondly, the proposed method avoids the need to convert each set into a vector, thereby simplifying the process of sorting and removing duplicates. This leads to a reduction in memory overhead and enhances the readability and maintainability of the algorithm, making it more straightforward and user-friendly, particularly for large datasets.
- 3.
- Compatibility: The proposed method exhibits high compatibility and flexibility, as it can be integrated with any sorting algorithm. This adaptability allows for tailored implementation and optimization based on specific requirements, making the method suitable for a wide range of applications. Furthermore, this compatibility facilitates the integration of the method with existing systems, enhancing its applicability across various domains.
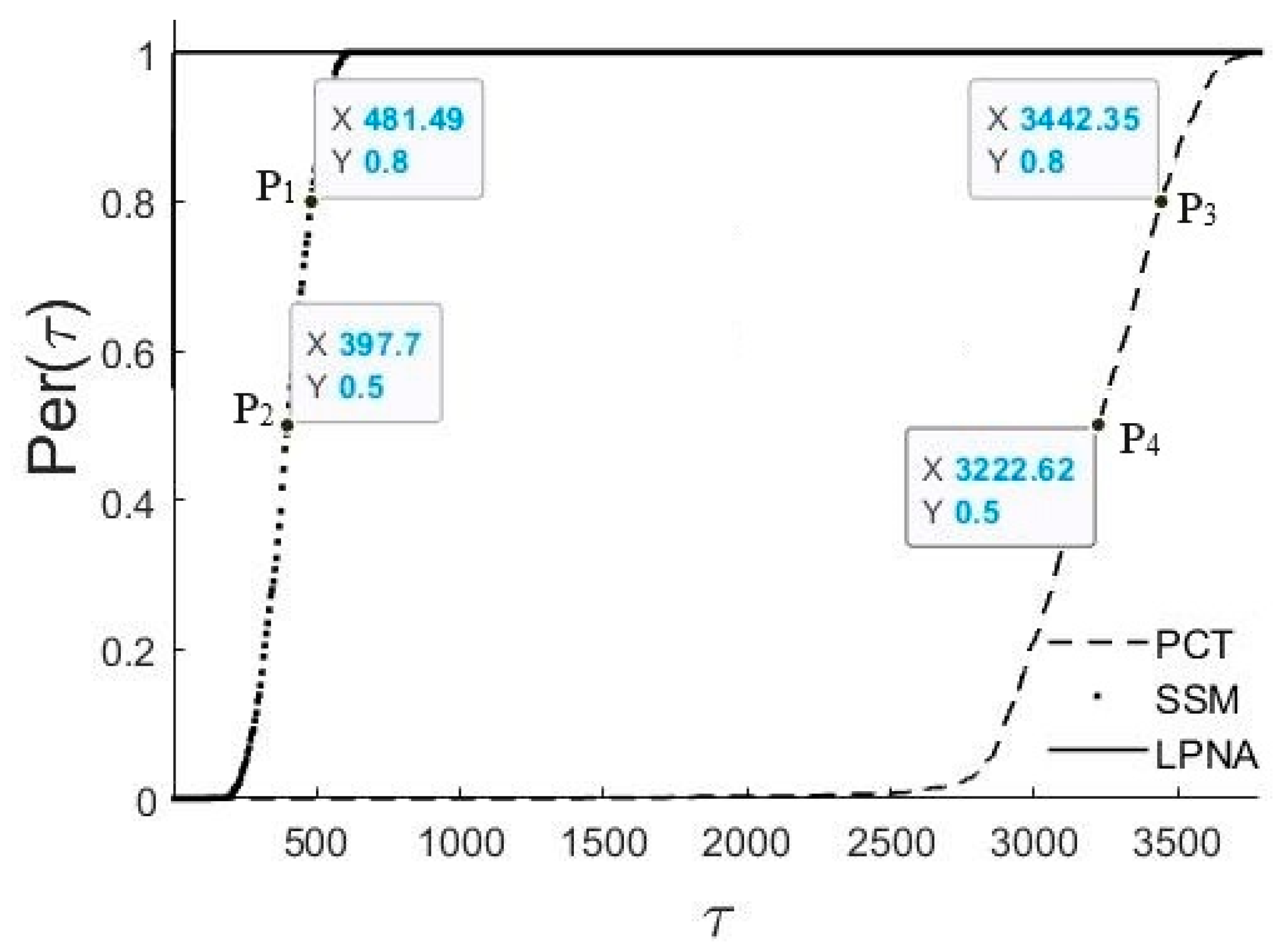
4. Experimental Results
4.1. Randomly Generated Test Problems
4.2. Potential Applications and Practical Examples
- In cybersecurity, analysts often deal with large datasets of malware samples. Duplicate samples can slow down the analysis and cause redundant work. By removing duplicates, the analysis becomes faster and more efficient, improving detection and mitigation strategies. This way, analysts can quickly identify unique threats. In Example 1, we compare the performance of LPNA and SSM in removing duplicates from malware datasets, showing the effectiveness and efficiency of our proposed algorithm.
- In bioinformatics, researchers frequently work with large datasets of genetic sequences, where duplicates can occur due to sequencing errors or redundant data submissions. Removing these duplicates is essential for accurate comparative analyses, like identifying unique genetic markers or mutations. Duplicates can lead to misleading results when comparing gene sequences to identify disease-related genes. Algorithms are used to filter out redundant sequences, ensuring the integrity of the analysis. In Example 2, we show how our proposed algorithm effectively detects and eliminates duplicate gene sequences, demonstrating its superiority over other algorithms from the literature.
- The method can identify and remove duplicate data entries in large databases, enhancing storage efficiency and data integrity.
- By efficiently sorting and removing duplicate user preferences or item similarities, our proposed method can improve the performance of recommendation algorithms, leading to more precise and relevant suggestions for users.
- The method can be utilized to sort and eliminate duplicate text fragments or documents, supporting tasks like plagiarism detection, document clustering, and information retrieval.
- Example 1. (Duplicate Removal in Malware Analysis)
- Example 2. (Duplicate Removal in Gene Sequence Analysis)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Selvi, P. An Analysis on Removal of Duplicate Records using Different Types of Data Mining Techniques: A Survey. Int. J. Comput. Sci. Mob. Comput. 2017, 6, 38–42. [Google Scholar]
- Forghani-Elahabad, M.; Francesquini, E. Usage of task and data parallelism for finding the lower boundary vectors in a stochastic-flow network. Reliab. Eng. Syst. Saf. 2023, 238, 109417. [Google Scholar] [CrossRef]
- Andriyanov, N.; Dementev, V.; Tashlinskiy, A.; Vasiliev, K. The Study of Improving the Accuracy of Convolutional Neural Networks in Face Recognition Tasks. In Pattern Recognition; ICPR International Workshops and Challenges. ICPR 2021. Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12665. [Google Scholar] [CrossRef]
- Marszałek, Z. Parallelization of Modified Merge Sort Algorithm. Symmetry 2017, 9, 176. [Google Scholar] [CrossRef]
- Raj, D.; Remya, R. An Efficient Technique for Removing Duplicates in A Dataset. Int. J. Eng. Res. Technol. 2013, 2, 3889–3893. [Google Scholar]
- Svitov, D.; Alyamkin, S. Margindistillation: Distillation for margin-based softmax. arXiv 2020, arXiv:2003.02586. [Google Scholar]
- Sadanandan, I.T.; Chitturi, B. Optimal Algorithms for Sorting Permutations with Brooms. Algorithms 2022, 15, 220. [Google Scholar] [CrossRef]
- Yeh, W.C. Novel Binary-Addition Tree Algorithm (BAT) for Binary-State Network Reliability Problem. Reliab. Eng. Syst. Saf. 2021, 208, 107448. [Google Scholar] [CrossRef]
- Niu, Y.F.; Shao, F.M. A practical bounding algorithm for computing two-terminal reliability based on decomposition technique. Comput. Math. Appl. 2011, 61, 2241–2246. [Google Scholar] [CrossRef]
- Dhivyabharathi, G.V.; Kumaresan, S. A survey on duplicate record detection in real world data. In Proceedings of the 2016 3rd International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 22–23 January 2016. [Google Scholar] [CrossRef]
- Dong, H.; Ge, Y.; Zhou, R.; Wang, H. An Improved Sorting Algorithm for Periodic PRI Signals Based on Congruence Transform. Symmetry 2024, 16, 398. [Google Scholar] [CrossRef]
- Huang, D.H. An algorithm to generate all d-lower boundary points for a stochastic flow network using dynamic flow constraints. Reliab. Eng. Syst. Saf. 2024, 249, 110217. [Google Scholar] [CrossRef]
- Forghani-elahabad, M.; Alsalami, O.M. Using a Node–Child Matrix to Address the Quickest Path Problem in Multistate Flow Networks under Transmission Cost Constraints. Mathematics 2023, 11, 4889. [Google Scholar] [CrossRef]
- Xu, X.Z.; Niu, Y.F.; Song, Y.F. Computing the reliability of a stochastic distribution network subject to budget constraint. Reliab. Eng. Syst. Saf. 2021, 216, 107947. [Google Scholar] [CrossRef]
- Yeh, W.C. Search for All d-Mincuts of a Limited-Flow Network. Comput. Oper. Res. 2002, 29, 1843–1858. [Google Scholar] [CrossRef]
- Niu, Y.F.; Wei, J.H.; Xu, X.Z. Computing the Reliability of a Multistate Flow Network with Flow Loss Effect. IEEE Trans. Reliab. 2023, 72, 1432–1441. [Google Scholar] [CrossRef]
- Wang, Q.; Jaffres-Runser, K.; Xu, Y.; Scharbarg, J.-L.; An, Z.; Fraboul, C. TDMA Versus CSMA/CA for Wireless Multihop Communications: A Stochastic Worst-Case Delay Analysis. IEEE Trans. Ind. Inform. 2017, 13, 877–887. [Google Scholar] [CrossRef]
- Sosa-Holwerda, A.; Park, O.-H.; Albracht-Schulte, K.; Niraula, S.; Thompson, L.; Oldewage-Theron, W. The Role of Artificial Intelligence in Nutrition Research: A Scoping Review. Nutrients 2024, 16, 2066. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.; Valeske, B.; Rabe, U. Efficient Detection of Defective Parts with Acoustic Resonance Testing Using Synthetic Training Data. Appl. Sci. 2022, 12, 7648. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Shu, J.; Wu, C.H.; Zhou, L.-H.; Song, X.R. Island microgrid based on distributed photovoltaic generation. Power Syst. Prot. Control 2014, 42, 55–61. [Google Scholar]
- Deb, K.; Jain, H. An evolutionary many-objective optimization algorithm using reference-point-based nondominated sorting approach, part I: Solving problems with box constraints. IEEE Trans. Evol. Comput. 2013, 18, 577–601. [Google Scholar] [CrossRef]
- Narushynska, O.; Teslyuk, V.; Doroshenko, A.; Arzubov, M. Data Sorting Influence on Short Text Manual Labeling Quality for Hierarchical Classification. Big Data Cogn. Comput. 2024, 8, 41. [Google Scholar] [CrossRef]
- Bureš, V.; Cabal, J.; Čech, P.; Mls, K.; Ponce, D. The Influence of Criteria Selection Method on Consistency of Pairwise Comparison. Mathematics 2020, 8, 2200. [Google Scholar] [CrossRef]
- Basheer Ahmed, M.I.; Zaghdoud, R.; Ahmed, M.S.; Sendi, R.; Alsharif, S.; Alabdulkarim, J.; Albin Saad, B.A.; Alsabt, R.; Rahman, A.; Krishnasamy, G. A Real-Time Computer Vision Based Approach to Detection and Classification of Traffic Incidents. Big Data Cogn. Comput. 2023, 7, 22. [Google Scholar] [CrossRef]
- Krivulin, N.; Prinkov, A.; Gladkikh, I. Using Pairwise Comparisons to Determine Consumer Preferences in Hotel Selection. Mathematics 2022, 10, 730. [Google Scholar] [CrossRef]
- Huang, D.-H.; Huang, C.-F.; Lin, Y.-K. Reliability Evaluation for a Stochastic Flow Network Based on Upper and Lower Boundary Vectors. Mathematics 2019, 7, 1115. [Google Scholar] [CrossRef]
- Dodevska, Z.; Radovanović, S.; Petrović, A.; Delibašić, B. When Fairness Meets Consistency in AHP Pairwise Comparisons. Mathematics 2023, 11, 604. [Google Scholar] [CrossRef]
- Cheon, J.; Son, J.; Ahn, Y. Economic and environmental factor-integrated optimal model for plastic-waste sorting. J. Ind. Eng. Chem. 2024; in press. [Google Scholar] [CrossRef]
- Qian, K.; Fachrizal, R.; Munkhammar, J.; Ebel, T.; Adam, R. Large-scale EV charging scheduling considering on-site PV generation by combining an aggregated model and sorting-based methods. Sustain. Cities Soc. 2024, 107, 105453. [Google Scholar] [CrossRef]
- Liu, T.; Chen, X.; Peng, Q.; Peng, J.; Meng, J. An enhanced sorting method for retired battery with feature selection and multiple clustering. J. Energy Storage 2024, 87, 111422. [Google Scholar] [CrossRef]
- Carbó-Dorca, R. On Prime Numbers Generation and Pairing. Int. J. Innov. Res. Sci. Eng. Stud. (IJIRSES) 2023, 3, 12–17. [Google Scholar]
- Dolan, E.D.; Moré, J.J. Benchmarking optimization software with performance profiles. Math. Program. 2002, 91, 201–213. [Google Scholar] [CrossRef]
- Ramamoorthy, J.; Gupta, K.; Shashidhar, N.K.; Varol, C. Linux IoT Malware Variant Classification Using Binary Lifting and Opcode Entropy. Electronics 2024, 13, 2381. [Google Scholar] [CrossRef]
- Brown, T.A. Gene Cloning and DNA Analysis: An Introduction; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Laforgia, A.; Inchingolo, A.D.; Piras, F.; Colonna, V.; Giorgio, R.V.; Carone, C.; Rapone, B.; Malcangi, G.; Inchingolo, A.M.; Inchingolo, F.; et al. Therapeutic Strategies and Genetic Implications for Periodontal Disease Management: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 7217. [Google Scholar] [CrossRef] [PubMed]
| s1 | s2 | s3 | s4 | s5 | s6 | s7 | |
|---|---|---|---|---|---|---|---|
| s1 | ≠ | ≠ | ≠ | ≠ | ≠ | ≠ | |
| s2 | ≠ | = | ≠ | ≠ | = | ||
| s3 | ≠ | ≠ | |||||
| s5 | ≠ |
| i | e1 | e2 | e3 | e4 | e5 | e6 | |
|---|---|---|---|---|---|---|---|
| Before employing the SSM | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 2 | 1 | 0 | 0 | 1 | 0 | 1 | |
| 3 | 0 | 1 | 1 | 0 | 1 | 0 | |
| 4 | 1 | 0 | 0 | 1 | 0 | 1 | |
| 5 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 6 | 0 | 1 | 0 | 0 | 0 | 1 | |
| 7 | 1 | 0 | 0 | 1 | 0 | 1 | |
| After employing the SSM | 6 | 0 | 1 | 0 | 0 | 0 | 1 |
| 3 | 0 | 1 | 1 | 0 | 1 | 0 | |
| 5 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 2 | 1 | 0 | 0 | 1 | 0 | 1 | |
| 4 | 1 | 0 | 0 | 1 | 0 | 1 | |
| 7 | 1 | 0 | 0 | 1 | 0 | 1 |
| i | e1 | e2 | e3 | e4 | e5 | e6 | L(si) |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 0 | 0 | 0 | 1 | 0 | log(2) + log(11) = 1.342423 |
| 2 | 1 | 0 | 0 | 1 | 0 | 1 | log(2) + log(7) + log(13) = 2.260071 |
| 3 | 0 | 1 | 1 | 0 | 1 | 0 | log(3) + log(5) + log(11) = 2.217484 |
| 4 | 1 | 0 | 0 | 1 | 0 | 1 | log(2) + log(7) + log(13) = 2.260071 |
| 5 | 1 | 0 | 0 | 0 | 0 | 1 | log(2) + log(13) = 1.414973 |
| 6 | 0 | 1 | 0 | 0 | 0 | 1 | log(3) + log(13) = 1.591065 |
| 7 | 1 | 0 | 0 | 1 | 0 | 1 | log(2) + log(7) + log(13) = 2.260071 |
| Approaches | Time Complexities |
|---|---|
| PCT | O(n∙m2) |
| SSM | O(n∙m∙log(m)) |
| LPNA | O(m∙n +m∙log(m)) |
| ith One Hundred | m | r | tPCT | tSSM | tLPNA | tPCT/tLPNA | tSSM/tLPNA |
|---|---|---|---|---|---|---|---|
| 1 | 8860 | 22.71 | 11.8533 | 0.9996 | 0.0046 | 2566.8 | 216.45 |
| 2 | 9000.8 | 22.14 | 12.1828 | 0.9647 | 0.0046 | 2668.7 | 211.32 |
| 3 | 8938.3 | 22.48 | 11.8803 | 0.9717 | 0.0044 | 2676.5 | 218.92 |
| 4 | 9080.1 | 22.24 | 12.2871 | 1.0144 | 0.0046 | 2661.3 | 219.71 |
| 5 | 9027.5 | 22.68 | 12.0707 | 1.0307 | 0.0045 | 2683.1 | 229.11 |
| 6 | 9036 | 22.58 | 12.1253 | 1.0378 | 0.0045 | 2720.8 | 232.87 |
| 7 | 8977.2 | 22.49 | 12.0541 | 1.0078 | 0.0044 | 2745.3 | 229.52 |
| 8 | 9071 | 22.42 | 12.1596 | 1.0041 | 0.0044 | 2768.9 | 228.64 |
| 9 | 9043.4 | 22.59 | 12.1674 | 1.0479 | 0.0045 | 2681.5 | 230.95 |
| 10 | 9069.4 | 22.22 | 12.2237 | 0.9888 | 0.0045 | 2710 | 219.21 |
| m | r | tSSM | tLPNA | tSSM/tLPNA |
|---|---|---|---|---|
| 5 × 105 | 50 | 8641.8 | 0.28622 | 30,193 |
| m | r | tSSM | tLPNA | tSSM/tLPNA |
|---|---|---|---|---|
| 106 | 100 | 66,707 | 0.92685 | 71,971 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, W.-C.; Forghani-elahabad, M. An Efficient Algorithm for Sorting and Duplicate Elimination by Using Logarithmic Prime Numbers. Big Data Cogn. Comput. 2024, 8, 96. https://doi.org/10.3390/bdcc8090096
Yeh W-C, Forghani-elahabad M. An Efficient Algorithm for Sorting and Duplicate Elimination by Using Logarithmic Prime Numbers. Big Data and Cognitive Computing. 2024; 8(9):96. https://doi.org/10.3390/bdcc8090096
Chicago/Turabian StyleYeh, Wei-Chang, and Majid Forghani-elahabad. 2024. "An Efficient Algorithm for Sorting and Duplicate Elimination by Using Logarithmic Prime Numbers" Big Data and Cognitive Computing 8, no. 9: 96. https://doi.org/10.3390/bdcc8090096
APA StyleYeh, W.-C., & Forghani-elahabad, M. (2024). An Efficient Algorithm for Sorting and Duplicate Elimination by Using Logarithmic Prime Numbers. Big Data and Cognitive Computing, 8(9), 96. https://doi.org/10.3390/bdcc8090096








