Xc- System as a Possible Target for ConBr Lectin Interaction in Glioma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture of Rat Glioblastoma C6 and Treatment
2.2. Cell Viability
2.3. Reactive Species Production
2.4. Non-Protein Thiol (NPSH) Content Quantification
2.5. Statistical Analysis
2.6. Molecular Docking of ConBr with N-glycans Present in CD98hc and Construction of the ConBr-Xc- System Complex
2.7. Molecular Dynamics
3. Results
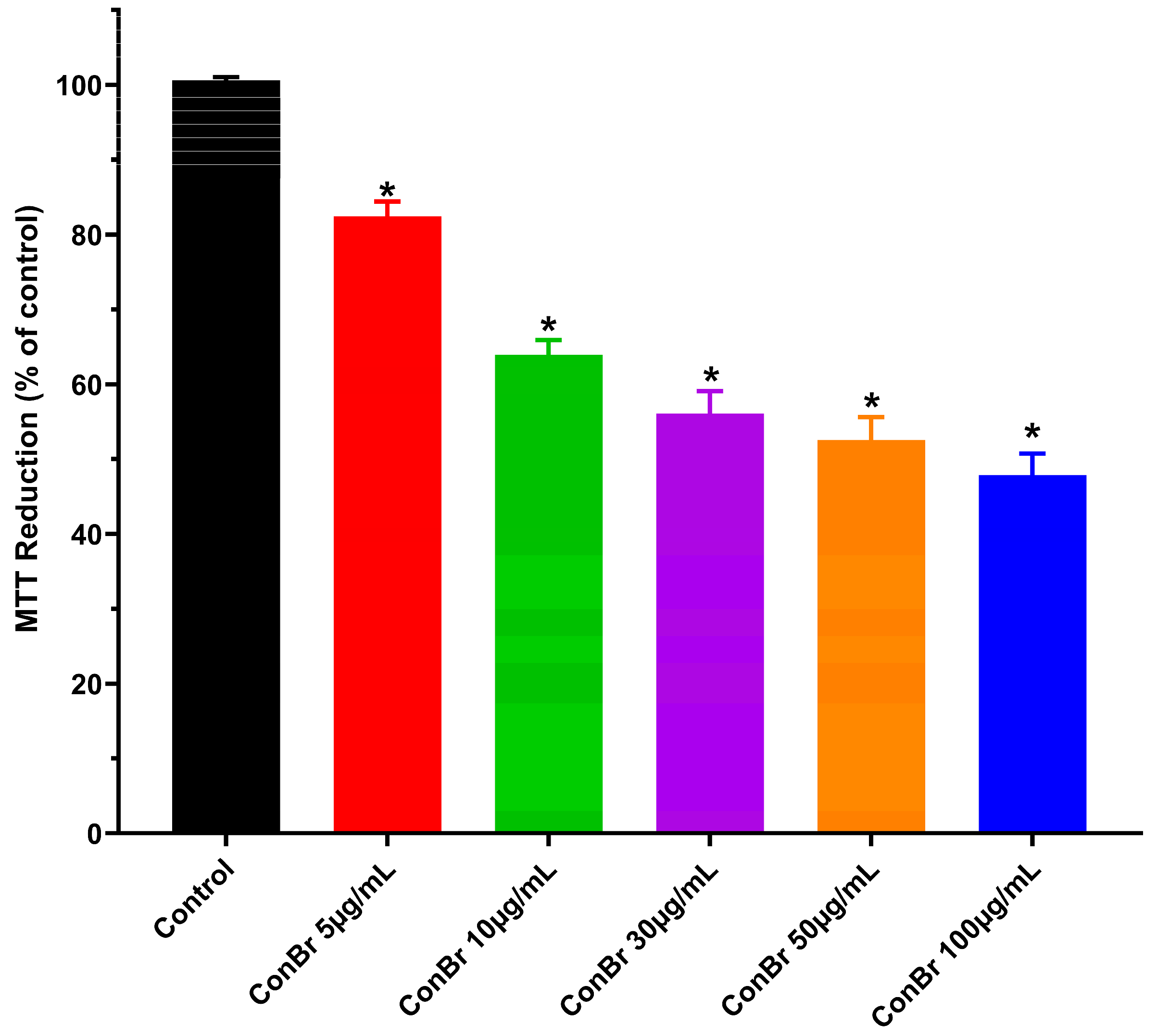
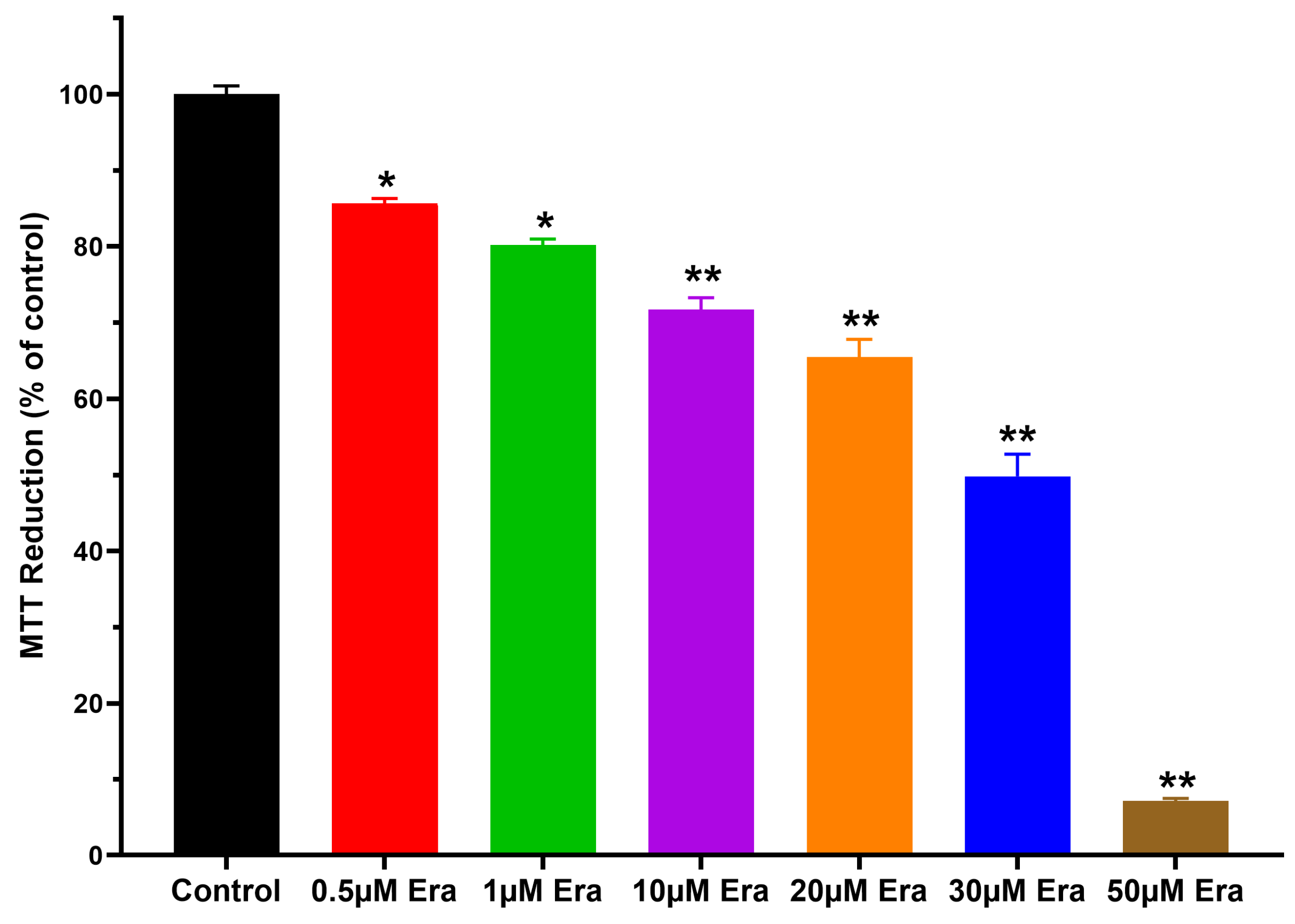
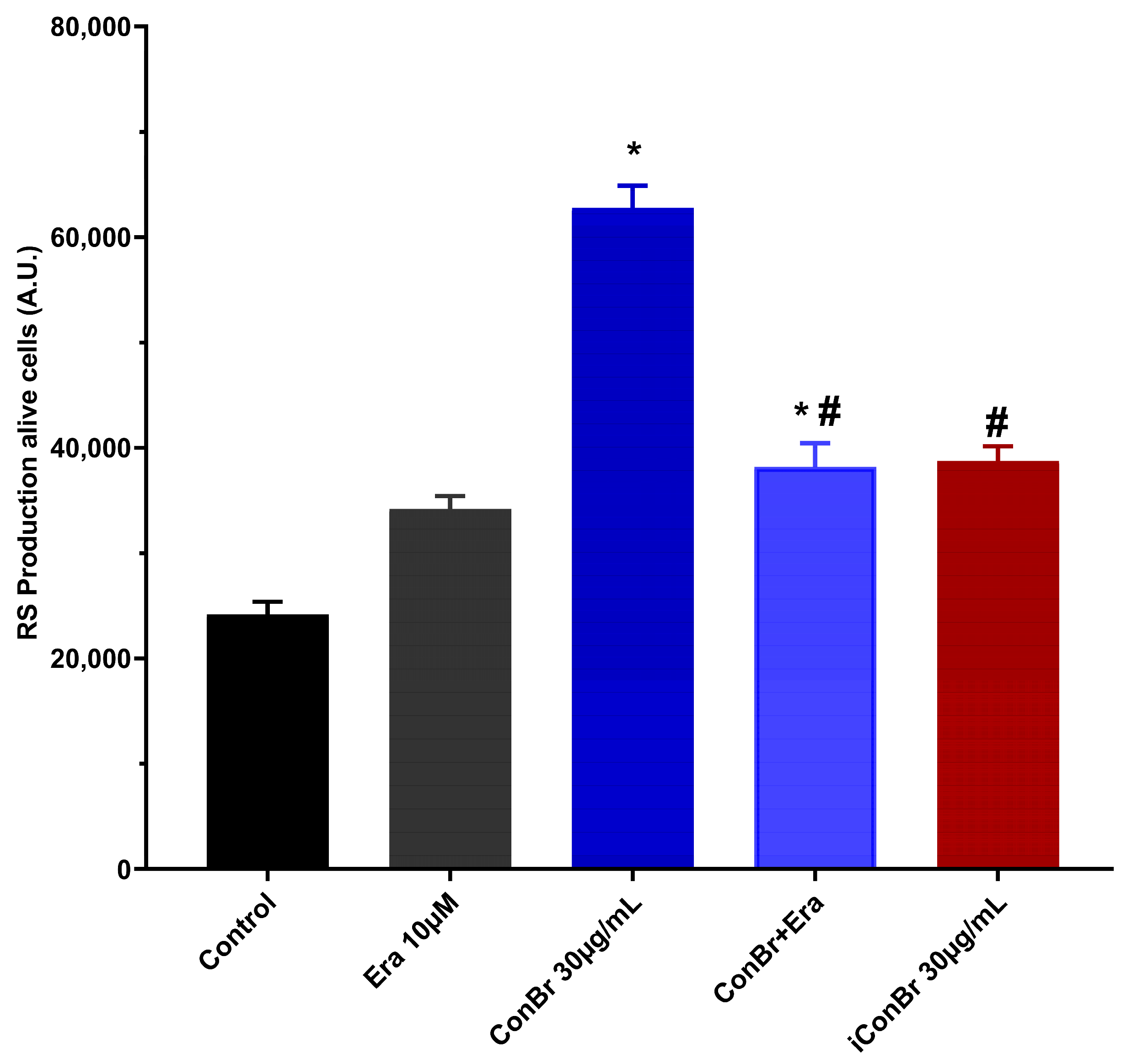
3.1. Effect of ConBr on C6 Glioma Cells via Modulation of Xc- System
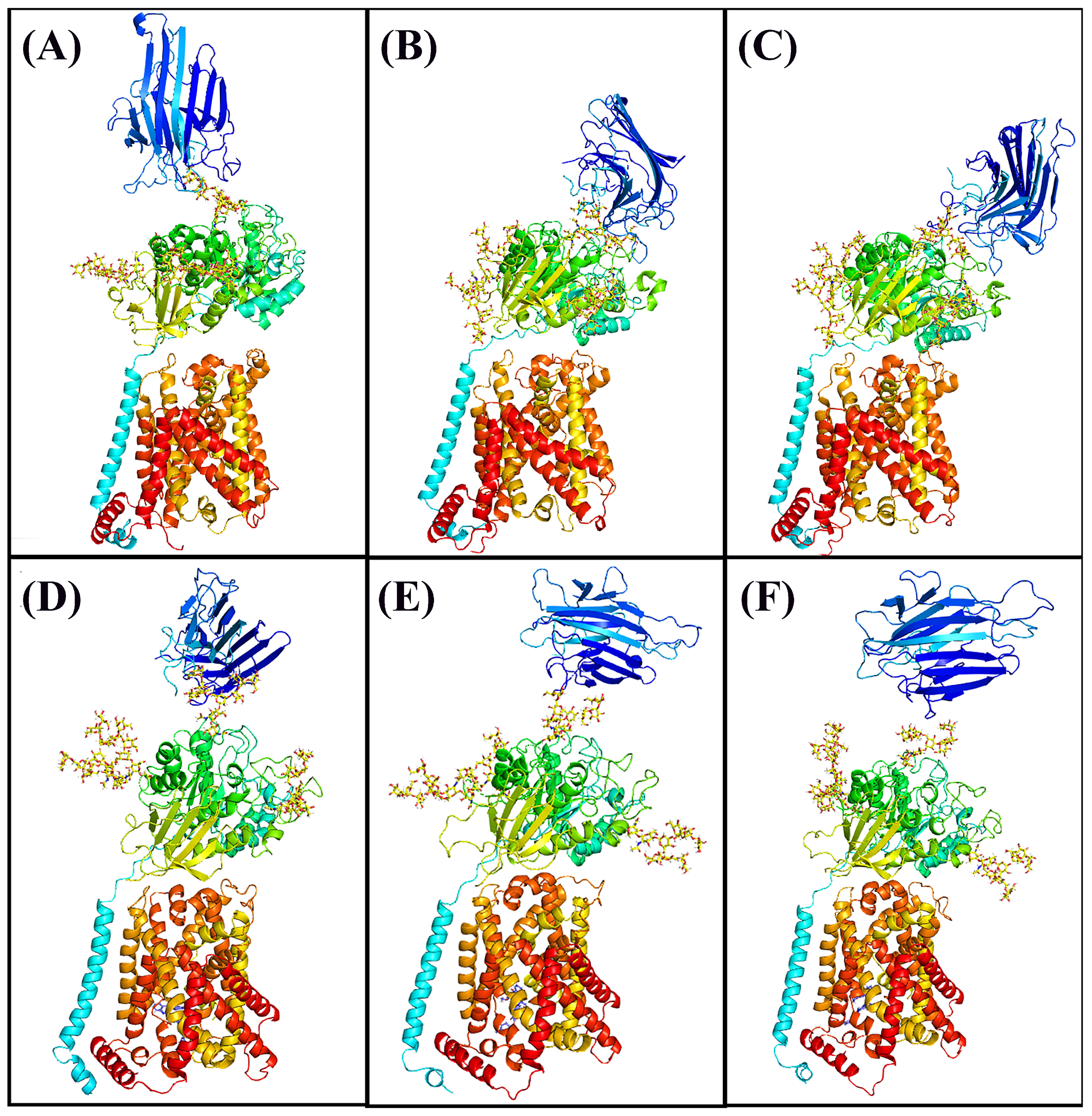
3.2. Molecular Docking
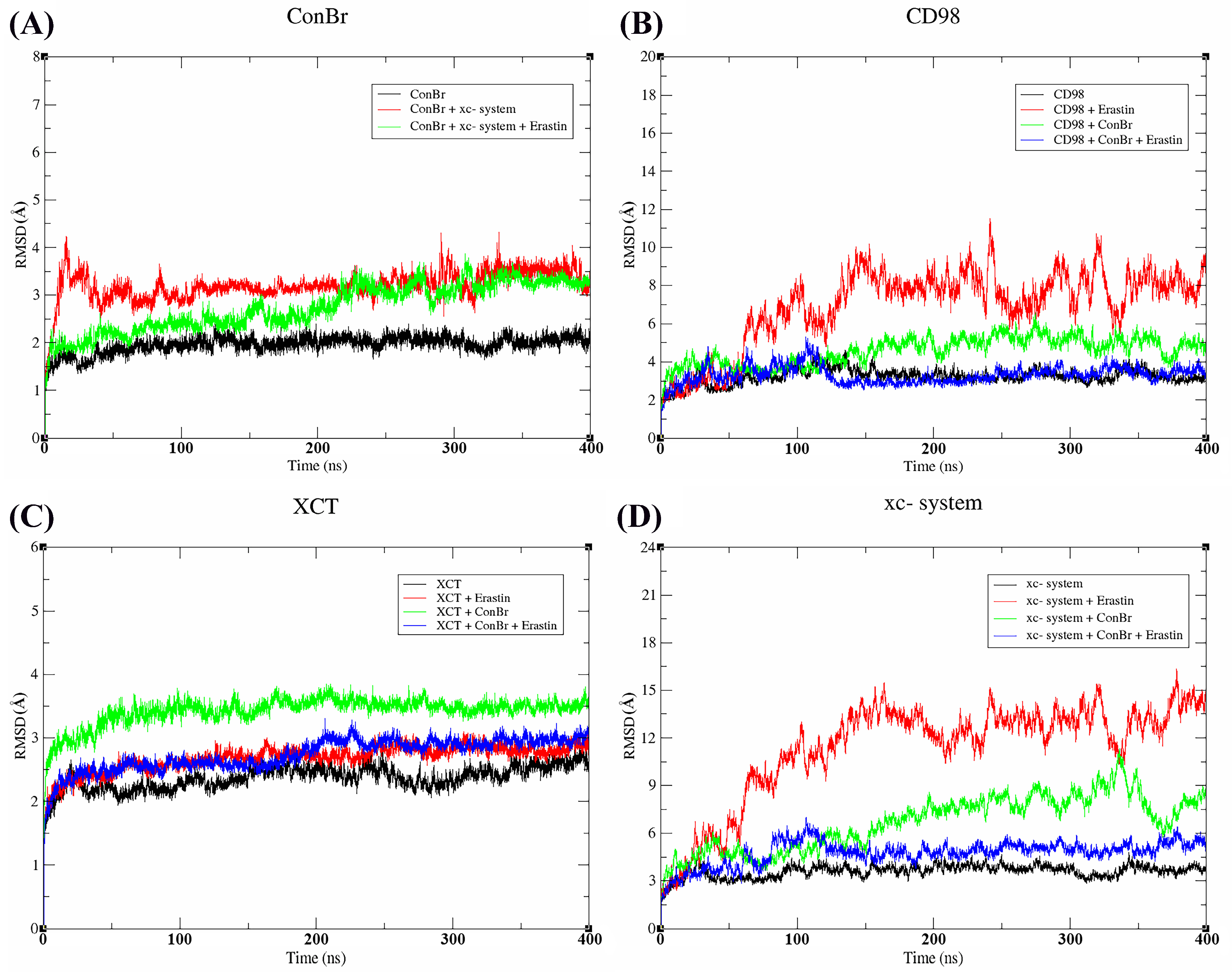
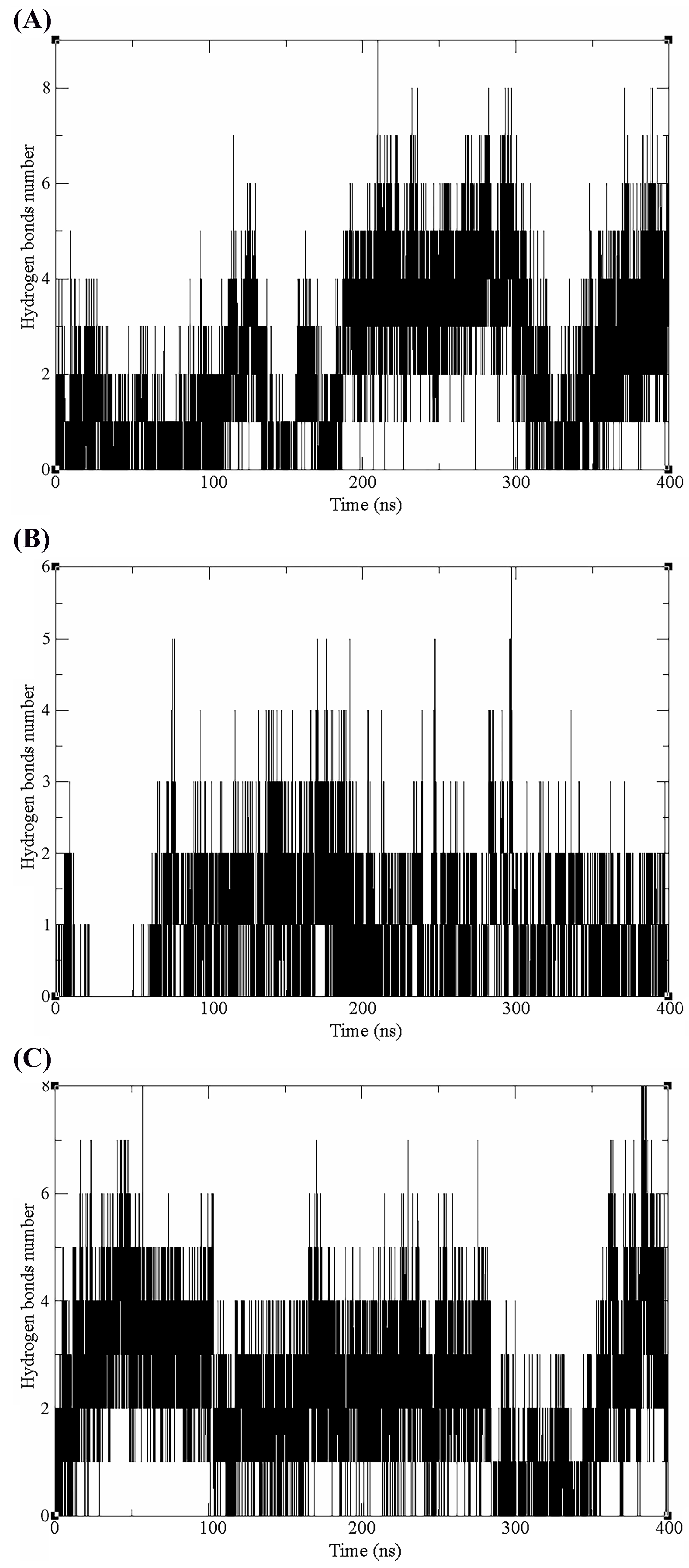
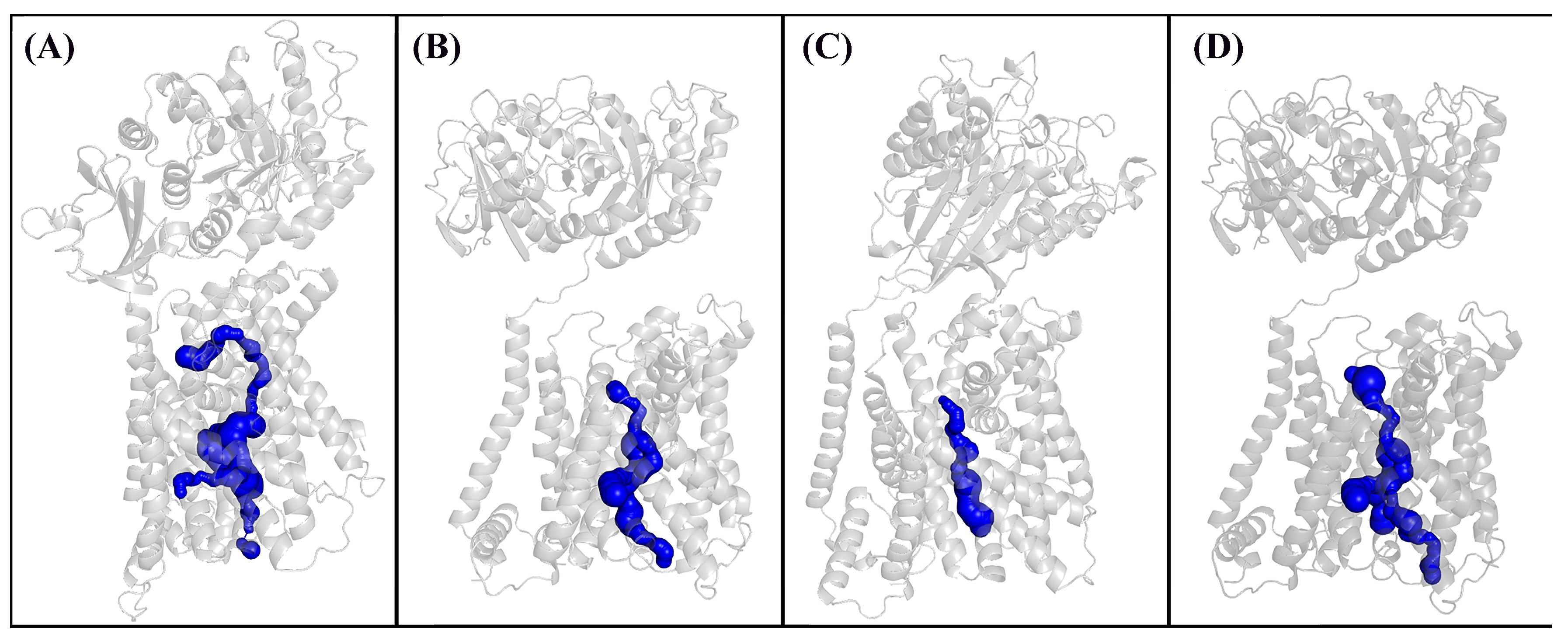
3.3. Molecular Dynamics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Prim. 2015, 1, 15017. [Google Scholar] [CrossRef]
- Horbinski, C.; Berger, T.; Packer, R.J.; Wen, P.Y. Clinical Implications of the 2021 Edition of the WHO Classification of Central Nervous System Tumours. Nat. Rev. Neurol. 2022, 18, 515–529. [Google Scholar] [CrossRef]
- Reifenberger, G.; Wirsching, H.-G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the Molecular Genetics of Gliomas—Implications for Classification and Therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
- Yool, A.J.; Ramesh, S. Molecular Targets for Combined Therapeutic Strategies to Limit Glioblastoma Cell Migration and Invasion. Front. Pharmacol. 2020, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, J.-I.; Tsuda, M.; Okada, K.; Kimura, T.; Piao, J.; Tanaka, S.; Shinohara, Y. Comprehensive Glycomics of a Multistep Human Brain Tumor Model Reveals Specific Glycosylation Patterns Related to Malignancy. PLoS ONE 2015, 10, e0128300. [Google Scholar] [CrossRef]
- Yue, J.; Huang, R.; Lan, Z.; Xiao, B.; Luo, Z. Abnormal Glycosylation in Glioma: Related Changes in Biology, Biomarkers and Targeted Therapy. Biomark. Res. 2023, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- van Solinge, T.S.; Nieland, L.; Chiocca, E.A.; Broekman, M.L.D. Advances in Local Therapy for Glioblastoma—Taking the Fight to the Tumour. Nat. Rev. Neurol. 2022, 18, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, S.K.; Panda, P.K.; Sinha, N.; Praharaj, P.P.; Bhol, C.S.; Panigrahi, D.P.; Mahapatra, K.K.; Saha, S.; Patra, S.; Mishra, S.R.; et al. Plant Lectins in Cancer Therapeutics: Targeting Apoptosis and Autophagy-Dependent Cell Death. Pharmacol. Res. 2019, 144, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Cavada, B.S.; Osterne, V.J.S.; Pinto-Junior, V.R.; Nascimento, K.S. ConBr, the Lectin from Canavalia Brasiliensis Mart. Seeds: Forty Years of Research. Curr. Protein Pept. Sci. 2019, 20, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.K.; Sharma, D.; Sharma, J.; Saini, K.C. Legume Lectins: Potential Use as a Diagnostics and Therapeutics against the Cancer. Int. J. Biol. Macromol. 2020, 142, 474–483. [Google Scholar] [CrossRef]
- Mazalovska, M.; Kouokam, J.C. Plant-Derived Lectins as Potential Cancer Therapeutics and Diagnostic Tools. BioMed Res. Int. 2020, 2020, 1631394. [Google Scholar] [CrossRef] [PubMed]
- Pace, A.; Scirocchi, F.; Napoletano, C.; Zizzari, I.G.; D’Angelo, L.; Santoro, A.; Nuti, M.; Rahimi, H.; Rughetti, A. Glycan-Lectin Interactions as Novel Immunosuppression Drivers in Glioblastoma. Int. J. Mol. Sci. 2022, 23, 6312. [Google Scholar] [CrossRef] [PubMed]
- Wolin, I.A.V.; Heinrich, I.A.; Nascimento, A.P.M.; Welter, P.G.; Sosa, L.D.V.; De Paul, A.L.; Zanotto-Filho, A.; Nedel, C.B.; Lima, L.D.; Osterne, V.J.S.; et al. ConBr Lectin Modulates MAPKs and Akt Pathways and Triggers Autophagic Glioma Cell Death by a Mechanism Dependent upon Caspase-8 Activation. Biochimie 2021, 180, 186–204. [Google Scholar] [CrossRef] [PubMed]
- Leal, R.B.; Mann, J.; Pinto-Junior, V.R.; Oliveira, M.V.; Osterne, V.J.S.; Wolin, I.A.V.; Nascimento, A.P.M.; Welter, P.G.; Ferreira, V.M.S.; Silva, A.A.; et al. Structural Prediction and Characterization of Canavalia Grandiflora (ConGF) Lectin Complexed with MMP1: Unveiling the Antiglioma Potential of Legume Lectins. Molecules 2022, 27, 7089. [Google Scholar] [CrossRef] [PubMed]
- Leal, R.B.; Pinto-Junior, V.R.; Osterne, V.J.S.; Wolin, I.A.V.; Nascimento, A.P.M.; Neco, A.H.B.; Araripe, D.A.; Welter, P.G.; Neto, C.C.; Correia, J.L.A.; et al. Crystal Structure of DlyL, a Mannose-Specific Lectin from Dioclea Lasiophylla Mart. Ex Benth Seeds That Display Cytotoxic Effects against C6 Glioma Cells. Int. J. Biol. Macromol. 2018, 114, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.P.M.; Knaut, J.L.; Rieger, D.K.; Wolin, I.A.V.; Heinrich, I.A.; Mann, J.; Juarez, A.V.; Sosa, L.D.V.; De Paul, A.L.; Moreira, C.G.; et al. Anti-Glioma Properties of DVL, a Lectin Purified from Dioclea Violacea. Int. J. Biol. Macromol. 2018, 120, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, K.S.; Andrade, M.L.L.; Silva, I.B.; Domingues, D.L.; Chicas, L.S.; Silva, M.T.L.; Bringel, P.H.S.F.; Marques, G.F.O.; Martins, M.G.Q.; Lóssio, C.F.; et al. Heterologous Production of α-Chain of Dioclea sclerocarpa Lectin: Enhancing the Biological Effects of a Wild-Type Lectin. Int. J. Biol. Macromol. 2020, 156, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.; Choudhury, A.; Keough, M.B.; Seo, K.; Ni, L.; Kakaizada, S.; Lee, A.; Aabedi, A.; Popova, G.; Lipkin, B.; et al. Glioblastoma Remodelling of Human Neural Circuits Decreases Survival. Nature 2023, 617, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.S.L.; Klamt, F.; Thomé, C.C.; Worm, P.V.; de Oliveira, D.L. Metabotropic Glutamate Receptors as a New Therapeutic Target for Malignant Gliomas. Oncotarget 2017, 8, 22279–22298. [Google Scholar] [CrossRef]
- Venkataramani, V.; Tanev, D.I.; Strahle, C.; Studier-Fischer, A.; Fankhauser, L.; Kessler, T.; Körber, C.; Kardorff, M.; Ratliff, M.; Xie, R.; et al. Glutamatergic Synaptic Input to Glioma Cells Drives Brain Tumour Progression. Nature 2019, 573, 532–538. [Google Scholar] [CrossRef]
- Venkatesh, H.S.; Morishita, W.; Geraghty, A.C.; Silverbush, D.; Gillespie, S.M.; Arzt, M.; Tam, L.T.; Espenel, C.; Ponnuswami, A.; Ni, L.; et al. Electrical and Synaptic Integration of Glioma into Neural Circuits. Nature 2019, 573, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.; Annabi, B. Induction of Autophagy Biomarker BNIP3 Requires a JAK2/STAT3 and MT1-MMP Signaling Interplay in Concanavalin-A-Activated U87 Glioblastoma Cells. Cell. Signal. 2014, 26, 917–924. [Google Scholar] [CrossRef]
- Pratt, J.; Iddir, M.; Bourgault, S.; Annabi, B. Evidence of MTCBP-1 Interaction with the Cytoplasmic Domain of MT1-MMP: Implications in the Autophagy Cell Index of High-Grade Glioblastoma. Mol. Carcinog. 2016, 55, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Deme, J.C.; Kolokouris, D.; Kuteyi, G.; Biggin, P.C.; Lea, S.M.; Newstead, S. Molecular Basis for Redox Control by the Human Cystine/glutamate Antiporter System Xc. Nat. Commun. 2021, 12, 7147. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Xie, E.; Li, Y.; Li, J.; Zhang, Y.; Chi, X.; Hu, X.; Xu, L.; Hou, T.; Stockwell, B.R.; et al. The Structure of Erastin-Bound xCT-4F2hc Complex Reveals Molecular Mechanisms Underlying Erastin-Induced Ferroptosis. Cell Res. 2022, 32, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, X.; Huang, P. xCT: A Critical Molecule That Links Cancer Metabolism to Redox Signaling. Mol. Ther. 2020, 28, 2358–2366. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-R.; Zhu, W.-T.; Pei, D.-S. System Xc-: A Key Regulatory Target of Ferroptosis in Cancer. Investig. New Drugs 2021, 39, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.; Judkins, J.; Salomonis, N.; Matlaf, L.; Soteropoulos, P.; McAllister, S.; Soroceanu, L. Reactive Oxygen Species-Mediated Therapeutic Response and Resistance in Glioblastoma. Cell Death Dis. 2015, 6, e1601. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, Biology and Role in Disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Dixon, S.J.; Patel, D.N.; Welsch, M.; Skouta, R.; Lee, E.D.; Hayano, M.; Thomas, A.G.; Gleason, C.E.; Tatonetti, N.P.; Slusher, B.S.; et al. Pharmacological Inhibition of Cystine–glutamate Exchange Induces Endoplasmic Reticulum Stress and Ferroptosis. eLife 2014, 3, e02523. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, I.; Yoshimura, S.H.; Katoh, H. High Cell Density Increases Glioblastoma Cell Viability under Glucose Deprivation via Degradation of the Cystine/glutamate Transporter xCT (SLC7A11). J. Biol. Chem. 2020, 295, 6936–6945. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, X.; Yu, K.; Xu, X.; Chen, T.; Shi, Y.; Wang, Y.; Qiu, S.; Guo, S.; Cui, J.; et al. Targeting SLC3A2 Subunit of System XC- Is Essential for m6A Reader YTHDC2 to Be an Endogenous Ferroptosis Inducer in Lung Adenocarcinoma. Free Radic. Biol. Med. 2021, 168, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Console, L.; Scalise, M.; Salerno, S.; Scanga, R.; Giudice, D.; De Bartolo, L.; Tonazzi, A.; Indiveri, C. N-Glycosylation Is Crucial for Trafficking and Stability of SLC3A2 (CD98). Sci. Rep. 2022, 12, 14570. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Ikeda, S.; Yaga, M.; Watanabe, K.; Urakawa, R.; Iehara, A.; Iwai, M.; Hashiguchi, S.; Morimoto, S.; Fujiki, F.; et al. Selective Targeting of Multiple Myeloma Cells with a Monoclonal Antibody Recognizing the Ubiquitous Protein CD98 Heavy Chain. Sci. Transl. Med. 2022, 14, eaax7706. [Google Scholar] [CrossRef] [PubMed]
- Cavada, B.S.; de Oliveira, M.V.; Osterne, V.J.S.; Pinto-Junior, V.R.; Martins, F.W.V.; Correia-Neto, C.; Pinheiro, R.F.; Leal, R.B.; Nascimento, K.S. Recent Advances in the Use of Legume Lectins for the Diagnosis and Treatment of Breast Cancer. Biochimie 2023, 208, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Jiang, X. The Chemistry and Biology of Ferroptosis. Cell Chem. Biol. 2020, 27, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L. Tissue Sulfhydryl Groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Powlesland, A.S.; Hitchen, P.G.; Parry, S.; Graham, S.A.; Barrio, M.M.; Elola, M.T.; Mordoh, J.; Dell, A.; Drickamer, K.; Taylor, M.E. Targeted Glycoproteomic Identification of Cancer Cell Glycosylation. Glycobiology 2009, 19, 899–909. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Aparicio, J.; Hermoso, J.; Grangeiro, T.B.; Calvete, J.J.; Cavada, B.S. The Crystal Structure of Canavalia Brasiliensis Lectin Suggests a Correlation between Its Quaternary Conformation and Its Distinct Biological Properties from Concanavalin A. FEBS Lett. 1997, 405, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical Scoring Functions for Advanced Protein-Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.M.; Heck, G.S.; de Avila, M.B.; Levin, N.M.B.; Pintro, V.O.; Carvalho, N.L.; Azevedo, W.F. de SAnDReS a Computational Tool for Statistical Analysis of Docking Results and Development of Scoring Functions. Comb. Chem. High. Throughput Screen. 2016, 19, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hitzenberger, M.; Rieger, M.; Kern, N.R.; Zacharias, M.; Im, W. CHARMM-GUI Supports the Amber Force Fields. J. Chem. Phys. 2020, 153, 035103. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Loncharich, R.J.; Brooks, B.R.; Pastor, R.W. Langevin Dynamics of Peptides: The Frictional Dependence of Isomerization Rates of N-Acetylalanyl-N’-Methylamide. Biopolymers 1992, 32, 523–535. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Kräutler, V.; van Gunsteren, W.F.; Hünenberger, P.H. A Fast SHAKE Algorithm to Solve Distance Constraint Equations for Small Molecules in Molecular Dynamics Simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Stourac, J.; Vavra, O.; Kokkonen, P.; Filipovic, J.; Pinto, G.; Brezovsky, J.; Damborsky, J.; Bednar, D. Caver Web 1.0: Identification of Tunnels and Channels in Proteins and Analysis of Ligand Transport. Nucleic Acids Res. 2019, 47, W414–W422. [Google Scholar] [CrossRef] [PubMed]
- Duhovny, D.; Nussinov, R.; Wolfson, H.J. Efficient Unbound Docking of Rigid Molecules. In Algorithms in Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2002; pp. 185–200. [Google Scholar]
- Pratt, J.; Roy, R.; Annabi, B. Concanavalin-A-Induced Autophagy Biomarkers Requires Membrane Type-1 Matrix Metalloproteinase Intracellular Signaling in Glioblastoma Cells. Glycobiology 2012, 22, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Mohanam, S.; Sawaya, R.; Fuller, G.N.; Seiki, M.; Sato, H.; Gokaslan, Z.L.; Liotta, L.A.; Nicolson, G.L.; Rao, J.S. Differential Expression of Membrane-Type Matrix Metalloproteinase and Its Correlation with Gelatinase A Activation in Human Malignant Brain Tumors In Vivo and In Vitro. Cancer Res. 1996, 56, 384–392. [Google Scholar] [PubMed]
- Osterne, V.J.S.; Pinto-Junior, V.R.; Oliveira, M.V.; Nascimento, K.S.; Van Damme, E.J.M.; Cavada, B.S. Computational Insights into the Circular Permutation Roles on ConA Binding and Structural Stability. Curr. Res. Struct. Biol. 2024, 7, 100140. [Google Scholar] [CrossRef] [PubMed]
- Cavada, B.S.; Osterne, V.J.S.; Lossio, C.F.; Pinto-Junior, V.R.; Oliveira, M.V.; Silva, M.T.L.; Leal, R.B.; Nascimento, K.S. One Century of ConA and 40 Years of ConBr Research: A Structural Review. Int. J. Biol. Macromol. 2019, 134, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Polewski, M.D.; Reveron-Thornton, R.F.; Cherryholmes, G.A.; Marinov, G.K.; Cassady, K.; Aboody, K.S. Increased Expression of System Xc- in Glioblastoma Confers an Altered Metabolic State and Temozolomide Resistance. Mol. Cancer Res. 2016, 14, 1229–1242. [Google Scholar] [CrossRef]
- An, S.; Lu, X.; Zhao, W.; Sun, T.; Zhang, Y.; Lu, Y.; Jiang, C. Amino Acid Metabolism Abnormity and Microenvironment Variation Mediated Targeting and Controlled Glioma Chemotherapy. Small 2016, 12, 5633–5645. [Google Scholar] [CrossRef] [PubMed]
- Chew, K.S.; Wells, R.C.; Moshkforoush, A.; Chan, D.; Lechtenberg, K.J.; Tran, H.L.; Chow, J.; Kim, D.J.; Robles-Colmenares, Y.; Srivastava, D.B.; et al. CD98hc Is a Target for Brain Delivery of Biotherapeutics. Nat. Commun. 2023, 14, 5053. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Stockwell, B.R. The Hallmarks of Ferroptosis. Annu. Rev. Cancer Biol. 2019, 3, 35–54. [Google Scholar] [CrossRef]
- Kar, F.; Kacar, S.; Hacioglu, C.; Kanbak, G.; Sahinturk, V. Concanavalin A Induces Apoptosis in a Dose-Dependent Manner by Modulating Thiol/disulfide Homeostasis in C6 Glioblastoma Cells. J. Biochem. Mol. Toxicol. 2021, 35, e22742. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Deng, G.; Zhong, W.; Gao, Z.; Ma, S.; Mo, C.; Li, Y.; Huang, S.; Zhou, C.; Lai, Y.; et al. Indoleamine 2, 3-Dioxygenase 1enhanceshepatocytes Ferroptosis in Acute Immune Hepatitis Associated with Excess Nitrative Stress. Free Radic. Biol. Med. 2020, 152, 668–679. [Google Scholar] [CrossRef]
- Yan, B.; Ai, Y.; Sun, Q.; Ma, Y.; Cao, Y.; Wang, J.; Zhang, Z.; Wang, X. Membrane Damage during Ferroptosis Is Caused by Oxidation of Phospholipids Catalyzed by the Oxidoreductases POR and CYB5R1. Mol. Cell 2021, 81, 355–369.e10. [Google Scholar] [CrossRef]





| Glycan | Structure | Docking Score |
|---|---|---|
| GlcNAc2Man7 |  | −58.42 |
| GlcNAc2Man8 |  | −49.30 |
| GlcNAc2Man9 |  | −51.31 |
| GlcNAc2Man3GlcNAc2Gal2Fuc |  | −43.19 |
| GlcNAc2Man3GlcNAc3Gal3Fuc | 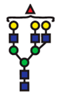 | −48.11 |
| FucGlcNAc2Man3GlcNAc2Gal2Fuc2 | 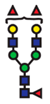 | - |
| FucGlcNAc2Man3GlcNAc2Gal2Fuc | 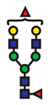 | - |
| FucGlcNAc2Man3GlcNAc3Gal3Fuc |  | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto-Junior, V.R.; Seeger, R.L.; Souza-Filho, C.H.D.; França, A.P.; Sartori, N.; Oliveira, M.V.; Osterne, V.J.S.; Nascimento, K.S.; Leal, R.B.; Cavada, B.S. Xc- System as a Possible Target for ConBr Lectin Interaction in Glioma Cells. Neuroglia 2024, 5, 202-222. https://doi.org/10.3390/neuroglia5030015
Pinto-Junior VR, Seeger RL, Souza-Filho CHD, França AP, Sartori N, Oliveira MV, Osterne VJS, Nascimento KS, Leal RB, Cavada BS. Xc- System as a Possible Target for ConBr Lectin Interaction in Glioma Cells. Neuroglia. 2024; 5(3):202-222. https://doi.org/10.3390/neuroglia5030015
Chicago/Turabian StylePinto-Junior, Vanir Reis, Rodrigo Lopes Seeger, Cláudio Henrique Dahne Souza-Filho, Angela Patricia França, Nicole Sartori, Messias Vital Oliveira, Vinicius Jose Silva Osterne, Kyria Santiago Nascimento, Rodrigo Bainy Leal, and Benildo Sousa Cavada. 2024. "Xc- System as a Possible Target for ConBr Lectin Interaction in Glioma Cells" Neuroglia 5, no. 3: 202-222. https://doi.org/10.3390/neuroglia5030015
APA StylePinto-Junior, V. R., Seeger, R. L., Souza-Filho, C. H. D., França, A. P., Sartori, N., Oliveira, M. V., Osterne, V. J. S., Nascimento, K. S., Leal, R. B., & Cavada, B. S. (2024). Xc- System as a Possible Target for ConBr Lectin Interaction in Glioma Cells. Neuroglia, 5(3), 202-222. https://doi.org/10.3390/neuroglia5030015








