Coming-of-Age Characterization of Soil Viruses: A User’s Guide to Virus Isolation, Detection within Metagenomes, and Viromics
Abstract
:1. Introduction
Virus-Like Particles (VLPs), Viruses, Microbes, and Other Terms
2. Potential Roles and Forms of Viruses in Soils
2.1. Importance of the Soil Virosphere
2.2. Viral Lysis of Soil Microbes
2.3. Viral Modification of Host Metabolism During Lytic Infections
2.4. Virus-Mediated Horizontal Gene Transfer
2.5. Many Environmental Viral States
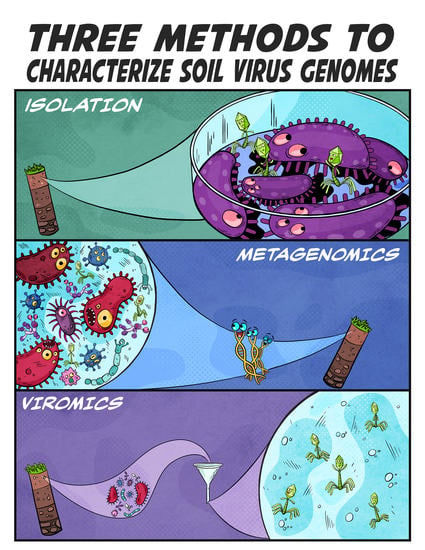
3. Three Ways to Characterize Soil Viruses
3.1. Virus Isolation
3.1.1. Techniques for Isolating Viruses
3.1.2. Well-Developed Soil-Virus Systems
3.1.3. Isolation of RNA Fungal Viruses
3.2. Metagenomics
3.2.1. Losing Sight of Virus Genes in a ‘Sea’ of Sequence
3.2.2. Vertical Coverage
3.2.3. Drawing Information from Bulk Sequence
3.2.4. Outlook
3.3. Viromics
3.3.1. Utility and Drawbacks of Viromes
3.3.2. The Challenge of Separating Virions from Soils
3.3.3. Additional Sources of VLP Losses
3.3.4. Efficiency of Virus Resuspension from Soils
3.3.5. Outlook
4. Metagenomic Dataset Contaminants
4.1. Non-Infectious Virus-Like Particles (niVLPs)
4.2. Extracellular DNA (eDNA/relic DNA)
Removing eDNA from Environmental Samples
4.3. Microbe-Derived Virome Contamination
4.3.1. Ultrasmall Microbes
4.3.2. Gene Transfer Agents (GTAs)
4.3.3. Plasmids
4.4. Amplification Artifacts
4.5. Ecologically Inactive Viruses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Glossary of Terms
References
- Hyman, P.; Abedon, S.T. Viruses of Microorganisms; Caister Academic Press: Norwich, UK, 2018. [Google Scholar]
- Witzany, G. Viruses: Essential Agents of Life; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Mushegian, A.R. Are there 1031 virus particles on Earth, or more, or less? J. Bacteriol. 2020. [Google Scholar] [CrossRef]
- Williamson, K.E.; Fuhrmann, J.J.; Wommack, K.E.; Radosevich, M. Viruses in soil ecosystems: An unknown quantity within an unexplored territory. Annu. Rev. Virol. 2017, 4, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.A.; van Elsas, J.D. The ‘neglected’ soil virome-potential role and impact. Trends Microbiol. 2018, 26, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Mason-Jones, K. Viruses in soil: Nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol. Biochem. 2018, 127, 305–317. [Google Scholar] [CrossRef]
- Williamson, K.E. Viruses of microorganisms in soil ecosystems. In Viruses of Microorganisms; Hyman, P., Abedon, S.T., Eds.; Caister Academic Press: Norwich, UK, 2018; pp. 77–93. [Google Scholar]
- Abedon, S.T.; Murray, K.L. Archaeal viruses, not archaeal phages: An archaeological dig. Archaea 2013, 2013, 251245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, P.; Abedon, S.T. Smaller fleas: Viruses of microorganisms. Scientifica 2012, 2012, 734023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Steward, G.F.; Culley, A.I.; Mueller, J.A.; Wood-Charlson, E.M.; Belcaid, M.; Poisson, G. Are we missing half of the viruses in the ocean? ISME J. 2013, 7, 672–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weitz, J.S.; Li, G.; Gulbudak, H.; Cortez, M.H.; Whitaker, R.J. Viral invasion fitness across a continuum from lysis to latency. Virus Evol. 2019, 5, vez006. [Google Scholar] [CrossRef] [Green Version]
- Raoult, D.; Forterre, P. Redefining viruses: Lessons from Mimivirus. Nat. Rev. Microbiol. 2008, 6, 315–319. [Google Scholar] [CrossRef]
- Dupré, J.; O’Malley, M.A. Varieties of living things: Life at the intersection of lineage and metabolism. Philos. Theor. Biol. 2009, 1, e003. [Google Scholar] [CrossRef] [Green Version]
- Forterre, P. Defining life: The virus viewpoint. Orig. Life Evol. Biosph. 2010, 40, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Kozloff, L.M. Biochemical studies of virus reproduction. VII. The appearance of parent nitrogen and phosphorus in the progeny. J. Biol. Chem. 1952, 194, 95–108. [Google Scholar] [PubMed]
- Thingstad, T.F.; Bratbak, G.; Heldal, M. Aquatic phage ecology. In Bacteriophage Ecology; Abedon, S.T., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 251–280. [Google Scholar]
- Mahmoudabadi, G.; Milo, R.; Phillips, R. Energetic cost of building a virus. Proc. Natl. Acad. Sci. USA 2017, 114, E4324–E4333. [Google Scholar] [CrossRef] [Green Version]
- Jacquet, S.; Zhong, X.; Peduzzi, P.; Thingstad, T.F.; Parikka, K.J.; Weinbauer, M.G. Virus interactions in the aquatic world. In Viruses of Microorganisms; Hyman, P., Abedon, S.T., Eds.; Caister Academic Press: Norwich, UK, 2018; pp. 115–141. [Google Scholar]
- Hobbs, Z.; Abedon, S.T. Diversity of phage infection types and associated terminology: The problem with ‘Lytic or lysogenic’. FEMS Microbiol. Lett. 2016, 363, fnw047. [Google Scholar] [CrossRef] [Green Version]
- Forde, S.E.; Thompson, J.N.; Bohannan, B.J.M. Adaptation varies through space and time in a coevolving host–parasitoid interaction. Nature 2004, 431, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.; Levin, B.R.; Stewart, F.M. A complex community in a simple habitat: An experimental study with bacteria and phage. Ecology 1977, 58, 369–378. [Google Scholar] [CrossRef]
- Heilmann, S.; Sneppen, K.; Krishna, S. Sustainability of virulence in a phage-bacterial ecosystem. J. Virol. 2010, 84, 3016–3022. [Google Scholar] [CrossRef] [Green Version]
- Suttle, C.A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Keen, E.C. A century of phage research: Bacteriophages and the shaping of modern biology. BioEssays 2015, 37, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.W.; Suttle, C.A. Viruses and nutrient cycles in the sea: Viruses play critical roles in the structure and function of aquatic food webs. BioScience 1999, 49, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Schimel, J.P.; Jastrow, J.D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2017, 2, 17105. [Google Scholar] [CrossRef] [PubMed]
- Barnard, R.L.; Blazewicz, S.J.; Firestone, M.K. Rewetting of soil: Revisiting the origin of soil CO2 emissions. Soil Biol. Biochem. 2020, 107819. [Google Scholar] [CrossRef]
- Hurst, C.J.; Gerba, C.P.; Cech, I. Effects of environmental variables and soil characteristics on virus survival in soil. Appl. Environ. Microbiol. 1980, 40, 1067–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abedon, S.T. Ecology of anti-biofilm agents II. bacteriophage exploitation and biocontrol of biofilm bacteria. Pharmaceuticals 2015, 8, 559–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barksdale, L.; Arden, S.B. Persisting bacteriophage infections, lysogeny, and phage conversions. Ann. Rev. Microbiol. 1974, 28, 265–299. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; U’Ren, J.M. Viral metabolic reprogramming in marine ecosystems. Curr. Opin. Microbiol. 2016, 31, 161–168. [Google Scholar] [CrossRef]
- Emerson, J.B.; Roux, S.; Brum, J.R.; Bolduc, B.; Woodcroft, B.J.; Jang, H.B.; Singleton, C.M.; Solden, L.M.; Naas, A.E.; Boyd, J.A.; et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 2018, 3, 870–880. [Google Scholar] [CrossRef]
- Trubl, G.; Jang, H.B.; Roux, S.; Emerson, J.B.; Solonenko, N.; Vik, D.R.; Solden, L.; Ellenbogen, J.; Runyon, A.T.; Bolduc, B.; et al. Soil viruses are underexplored players in ecosystem carbon processing. mSystems 2018, 3, e00076-18. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Guo, X.; Zhang, R.; Qu, W.; Gao, B.; Zeng, R. Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome 2019, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Zeph, L.R.; Onaga, M.A.; Stotzky, G. Transduction of Escherichia coli by bacteriophage P1 in soil. Appl. Environ. Microbiol. 1988, 54, 1731–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, D.; Roy, K.; Williamson, K.E.; White, D.C.; Wommack, K.E.; Sublette, K.L.; Radosevich, M. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl. Environ. Microbiol. 2008, 74, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redfield, R.J. Do bacteria have sex? Nat. Rev. Genet. 2001, 2, 634–639. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.L. Bacteriophage-mediated horizontal gene transfer: Transduction. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B., McConville, M., Eds.; Springer: New York City, NY, USA, 2017; Available online: https://link.springer.com/referenceworkentry/10.1007/978-3-319-40598-8_4-1 (accessed on 18 April 2020).
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penades, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Los, M.; Kuzio, J.; McConnell, M.R.; Kropinski, A.M.; Wegrzyn, G.; Christie, G.E. Lysogenic conversion in bacteria of importance to the food industry. In Bacteriophages in the Control of Food- and Waterborne Pathogens; Sabour, P.M., Griffiths, M.W., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 157–198. [Google Scholar]
- Christie, G.E.; Allison, H.A.; Kuzio, J.; McShan, M.; Waldor, M.K.; Kropinski, A.M. Prophage-induced changes in cellular cytochemistry and virulence. In Bacteriophages in Health and Disease; Hyman, P., Abedon, S.T., Eds.; CABI Press: Wallingford, UK, 2012; pp. 33–60. [Google Scholar]
- Hendrix, R.W.; Lawrence, J.G.; Hatfull, G.F.; Casjens, S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000, 8, 504–508. [Google Scholar] [CrossRef]
- Cumby, N.; Davidson, A.R.; Maxwell, K.L. The moron comes of age. Bacteriophage 2012, 2, 225–228. [Google Scholar] [CrossRef]
- Taylor, V.L.; Fitzpatrick, A.D.; Islam, Z.; Maxwell, K.L. The diverse impacts of phage morons on bacterial fitness and virulence. Adv. Virus Res. 2019, 103, 1–31. [Google Scholar] [PubMed]
- Forterre, P. The virocell concept and environmental microbiology. ISME J. 2013, 7, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Delbrück, M. Bacterial viruses or bacteriophages. Biol. Rev. 1946, 21, 30–40. [Google Scholar] [CrossRef]
- Mohiuddin, M.; Schellhorn, H.E. Spatial and temporal dynamics of virus occurrence in two freshwater lakes captured through metagenomic analysis. Front. Microbiol. 2015, 6, 960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiffer, J.T.; Aubert, M.; Weber, N.D.; Mintzer, E.; Stone, D.; Jerome, K.R. Targeted DNA mutagenesis for the cure of chronic viral infections. J. Virol. 2012, 86, 8920–8936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.V.; Day, M. Contribution of lysogeny, pseudolysogeny, and starvation to phage ecology. In Bacteriophage Ecology; Abedon, S.T., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 114–143. [Google Scholar]
- Abedon, S.T. Disambiguating bacteriophage pseudolysogeny: An historical analysis of lysogeny, pseudolysogeny, and the phage carrier state. In Contemporary Trends in Bacteriophage Research; Adams, H.T., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2009; pp. 285–307. [Google Scholar]
- Los, M.; Wegrzyn, G. Pseudolysogeny. Adv. Virus Res. 2012, 82, 339–349. [Google Scholar] [PubMed]
- Soler, N.; Marguet, E.; Verbavatz, J.M.; Forterre, P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 2008, 159, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t, H.E.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vainio, E.J.; Hantula, J. Fungal viruses. In Viruses of Microorganisms; Hyman, P., Abedon, S.T., Eds.; Caister Academic Press: Norwich, UK, 2018; pp. 193–209. [Google Scholar]
- Sutela, S.; Poimala, A.; Vainio, E.J. Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol. Ecol. 2019, 95, fiz119. [Google Scholar] [CrossRef] [Green Version]
- Stahl, F.W. Amber mutants of bacteriophage T4D: Their isolation and genetic characterization. Genetics 2012, 190, 831–832. [Google Scholar]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Meth. Mol. Biol. 2009, 501, 15–21. [Google Scholar]
- Wommack, K.E.; Williamson, K.E.; Helton, R.R.; Bench, S.R.; Winget, D.M. Methods for the isolation of viruses from environmental samples. Meth. Mol. Biol. 2009, 501, 3–14. [Google Scholar]
- Lobocka, M.; Hejnowicz, M.S.; Gagala, U.; Weber-Dabrowska, B.; Wegrzyn, G.; Dadlez, M. The first step to bacteriophage therapy: How to choose the correct phage. In Phage Therapy: Current Research and Applications; Borysowski, J., Miêdzybrodzki, R., Górski, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 23–67. [Google Scholar]
- Hyman, P. Phages for phage therapy: Isolation, characterization, and host range breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czajkowski, R.; Ozymko, Z.; Lojkowska, E. Isolation and characterization of novel soilborne lytic bacteriophages infecting Dickeya spp. biovar 3 (‘D. solani’). Plant Pathol. 2013, 63, 758–772. [Google Scholar] [CrossRef]
- Anne, J.; Wohlleben, W.; Burkardt, H.J.; Springer, R.; Puhler, A. Morphological and molecular characterization of several actinophages isolated from soil which lyse Streptomyces cattleya or S. venezuelae. J Gen. Microbiol. 1984, 130, 2639–2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages. Methods and Protocols. Volume 1: Isolation, Characterization, and Interactions; Humana Press: New York, NY, USA, 2009; Volume 501. [Google Scholar]
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages. Methods and Protocols. Volume 2: Molecular and Applied Aspects; Humana Press: New York, NY, USA, 2009; Volume 502. [Google Scholar]
- Clokie, M.R.J.; Kropinski, A.M.; Lavigne, R. Bacteriophages: Methods and Protocols. Volume 3. Methods and Protocols; Springer protocols (Series); Humana Press; Springer: New York, NY, USA, 2018; Volume 1681. [Google Scholar]
- The actinobacteriophage database at PhagesDB.org. 2020. Available online: https://www.phagesdb.org (accessed on 18 April 2020).
- Dhar, B.; Singh, B.D.; Singh, R.B.; Srivastava, J.S.; Singh, V.P.; Singh, R.M. Occurrence and distribution of rhizobiophages in Indian soils. Acta Microbiol. Pol. 1979, 28, 319–324. [Google Scholar] [PubMed]
- Rombouts, S.; Volckaert, A.; Venneman, S.; Declercq, B.; Vandenheuvel, D.; Allonsius, C.N.; Van Malderghem, C.; Jang, H.B.; Briers, Y.; Noben, J.P. Characterization of novel bacteriophages for biocontrol of bacterial blight in leek caused by Pseudomonas syringae pv. porri. Front. Microbiol. 2016, 7, 279. [Google Scholar] [CrossRef] [Green Version]
- Russell, D.A.; Hatfull, G.F. PhagesDB: The actinobacteriophage database. Bioinformatics 2017, 33, 784–786. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, H.-W.; DuBow, M.S. Bacteriophage Taxonomy. In Viruses of Prokaryotes. Volume I. General Properties of Bacteriophages; Ackermann, H.-W., DuBow, M.S., Eds.; CRC Press: Boca Raton, FL, USA, 1987; pp. 13–28. [Google Scholar]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.; Mathieu, J.; Li, M.; Dai, Z.; Alvarez, P.J. Isolation of polyvalent bacteriophages by sequential multiple-host approaches. Appl. Environ. Microbiol. 2016, 82, 808–815. [Google Scholar] [CrossRef] [Green Version]
- Williamson, K.E.; Schnitker, J.B.; Radosevich, M.; Smith, D.W.; Wommack, K.E. Cultivation-based assessment of lysogeny among soil bacteria. Microb. Ecol. 2008, 56, 437–447. [Google Scholar] [CrossRef]
- Chen, F.; Wang, K.; Stewart, J.; Belas, R. Induction of multiple prophages from a marine bacterium: A genomic approach. Appl. Environ. Microbiol. 2006, 72, 4995–5001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, R.A.; Roux, S.; Borton, M.A.; Morgan, D.M.; Johnston, M.D.; Booker, A.E.; Hoyt, D.W.; Meulia, T.; Wolfe, R.A.; Hanson, A.J.; et al. Viruses control dominant bacteria colonizing the terrestrial deep biosphere after hydraulic fracturing. Nat. Microbiol. 2019, 4, 352–361. [Google Scholar] [CrossRef]
- Sinha, V.; Goyal, A.; Svenningsen, S.L.; Semsey, S.; Krishna, S. In silico evolution of lysis-lysogeny strategies reproduces observed lysogeny propensities in temperate bacteriophages. Front. Microbiol. 2017, 8, 1386. [Google Scholar] [CrossRef] [PubMed]
- Avlund, M.; Dodd, I.B.; Semsey, S.; Sneppen, K.; Krishna, S. Why does phage play dice? J. Virol. 2009, 83, 11416–11420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waller, A.S.; Yamada, T.; Kristensen, D.M.; Kultima, J.R.; Sunagawa, S.; Koonin, E.V.; Bork, P. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014, 8, 1391–1402. [Google Scholar] [CrossRef]
- Maslov, S.; Sneppen, K. Well-temperate phage: Optimal bet-hedging against local environmental collapses. Sci. Rep. 2015, 5, 10523. [Google Scholar] [CrossRef] [Green Version]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef]
- Abedon, S.T. Look who’s talking: T-even phage lysis inhibition, the granddaddy of virus-virus intercellular communication research. Viruses 2019, 11, 951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, D.; Roy, K.; Williamson, K.E.; Srinivasiah, S.; Wommack, K.E.; Radosevich, M. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: An alternative paradigm for prophage induction. Appl. Environ. Microbiol. 2009, 75, 7142–7152. [Google Scholar] [CrossRef] [Green Version]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R. What does the talking? quorum sensing signalling genes discovered in a bacteriophage genome. PLoS ONE 2014, 9, e85131. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Commentary: Communication between viruses guides lysis-lysogeny decisions. Front. Microbiol. 2017, 8, 983. [Google Scholar] [CrossRef] [PubMed]
- Trinh, J.T.; Szekely, T.; Shao, Q.; Balazsi, G.; Zeng, L. Cell fate decisions emerge as phages cooperate or compete inside their host. Nat. Commun. 2017, 8, 14341. [Google Scholar] [CrossRef] [PubMed]
- Igler, C.; Abedon, S.T. Commentary: A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Front. Microbiol. 2019, 10, 1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silpe, J.E.; Bassler, B.L. A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 2019, 176, 268–280. [Google Scholar] [CrossRef] [Green Version]
- Hynes, A.P.; Moineau, S. Phagebook: The social network. Mol. Cell 2017, 65, 963–964. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically novel uncultured microbial cells dominate Earth microbiomes. mSystems 2018, 3, e00055-18. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Novick, R.P. Phage-mediated intergeneric transfer of toxin genes. Science 2009, 323, 139–141. [Google Scholar] [CrossRef] [Green Version]
- Willner, D.; Hugenholtz, P. From deep sequencing to viral tagging: Recent advances in viral metagenomics. BioEssays 2013, 35, 436–442. [Google Scholar] [CrossRef]
- Sullivan, M.B.; Waterbury, J.B.; Chisholm, S.W. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 2003, 424, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Waterbury, J.B.; Valois, F.W. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 1993, 59, 3393–3399. [Google Scholar] [CrossRef] [Green Version]
- Thomas, E.L.; Corbel, M.J. Isolation of a phage lytic for several Brucella species following propagation of Tbilisi phage in the presence of mitomycin C. Arch. Virol. 1977, 54, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.; Biswas, B.; Vincent, L.; Mokashi, V.; Schuch, R.; Bishop-Lilly, K.A.; Sozhamannan, S. Development of a high throughput assay for indirectly measuring phage growth using the OmniLog(TM) system. Bacteriophage 2012, 2, 159–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard-Varona, C.; Roux, S.; Dore, H.; Solonenko, N.E.; Holmfeldt, K.; Markillie, L.M.; Orr, G.; Sullivan, M.B. Regulation of infection efficiency in a globally abundant marine Bacteriodetes virus. ISME J. 2017, 11, 284–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enav, H.; Kirzner, S.; Lindell, D.; Mandel-Gutfreund, Y.; Beja, O. Adaptation to sub-optimal hosts is a driver of viral diversification in the ocean. Nat. Commun. 2018, 9, 4698. [Google Scholar] [CrossRef] [Green Version]
- Buckling, A.; Rainey, P.B. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. Lond. B Biol. Sci. 2002, 269, 931–936. [Google Scholar] [CrossRef] [Green Version]
- Buckling, A.; Rainey, P.B. The role of parasites in sympatric and allopatric host diversification. Nature 2002, 420, 496–499. [Google Scholar] [CrossRef]
- Gomez, P.; Buckling, A. Real-time microbial adaptive diversification in soil. Ecol. Lett. 2013, 16, 650–655. [Google Scholar] [CrossRef]
- Poisot, T.; Lepennetier, G.; Martinez, E.; Ramsayer, J.; Hochberg, M.E. Resource availability affects the structure of a natural bacteria-bacteriophage community. Biol. Lett. 2011, 7, 201–204. [Google Scholar] [CrossRef] [Green Version]
- Vos, M.; Birkett, P.J.; Birch, E.; Griffiths, R.I.; Buckling, A. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 2009, 325, 833. [Google Scholar] [CrossRef] [Green Version]
- Hanauer, D.I.; Graham, M.J.; Betancur, L.; Bobrownicki, A.; Cresawn, S.G.; Garlena, R.A.; Jacobs-Sera, D.; Kaufmann, N.; Pope, W.H.; Russell, D.A.; et al. An inclusive research education community (iREC): Impact of the SEA-PHAGES program on research outcomes and student learning. Proc. Natl. Acad. Sci. USA 2017, 114, 13531–13536. [Google Scholar] [CrossRef] [Green Version]
- Sea Phages 2020. Available online: https://www.seaphages.org/ (accessed on 18 April 2020).
- Hatfull, G.F. Bacteriophage discovery and genomics. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–13. [Google Scholar]
- Hatfull Lab 2020. Available online: http://www.hatfull.org/courses (accessed on 18 April 2020).
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Caston, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479–480, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Li, M.; Redda, E.T.; Mei, J.; Zhang, J.; Wu, B.; Jiang, X. A novel double-stranded RNA mycovirus isolated from Trichoderma harzianum. Virol. J. 2019, 16, 113. [Google Scholar] [CrossRef]
- Arjona-Lopez, J.M.; Telengech, P.; Jamal, A.; Hisano, S.; Kondo, H.; Yelin, M.D.; Arjona-Girona, I.; Kanematsu, S.; Lopez-Herrera, C.J.; Suzuki, N. Novel, diverse RNA viruses from Mediterranean isolates of the phytopathogenic fungus, Rosellinia necatrix: Insights into evolutionary biology of fungal viruses. Environ. Microbiol. 2018, 20, 1464–1483. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, J.; Cheng, J.; Fu, Y.; Li, G.; Yi, X.; Jiang, D. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. USA 2014, 111, 12205–12210. [Google Scholar] [CrossRef] [Green Version]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef]
- Tringe, S.G.; von, M.C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C.; et al. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef]
- Roux, S. A viral ecogenomics framework to uncover the secrets of nature’s “Microbe whisperers”. mSystems 2019, 4, e00111-19. [Google Scholar] [CrossRef] [Green Version]
- Trevors, J.T. One gram of soil: A microbial biochemical gene library. Antonie van Leeuwenhoek J. Microbiol. 2010, 97, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.O.; Robe, P.; Clark, I.; Simonet, P.; Vogel, T.M. Metagenomic comparison of direct and indirect soil DNA extraction approaches. J. Microbiol. Methods 2011, 86, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Kunin, V.; Copeland, A.; Lapidus, A.; Mavromatis, K.; Hugenholtz, P. A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008, 72, 557–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, S.; Emerson, J.B.; Eloe-Fadrosh, E.A.; Sullivan, M.B. Benchmarking viromics: An in silico evaluation of metagenome-enabled estimates of viral community composition and diversity. PeerJ 2017, 5, e3817. [Google Scholar] [CrossRef] [Green Version]
- Vestergaard, G.; Schulz, S.; Schöler, A.; Schloter, M. Making big data smart—how to use metagenomics to understand soil quality. Biol. Fertil. Soils 2017, 53, 479–484. [Google Scholar] [CrossRef]
- McLaren, M.R.; Willis, A.D.; Callahan, B.J. Consistent and correctable bias in metagenomic sequencing experiments. eLife 2019, 8, e46923. [Google Scholar] [CrossRef]
- Martinez-Hernandez, F.; Fornas, O.; Lluesma, G.M.; Bolduc, B.; de la Cruz Peña, M.; Martinez, J.M.; Anton, J.; Gasol, J.M.; Rosselli, R.; Rodriguez-Valera, F.; et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 2017, 8, 15892. [Google Scholar] [CrossRef] [Green Version]
- Sieradzki, E.T.; Ignacio-Espinoza, J.C.; Needham, D.M.; Fichot, E.B.; Fuhrman, J.A. Dynamic marine viral infections and major contribution to photosynthetic processes shown by spatiotemporal picoplankton metatranscriptomes. Nat. Commun. 2019, 10, 1169. [Google Scholar] [CrossRef]
- Rédei, G.P. Encyclopedia of Genetics, Genomics, Proteomics, and Informatics; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Nomiya, T. Discussions on target theory: Past and present. J. Radiat. Res. 2013, 54, 1161–1163. [Google Scholar] [CrossRef] [Green Version]
- Sutton, T.D.S.; Clooney, A.G.; Ryan, F.J.; Ross, R.P.; Hill, C. Choice of assembly software has a critical impact on virome characterisation. Microbiome 2019, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Goordial, J.; Davila, A.; Greer, C.W.; Cannam, R.; DiRuggiero, J.; McKay, C.P.; Whyte, L.G. Comparative activity and functional ecology of permafrost soils and lithic niches in a hyper-arid polar desert. Environ. Microbiol. 2017, 19, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotech. 2017, 35, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Li, L.; Zhang, G. Coverage recommendation for genotyping analysis of highly heterologous species using next-generation sequencing technology. Sci. Rep. 2016, 6, 35736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaheer, R.; Noyes, N.; Ortega, P.R.; Cook, S.R.; Marinier, E.; Van, D.G.; Belk, K.E.; Morley, P.S.; McAllister, T.A. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci. Rep. 2018, 8, 5890. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef]
- Ren, J.; Song, K.; Deng, C.; Ahlgren, N.A.; Fuhrman, J.A.; Li, Y.; Xie, X.; Sun, F. Identifying viruses from metagenomic data by deep learning. arXiv 2018, arXiv:1806.07810. [Google Scholar] [CrossRef] [Green Version]
- CyVerse. 2020. Available online: https://www.cyverse.org/ (accessed on 18 April 2020).
- KBase Predictive Biology. 2020. Available online: https://www.kbase.us/ (accessed on 18 April 2020).
- Bolduc, B.; Youens-Clark, K.; Roux, S.; Hurwitz, B.L.; Sullivan, M.B. iVirus: Facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 2017, 11, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Benjamin, B. Welcome to iVirus’s Documentation. 2020. Available online: https://ivirus.readthedocs.io/en/latest/ (accessed on 18 April 2020).
- Hurwitz lab. VerveNet. 2020. Available online: https://www.protocols.io/groups/verve-net (accessed on 18 April 2020).
- Sangwan, N.; Xia, F.; Gilbert, J.A. Recovering complete and draft population genomes from metagenome datasets. Microbiome 2016, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Roux, S.; Adriaenssens, E.M.; Dutilh, B.E.; Koonin, E.V.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Lavigne, R.; Brister, J.R.; Varsani, A.; et al. Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotech. 2019, 37, 29–37. [Google Scholar] [CrossRef]
- Alrasheed, H.; Jin, R.; Weitz, J.S. Caution in inferring viral strategies from abundance correlations in marine metagenomes. Nat. Commun. 2019, 10, 501. [Google Scholar] [CrossRef] [PubMed]
- Starr, E.P.; Nuccio, E.E.; Pett-Ridge, J.; Banfield, J.F.; Firestone, M.K. Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc. Natl. Acad. Sci. USA 2019, 116, 25900–25908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Farias, S.T.; Dos Santos Junior, A.P.; Rego, T.G.; Jose, M.V. Origin and evolution of RNA-dependent RNA polymerase. Front. Genet. 2017, 8, 125. [Google Scholar] [CrossRef]
- Ponsero, A.J.; Hurwitz, B.L. The promises and pitfalls of machine learning for detecting viruses in aquatic metagenomes. Front. Microbiol. 2019, 10, 806. [Google Scholar] [CrossRef]
- Breitbart, M.; Salamon, P.; Andresen, B.; Mahaffy, J.M.; Segall, A.M.; Mead, D.; Azam, F.; Rohwer, F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 2002, 99, 14250–14255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, A.C.; Zayed, A.A.; Conceicao-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 2019, 177, 1109–1123. [Google Scholar] [CrossRef] [Green Version]
- Michen, B.; Graule, T. Isoelectric points of viruses. J. Appl. Microbiol. 2010, 109, 388–397. [Google Scholar] [CrossRef] [Green Version]
- Zablocki, O.; van, Z.L.; Adriaenssens, E.M.; Rubagotti, E.; Tuffin, M.; Cary, C.; Cowan, D. High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of Antarctic soils. Appl. Environ. Microbiol. 2014, 80, 6888–6897. [Google Scholar] [CrossRef] [Green Version]
- Segobola, J.; Adriaenssens, E.; Tsekoa, T.; Rashamuse, K.; Cowan, D. Exploring viral diversity in a unique South African soil habitat. Sci. Rep. 2018, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Trubl, G.; Roux, S.; Solonenko, N.; Li, Y.F.; Bolduc, B.; Rodriguez-Ramos, J.; Eloe-Fadrosh, E.A.; Rich, V.I.; Sullivan, M.B. Towards optimized viral metagenomes for double-stranded and single-stranded DNA viruses from challenging soils. PeerJ 2019, 7, e7265. [Google Scholar] [CrossRef]
- Mehlich, A. Determination of cation-and anion-exchange properties of soils. Soil Sci. 1948, 66, 429–446. [Google Scholar] [CrossRef]
- Kozlowski, L.P. Proteome-pI: Proteome isoelectric point database. Nucl. Acids Res. 2017, 45, D1112–D1116. [Google Scholar] [CrossRef]
- Thurber, R.V.; Haynes, M.; Breitbart, M.; Wegley, L.; Rohwer, F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009, 4, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Parras-Molto, M.; Rodriguez-Galet, A.; Suarez-Rodriguez, P.; Lopez-Bueno, A. Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses. Microbiome 2018, 6, 119. [Google Scholar] [CrossRef] [PubMed]
- Anesio, A.M.; Hollas, C.; Graneli, W.; Laybourn-Parry, J. Influence of humic substances on bacterial and viral dynamics in freshwaters. Appl. Environ. Microbiol. 2004, 70, 4848–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tebbe, C.C.; Vahjen, W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 1993, 59, 2657–2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alaeddini, R. Forensic implications of PCR inhibition—A review. Forensic Sci. Int. Genet. 2012, 6, 297–305. [Google Scholar] [CrossRef]
- Williamson, K.E.; Corzo, K.A.; Drissi, C.L.; Buckingham, J.M.; Thompson, C.P.; Helton, R.R. Estimates of viral abundance in soils are strongly influenced by extraction and enumeration methods. Biol. Fertil. Soils 2013, 49, 857–869. [Google Scholar] [CrossRef]
- Trubl, G.; Solonenko, N.; Chittick, L.; Solonenko, S.A.; Rich, V.I.; Sullivan, M.B. Optimization of viral resuspension methods for carbon-rich soils along a permafrost thaw gradient. PeerJ 2016, 4, e1999. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.; Noble, R.T.; Steele, J.A.; Schwalbach, M.S.; Hewson, I.; Fuhrman, J.A. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2007, 2, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Forterre, P.; Soler, N.; Krupovic, M.; Marguet, E.; Ackermann, H.-W. Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 2013, 21, 1–5. [Google Scholar] [PubMed]
- Cunningham, B.R.; Brum, J.R.; Schwenck, S.M.; Sullivan, M.B.; John, S.G. An inexpensive, accurate, and precise wet-mount method for enumerating aquatic viruses. Appl. Environ. Microbiol. 2015, 81, 2995–3000. [Google Scholar] [PubMed] [Green Version]
- Zhang, Y.; Hung, T.; Song, J.; He, J. Electron microscopy: Essentials for viral structure, morphogenesis and rapid diagnosis. Sci. China Life Sci. 2013, 56, 421–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burge, W.D.; Enkiri, N.K. Virus adsorption by five soils. J. Environ. Qual. 1978, 7, 73–76. [Google Scholar] [CrossRef]
- Rodriguez, R.A.; Pepper, I.L.; Gerba, C.P. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 2009, 75, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Farkas, K.; Hassard, F.; McDonald, J.E.; Malham, S.K.; Jones, D.L. Evaluation of molecular methods for the detection and quantification of pathogen-derived nucleic acids in sediment. Front. Microbiol. 2017, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, S.R.; Wang, D. Origins and challenges of viral dark matter. Virus Res. 2017, 239, 136–142. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Klimke, W.; Maglott, D.R. NCBI reference sequences: Current status, policy and new initiatives. Nucl. Acids Res. 2009, 37, D32–D36. [Google Scholar] [CrossRef] [Green Version]
- Paez-Espino, D.; Chen, I.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Szeto, E.; Pillay, M.; Huang, J.; Markowitz, V.M.; Nielsen, T.; et al. IMG/VR: A database of cultured and uncultured DNA Viruses and retroviruses. Nucl. Acids Res. 2017, 45, D457–D465. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; U’Ren, J.M.; Youens-Clark, K. Computational prospecting the great viral unknown. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [Green Version]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de, V.C.; Gasol, J.M.; et al. Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, R.; Constantinides, B.; Tapinos, A.; Robertson, D.L.; Prosperi, M. Challenges in the analysis of viral metagenomes. Virus Evol. 2016, 2, vew022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooke, C.B. Biological activities of ‘noninfectious’ influenza A virus particles. Future Virol. 2014, 9, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilpiszeski, R.L.; Aufrecht, J.A.; Retterer, S.T.; Sullivan, M.B.; Graham, D.E.; Pierce, E.M.; Zablocki, O.D.; Palumbo, A.V.; Elias, D.A. Soil aggregate microbial communities: Towards understanding microbiome interactions at biologically relevant scales. Appl. Environ. Microbiol. 2019, 85, e00324-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef] [Green Version]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2016, 2, 16242. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; Deng, L.; Poulos, B.T.; Sullivan, M.B. Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ. Microbiol. 2013, 15, 1428–1440. [Google Scholar] [CrossRef] [Green Version]
- Zolfo, M.; Pinto, F.; Asnicar, F.; Manghi, P.; Tett, A.; Bushman, F.D.; Segata, N. Detecting contamination in viromes using ViromeQC. Nat. Biotech. 2019, 37, 1408–1412. [Google Scholar] [CrossRef]
- Fox, K.R. DNase I footprinting. In Drug-DNA Interaction Protocols; Springer: Berlin/Heidelberg, Germany, 1997; pp. 1–22. [Google Scholar]
- Romanowski, G.; Lorenz, M.G.; Wackernagel, W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl. Environ. Microbiol. 1991, 57, 1057–1061. [Google Scholar] [CrossRef] [Green Version]
- Crecchio, C.; Stotzky, G. Binding of DNA on humic acids: Effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol. Biochem. 1998, 30, 1061–1067. [Google Scholar] [CrossRef]
- Cai, P.; Huang, Q.Y.; Zhang, X.W. Interactions of DNA with clay minerals and soil colloidal particles and protection against degradation by DNase. Environ. Sci. Technol. 2006, 40, 2971–2976. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Roman, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Lennon, J.T.; Muscarella, M.E.; Placella, S.A.; Lehmkuhl, B.K. How, when, and where relic DNA affects microbial diversity. MBio 2018, 9, e00637-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fittipaldi, M.; Rodriguez, N.J.; Codony, F.; Adrados, B.; Penuela, G.A.; Morato, J. Discrimination of infectious bacteriophage T4 virus by propidium monoazide real-time PCR. J. Virol. Methods 2010, 168, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Biotium. PMA & PMAxx™ Selected References. 2020. Available online: https://biotium.com/wp-content/uploads/2017/10/PMA-PMAxx-References.pdf (accessed on 18 April 2020).
- Mizuno, C.M.; Guyomar, C.; Roux, S.; Lavigne, R.; Rodriguez-Valera, F.; Sullivan, M.B.; Gillet, R.; Forterre, P.; Krupovic, M. Numerous cultivated and uncultivated viruses encode ribosomal proteins. Nat. Commun. 2019, 10, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Gao, M. Jumbo bacteriophages: An overview. Front. Microbiol. 2017, 8, 403. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, E.; Ichimura, A.S.; Peng, V.; Fritsen, C.H.; Trubl, G.; Doran, P.T.; Murray, A.E. Brine assemblages of ultrasmall microbial cells within the ice cover of Lake Vida, Antarctica. Appl. Environ. Microbiol. 2014, 80, 3687–3698. [Google Scholar] [CrossRef] [Green Version]
- Velimirov, B. Nanobacteria, ultramicrobacteria and starvation forms: A search for the smallest metabolizing bacterium. Microbes Environ. 2001, 16, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Ghuneim, L.J.; Jones, D.L.; Golyshin, P.N.; Golyshina, O.V. Nano-sized and filterable Bacteria and Archaea: Biodiversity and function. Front. Microbiol. 2018, 9, 1971. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, D.M.; Mushegian, A.R.; Dolja, V.V.; Koonin, E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010, 18, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurwitz, B.L.; Sullivan, M.B. The Pacific Ocean virome (POV): A marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE 2013, 8, e57355. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.S.; Westbye, A.B.; Beatty, J.T. The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Annu. Rev. Virol. 2017, 4, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Shakya, M.; Soucy, S.M.; Zhaxybayeva, O. Insights into origin and evolution of alpha-proteobacterial gene transfer agents. Virus Evol. 2017, 3, vex036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-Gonzalez, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Shintani, M.; Sanchez, Z.K.; Kimbara, K. Genomics of microbial plasmids: Classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 2015, 6, 242. [Google Scholar] [CrossRef] [PubMed]
- Ruhfel, R.E.; Robillard, N.J.; Thorne, C.B. Interspecies transduction of plasmids among Bacillus anthracis, B. cereus, and B. thuringiensis. J. Bacteriol. 1984, 157, 708–711. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Dolja, V.V. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 2014, 78, 278–303. [Google Scholar] [CrossRef] [Green Version]
- Kazlauskas, D.; Varsani, A.; Koonin, E.V.; Krupovic, M. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat. Commun. 2019, 10, 3425. [Google Scholar] [CrossRef] [Green Version]
- Roux, S.; Krupovic, M.; Debroas, D.; Forterre, P.; Enault, F. Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open. Biol. 2013, 3, 130160. [Google Scholar] [CrossRef] [Green Version]
- Rozov, R.; Brown, K.A.; Bogumil, D.; Shterzer, N.; Halperin, E.; Mizrahi, I.; Shamir, R. Recycler: An algorithm for detecting plasmids from de novo assembly graphs. Bioinformatics 2017, 33, 475–482. [Google Scholar] [PubMed] [Green Version]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting plasmid sequences in metagenomic data using genome signatures. Nucl. Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaulaurier, J.; Zhu, S.; Deikus, G.; Mogno, I.; Zhang, X.S.; Davis-Richardson, A.; Canepa, R.; Triplett, E.W.; Faith, J.J.; Sebra, R.; et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat. Biotech. 2018, 36, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Shulman, L.M.; Davidson, I. Viruses with circular single-stranded DNA genomes are everywhere! Annu. Rev. Virol. 2017, 4, 159–180. [Google Scholar] [CrossRef] [PubMed]
- Malathi, V.G.; Devi, P.R. ssDNA viruses: Key players in global virome. Virusdisease 2019, 30, 3–12. [Google Scholar] [CrossRef]
- Karlsson, O.E.; Belak, S.; Granberg, F. The effect of preprocessing by sequence-independent, single-primer amplification (SISPA) on metagenomic detection of viruses. Biosecur. Bioterror. 2013, 11 (Suppl. 1), S227–S234. [Google Scholar] [CrossRef]
- Roux, S.; Solonenko, N.E.; Dang, V.T.; Poulos, B.T.; Schwenck, S.M.; Goldsmith, D.B.; Coleman, M.L.; Breitbart, M.; Sullivan, M.B. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ 2016, 4, e2777. [Google Scholar] [CrossRef] [Green Version]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-throughput sequencing technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef] [Green Version]
- Gansauge, M.T.; Meyer, M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 2013, 8, 737–748. [Google Scholar] [CrossRef]
- Aigrain, L.; Gu, Y.; Quail, M.A. Quantitation of next generation sequencing library preparation protocol efficiencies using droplet digital PCR assays - a systematic comparison of DNA library preparation kits for Illumina sequencing. BMC Genom. 2016, 17, 458. [Google Scholar] [CrossRef] [Green Version]
- Bekliz, M.; Brandani, J.; Bourquin, M.; Battin, T.J.; Peter, H. Benchmarking protocols for the metagenomic analysis of stream biofilm viromes. PeerJ 2019, 7, e8187. [Google Scholar] [CrossRef] [PubMed]
- Reavy, B.; Swanson, M.M.; Cock, P.J.; Dawson, L.; Freitag, T.E.; Singh, B.K.; Torrance, L.; Mushegian, A.R.; Taliansky, M. Distinct circular single-stranded DNA viruses exist in different soil types. Appl. Environ. Microbiol. 2015, 81, 3934–3945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Haig, S.J.; Schirmer, M.; D’Amore, R.; Gibbs, J.; Davies, R.L.; Collins, G.; Quince, C. Stable-isotope probing and metagenomics reveal predation by protozoa drives E. coli removal in slow sand filters. ISME J. 2015, 9, 797–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trubl, G.; Hyman, P.; Roux, S.; Abedon, S.T. Coming-of-Age Characterization of Soil Viruses: A User’s Guide to Virus Isolation, Detection within Metagenomes, and Viromics. Soil Syst. 2020, 4, 23. https://doi.org/10.3390/soilsystems4020023
Trubl G, Hyman P, Roux S, Abedon ST. Coming-of-Age Characterization of Soil Viruses: A User’s Guide to Virus Isolation, Detection within Metagenomes, and Viromics. Soil Systems. 2020; 4(2):23. https://doi.org/10.3390/soilsystems4020023
Chicago/Turabian StyleTrubl, Gareth, Paul Hyman, Simon Roux, and Stephen T. Abedon. 2020. "Coming-of-Age Characterization of Soil Viruses: A User’s Guide to Virus Isolation, Detection within Metagenomes, and Viromics" Soil Systems 4, no. 2: 23. https://doi.org/10.3390/soilsystems4020023
APA StyleTrubl, G., Hyman, P., Roux, S., & Abedon, S. T. (2020). Coming-of-Age Characterization of Soil Viruses: A User’s Guide to Virus Isolation, Detection within Metagenomes, and Viromics. Soil Systems, 4(2), 23. https://doi.org/10.3390/soilsystems4020023