Effect of Pyroligneous Acid on the Microbial Community Composition and Plant Growth-Promoting Bacteria (PGPB) in Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Experimental Soil, and Treatment Description
2.2. Soil Dehydrogenase Activity (DHA)
2.3. Soil DNA Extraction
2.4. Illumina Miseq Analysis
2.5. Bioinformatics Analysis of Amplicon Sequences
3. Results
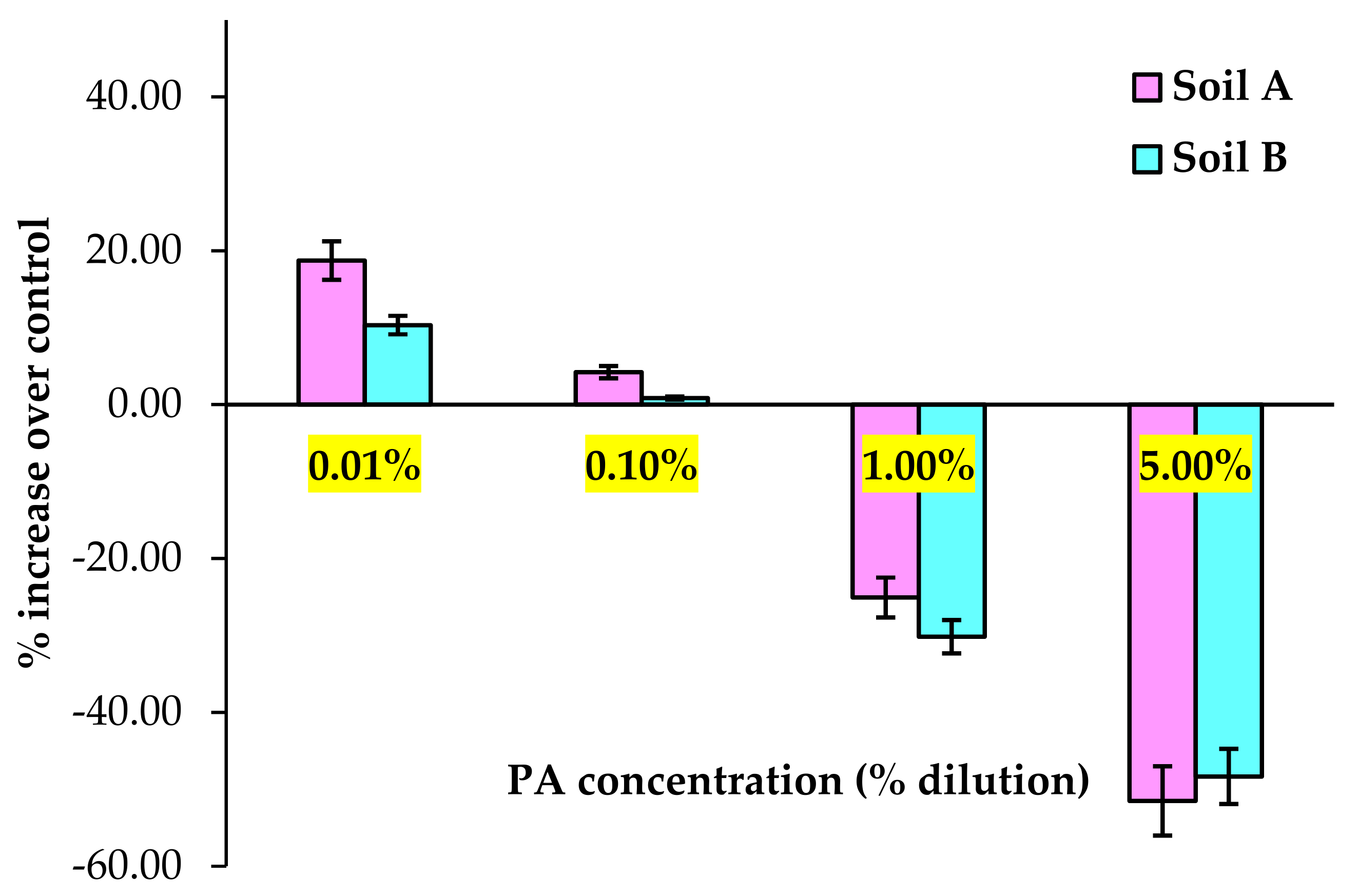
3.1. Soil Physicochemical Properties and Dehydrogenase Activity
3.2. Soil Microbial Diversity
3.3. Soil Bacterial Community Composition
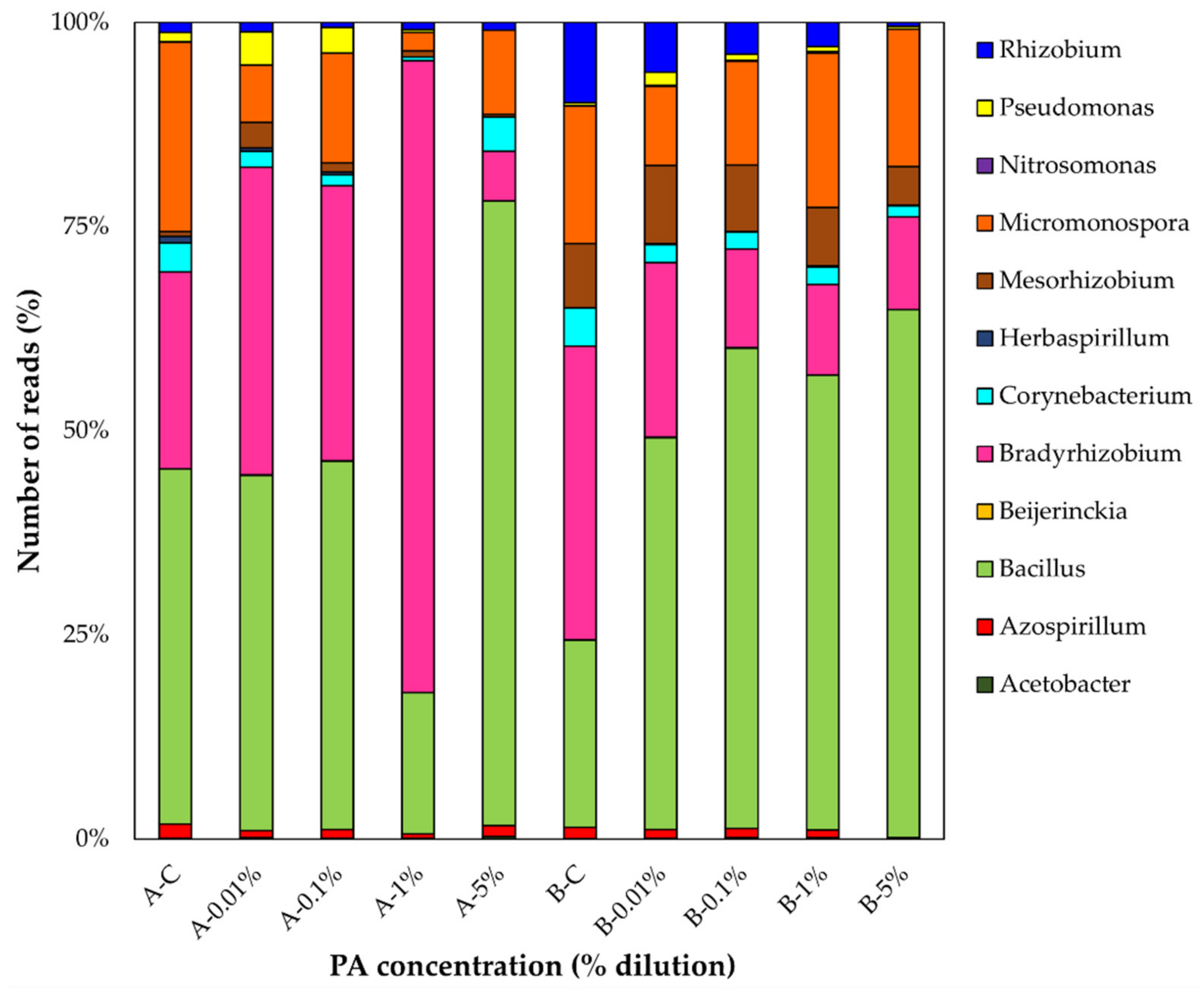
3.4. Changes in Plant Growth Promoting Bacteria (PGPB) in Response to PA
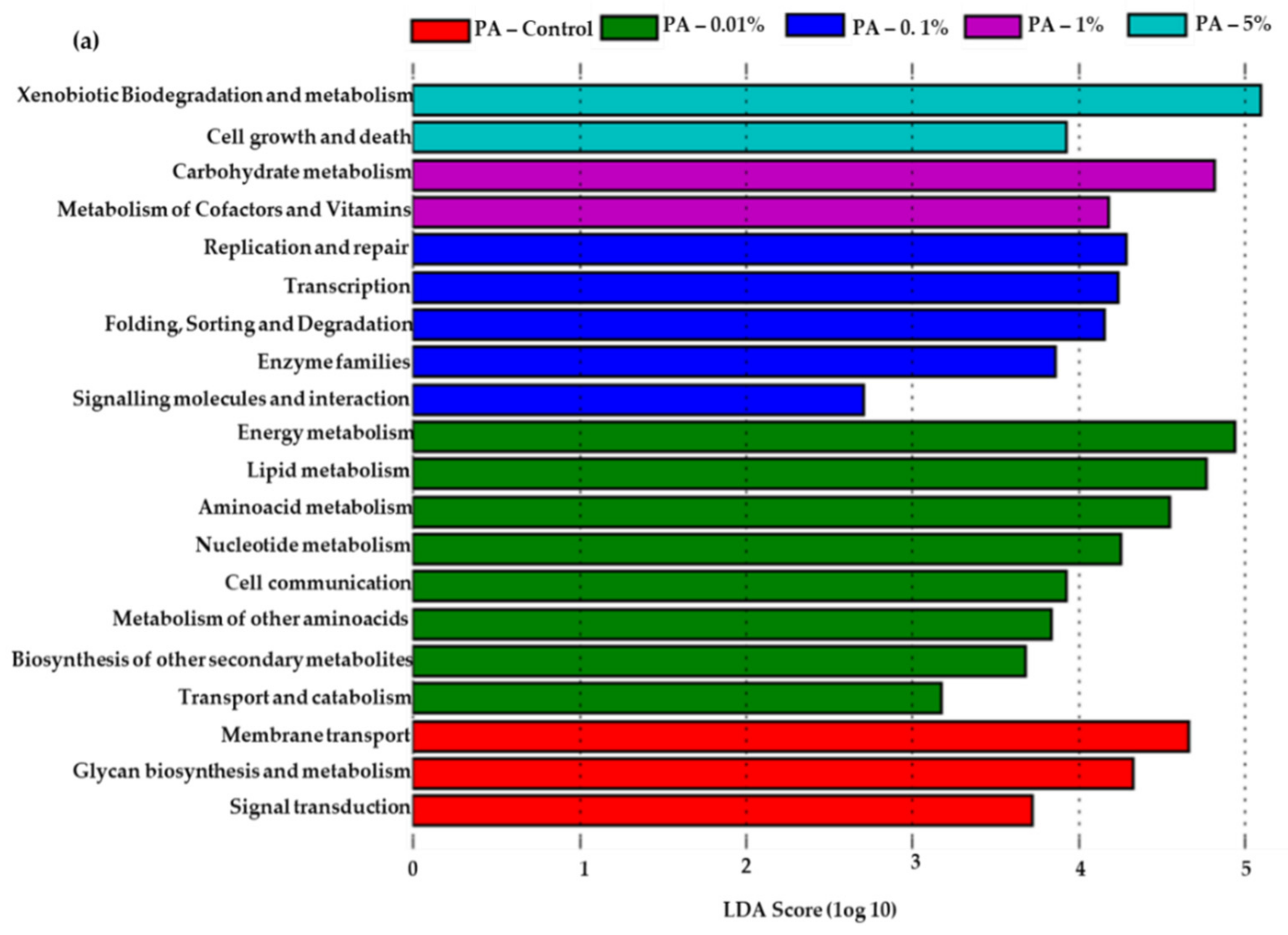
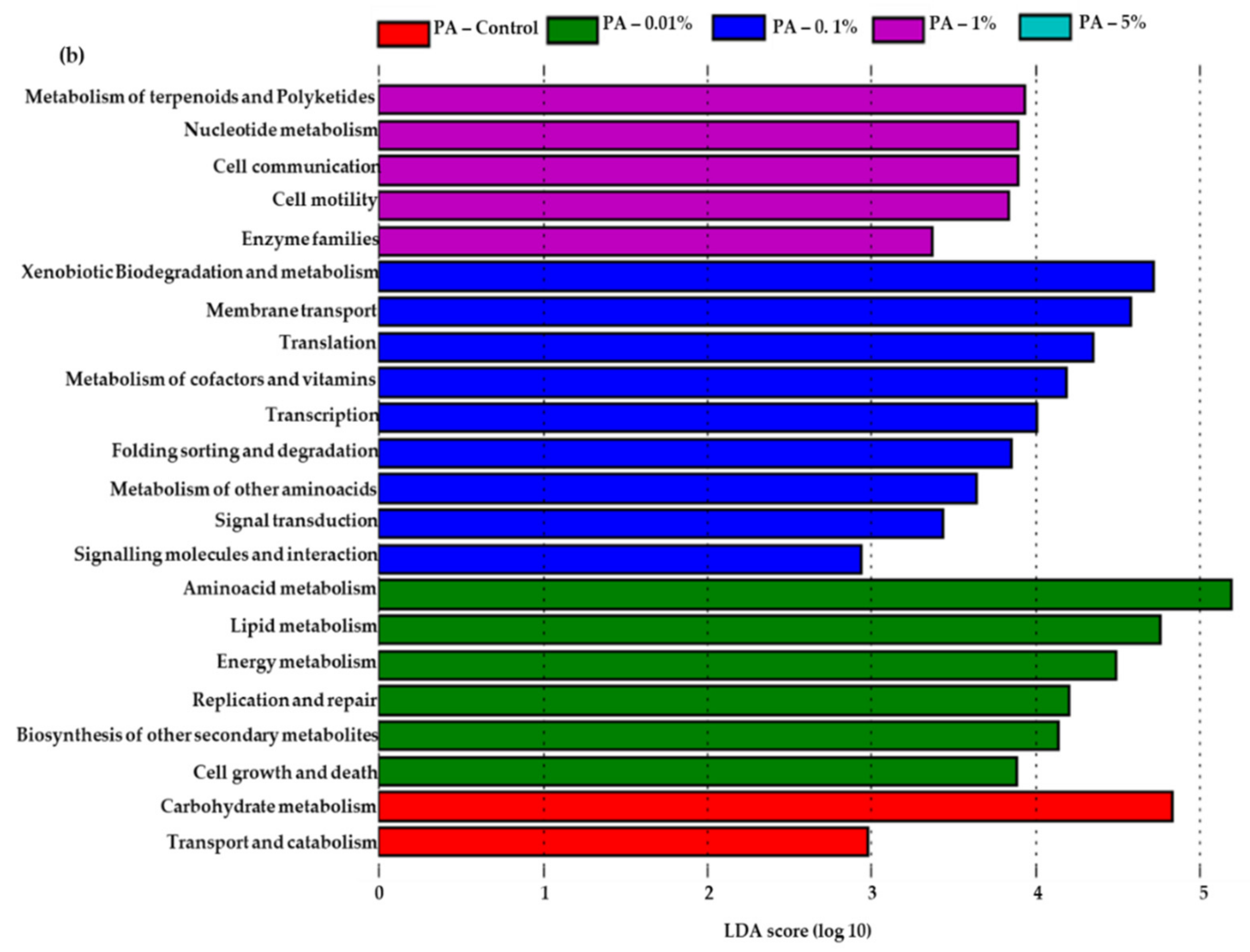
3.5. Predicted Functional Features in PA Amended Soils
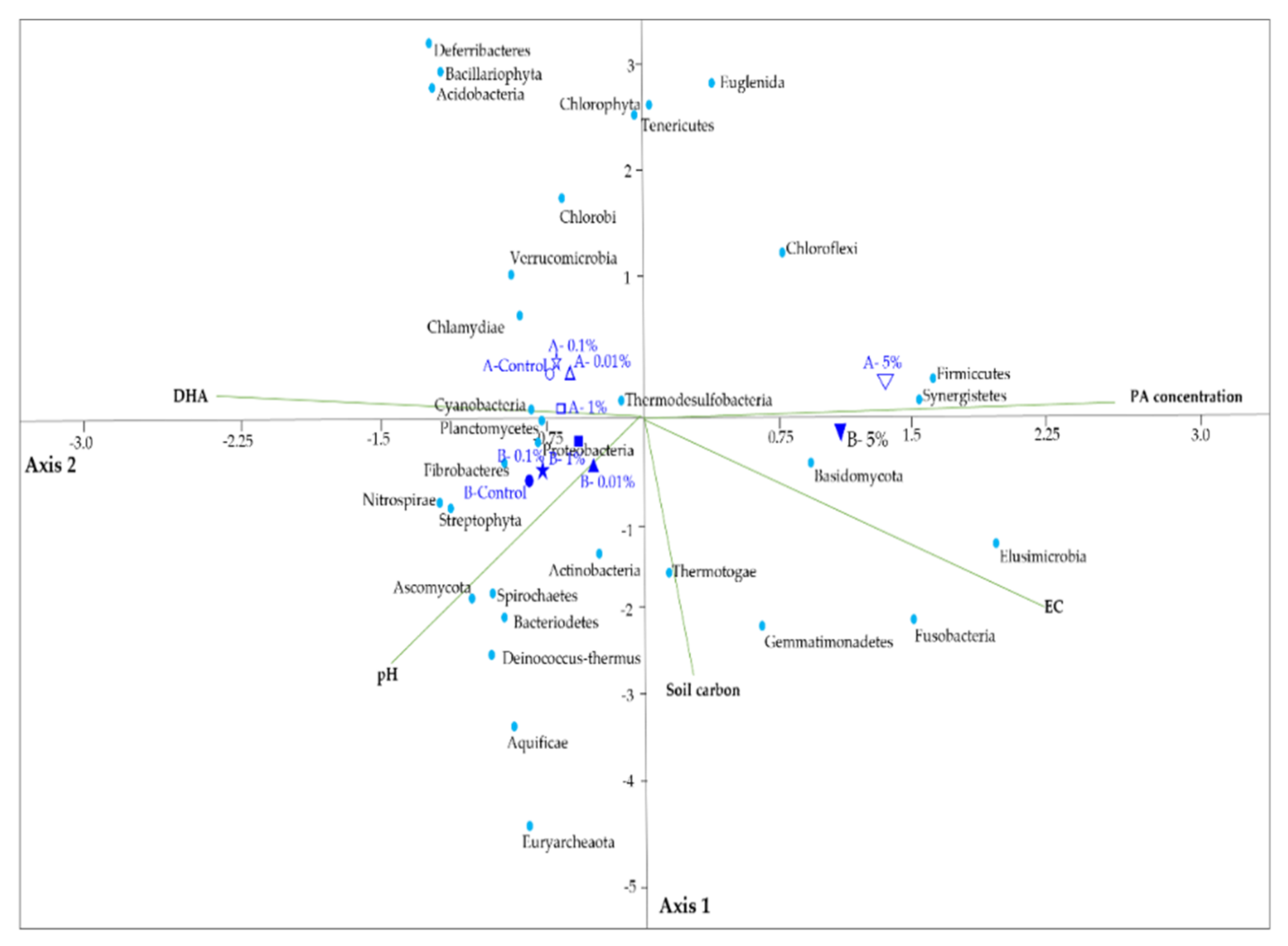
3.6. The Response of the Microbial Communities to Soil Properties
3.7. Changes in the Microbial Community Structure in Response to PA Concentration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, H.; Sun, C.; Hou, X.; Wu, M.; Yao, Y.; Li, F. Pyrolysis of Arundo donax L. to produce pyrolytic vinegar and its effect on the growth of dinoflagellate Karenia brevis. Bioresour Technol. 2018, 247, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, S.; Hou, B.; Zheng, H.; Deng, W.; Liu, D.; Tang, W. Study on the preparation of wood vinegar from biomass residues by carbonization process. Bioresour. Technol. 2015, 179, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, A.S.; Fasciotti, M.; Monteiro, T.V.C.; Lima, K.M.G. Chemical composition of pyroligneous acid obtained from Eucalyptus GG100 clone. Molecules 2018, 23, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza Araújo, E.; Pimenta, A.; Feijó, F.; Castro, R.; Fasciotti, M.; Monteiro, T.; De Lima, K. Antibacterial and antifungal activities of pyroligneous acid from wood of Eucalyptus urograndis and Mimosa tenuiflora. J. Appl. Microbiol. 2018, 124, 85–96. [Google Scholar] [CrossRef]
- Oramahi, H.A.; Yoshimura, T.; Diba, F.; Setyawati, D. Antifungal and antitermitic activities of wood vinegar from oil palm trunk. J. Wood Sci. 2018, 64, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Ma, X.; Dong, J. Preparation, chemical constituents and antimicrobial activity of pyroligneous acids from walnut-tree branches. J. Anal. Appl. Pyrolysis 2010, 87, 24–28. [Google Scholar] [CrossRef]
- Lashari, M.S.; Liu, Y.; Li, L.; Pan, W.; Fu, J.; Pan, G.; Zheng, J.; Zheng, J.; Zhang, X.; Yu, X. Effects of amendment of biochar-manure compost in conjunction with pyroligneous solution on soil quality and wheat yield of a salt-stressed cropland from Central China Great Plain. Field Crops Res. 2013, 144, 113–118. [Google Scholar] [CrossRef]
- Hagner, M.; Penttinen, O.P.; Tiilikkala, K.; Setälä, H. The effects of biochar, wood vinegar and plants on glyphosate leaching and degradation. Eur. J. Soil Biol. 2013, 58, 1–7. [Google Scholar] [CrossRef]
- Li, Y.; Pei, G.; Qiao, X.; Zhu, Y.; Li, H. Remediation of cadmium contaminated water and soil using vinegar residue biochar. Environ. Sci. Pollut. Res. 2018, 25, 15754–15764. [Google Scholar] [CrossRef]
- Luo, X.; Wang, Z.; Meki, K.; Wang, X.; Liu, B.; Zheng, H.; You, X.; Li, F. Effect of co-application of wood vinegar and biochar on seed germination and seedling growth. J. Soils Sediment 2019, 19, 3934–3944. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Ardakani, S.S.; Heydari, A.; Tayebi, L.; Mohammadi, M. Promotion of cotton seedlings growth characteristics by development and use of new bioformulations. Int. J. Bot. 2010, 6, 95–100. [Google Scholar] [CrossRef]
- Pandey, S.; Gupta, S. ACC deaminase producing bacteria with multifarious plant growth-promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front. Microbiol. 2019, 10, 1506. [Google Scholar]
- Tariq, M.; Noman, M.; Ahmed, T.; Hameed, A.; Manzoor, N.; Zafar, M. Antagonistic features displayed by plant growth-promoting rhizobacteria (PGPR): A review. J. Plant Sci. Phytopathol. 2017, 1, 38–43. [Google Scholar]
- Lin, Y.; Watts, D.B.; Kloepper, J.W.; Feng, Y.; Torbert, H.A. Influence of Plant Growth-Promoting Rhizobacteria on Corn Growth under Drought Stress. Commun. Soil Sci. Plant Anal. 2020, 51, 250–264. [Google Scholar] [CrossRef]
- Steiner, C.; Das, K.C.; Garcia, M.; Förster, B.; Zech, W. Charcoal and smoke extract stimulate the soil microbial community in a highly weathered xanthic Ferralsol. Pedobiologia 2008, 51, 359–366. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.; McGarrell, D.M.; Marsh, T.; Garrity, G.M. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef] [Green Version]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef]
- Miller, W.P.; Miller, D.M. A micro-pipette method for soil mechanical analysis. Commun. Soil Sci. Plant Anal. 1987, 18, 1–15. [Google Scholar] [CrossRef]
- Sivaram, A.K.; Logeshwaran, P.; Subashchandrabose, S.R.; Lockington, R.; Naidu, R.; Megharaj, M. Comparison of plants with C3 and C4 carbon fixation pathways for remediation of polycyclic aromatic hydrocarbon contaminated soils. Sci. Rep. 2018, 8, 2100. [Google Scholar] [CrossRef]
- Casida, L.; Klein, D.; Santoro, T. Soil dehydrogenase activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Meyer, F.; Bagchi, S.; Chaterji, S.; Gerlach, W.; Grama, A.; Harrison, T.; Paczian, T.; Trimble, W.L.; Wilke, A. MG-RAST version 4—Lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Brief. Bioinform. 2019, 20, 1151–1159. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST-palaeontological statistics, ver. 1.89. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Gupta, V.V.S.R. Beneficial microorganisms for sustainable agriculture. Microbiol. Aust. 2012, 33, 113–115. [Google Scholar] [CrossRef]
- Malik, A.A.; Puissant, J.; Buckeridge, K.M.; Goodall, T.; Jehmlich, N.; Chowdhury, S.; Gweon, H.S.; Peyton, J.M.; Mason, K.E.; van Agtmaal, M. Land use driven change in soil pH affects microbial carbon cycling processes. Nat. Commun. 2018, 9, 3591. [Google Scholar] [CrossRef]
- Sherene, T. Mobility and transport of heavy metals in polluted soil environment. Biol. Forum—Int. J. 2010, 2, 112–121. [Google Scholar]
- Yang, X.; Liu, J.; McGrouther, K.; Huang, H.; Lu, K.; Guo, X.; He, L.; Lin, X.; Che, L.; Ye, Z. Effect of biochar on the extractability of heavy metals (Cd, Cu, Pb, and Zn) and enzyme activity in soil. Environ. Sci. Pollut. Res. 2016, 23, 974–984. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Wolińska, A.; Stępniewska, Z. Dehydrogenase activity in the soil environment. In Dehydrogenases; Canuta, R.A., Ed.; IntechOpen: London, UK, 2012; pp. 183–210. [Google Scholar]
- Willis, A.D. Rarefaction, alpha diversity, and statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Xie, L.; Jin, D.; Mi, B.; Wang, D.; Li, X.; Dai, X.; Zou, X.; Zhang, Z.; Ma, Y. Bacterial community response to cadmium contamination of agricultural paddy soil. Appl. Soil Ecol. 2019, 139, 100–106. [Google Scholar] [CrossRef]
- Mahbub, K.R.; Subashchandrabose, S.R.; Krishnan, K.; Naidu, R.; Megharaj, M. Mercury alters the bacterial community structure and diversity in soil even at concentrations lower than the guideline values. Appl. Microbiol. Biotechnol. 2017, 101, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Mandakovic, D.; Rojas, C.; Maldonado, J.; Latorre, M.; Travisany, D.; Delage, E.; Bihouée, A.; Jean, G.; Díaz, F.P.; Fernández-Gómez, B. Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience. Sci. Rep. 2018, 8, 5875. [Google Scholar] [CrossRef]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity, and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Sánchez-Cañizares, C.; Jorrín, B.; Durán, D.; Nadendla, S.; Albareda, M.; Rubio-Sanz, L.; Lanza, M.; González-Guerrero, M.; Prieto, R.I.; Brito, B. Genomic diversity in the endosymbiotic bacterium Rhizobium leguminosarum. Genes 2018, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Zurdo, J.I.; Valverde, C.; Becker, A. Insights into the noncoding RNome of nitrogen-fixing endosymbiotic α-proteobacteria. Mol. Plant-Microbe Interact. 2013, 26, 160–167. [Google Scholar] [CrossRef] [Green Version]
- Nemergut, D.R.; Cleveland, C.C.; Wieder, W.R.; Washenberger, C.L.; Townsend, A.R. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol. Biochem. 2010, 42, 2153–2160. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef]
- Teixeira, L.C.R.S.; Peixoto, R.S.; Cury, J.C.; Sul, W.J.; Pellizari, V.H.; Tiedje, J.; Rosado, A.S. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J. 2010, 4, 989–1001. [Google Scholar] [CrossRef] [Green Version]
- Kloepper, J.; Gutierrez-Estrada, A.; McInroy, J. Photoperiod regulates elicitation of growth promotion but not induced resistance by plant growth-promoting rhizobacteria. Can. J. Microbiol. 2007, 53, 159–167. [Google Scholar] [CrossRef]
- García-Fraile, P.; Menéndez, E.; Rivas, R. Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng. 2015, 2, 183–205. [Google Scholar] [CrossRef]
- Kang, S.M.; Radhakrishnan, R.; Lee, I.J. Bacillus amyloliquefaciens subsp. plantarum GR53, a potent biocontrol agent resists Rhizoctonia disease on Chinese cabbage through hormonal and antioxidants regulation. World J. Microbiol. Biotechnol. 2015, 31, 1517–1527. [Google Scholar] [CrossRef]
- Queiroz Rego, C.H.; Cardoso, F.B.; Carina da Silva Cândido, A.; Teodoro, P.E.; Alves, C.Z. Co-inoculation with Bradyrhizobium and Azospirillum increases yield and quality of soybean seeds. J. Agron. 2018, 110, 2302–2309. [Google Scholar] [CrossRef]
- de Souza, J.T. Distribution, Diversity, and Activity of Antibiotic-Producing Pseudomonas spp. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2002. [Google Scholar]
- Louca, S.; Jacques, S.M.; Pires, A.P.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High taxonomic variability despite stable, functional structure across microbial communities. Nat. Ecol. Evol. 2016, 1, 0015. [Google Scholar] [CrossRef]
- Douglas, G.M.; Beiko, R.G.; Langille, M.G. Predicting the functional potential of the microbiome from marker genes using PICRUSt. In Microbiome Analysis; Humana Press: New York, NY, USA, 2018; pp. 169–177. [Google Scholar]
- Ortiz-Estrada, Á.M.; Gollas-Galván, T.; Martínez-Córdova, L.R.; Martínez-Porchas, M. Predictive functional profiles using metagenomic 16S rRNA data: A novel approach to understanding the microbial ecology of aquaculture systems. Rev. Aquac. 2019, 11, 234–245. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Tian, W.; Shao, Y.; Li, Y.J.; Lin, L.A.; Zhang, Y.J.; Han, H.; Chen, Z.J. Miscanthus cultivation shapes rhizosphere microbial community structure and function as assessed by Illumina MiSeq sequencing combined with PICRUSt and FUNGUIld analyses. Arch. Microbiol. 2020, 202, 1157–1171. [Google Scholar] [CrossRef]
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front. Plant Sci. 2017, 8, 132. [Google Scholar] [CrossRef] [Green Version]
- Pii, Y.; Borruso, L.; Brusetti, L.; Crecchio, C.; Cesco, S.; Mimmo, T. The interaction between iron nutrition, plant species, and soil type shapes the rhizosphere microbiome. Plant Physiol. Biochem. 2016, 99, 39–48. [Google Scholar] [CrossRef]


| Treatments | Soil A | Soil B | ||
|---|---|---|---|---|
| pH | EC (µS/cm) | pH | EC (µS/cm) | |
| Control | 5.9 | 39.5 | 6.6 | 145.7 |
| 0.01% PA | 5.9 | 39.2 | 6.6 | 142.5 |
| 0.1% PA | 5.5 | 44.2 | 6.5 | 137.1 |
| 1% PA | 4.6 | 85.9 | 6.0 | 194.9 |
| 5% PA | 4.0 | 146.7 | 5.1 | 373.7 |
| Treatment | Taxa_S | Alpha Diversity | Chao-1 | Shannon Index | ||||
|---|---|---|---|---|---|---|---|---|
| Soil A | Soil B | Soil A | Soil B | Soil A | Soil B | Soil A | Soil B | |
| Control | 1312 | 2167 | 259 | 404.9 | 1945 | 2933 | 3.15 | 4.66 |
| 0.01% PA | 1734 | 2302 | 299.4 | 426.2 | 2349 | 3151 | 3.67 | 5.17 |
| 0.1% PA | 1426 | 2174 | 253.5 | 423.5 | 2085 | 2938 | 3.34 | 4.89 |
| 1% PA | 1375 | 1929 | 235.9 | 392.0 | 1947 | 2629 | 2.79 | 4.65 |
| 5% PA | 1305 | 1535 | 216 | 263.5 | 1807 | 2225 | 2.51 | 4.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivaram, A.K.; Panneerselvan, L.; Mukunthan, K.; Megharaj, M. Effect of Pyroligneous Acid on the Microbial Community Composition and Plant Growth-Promoting Bacteria (PGPB) in Soils. Soil Syst. 2022, 6, 10. https://doi.org/10.3390/soilsystems6010010
Sivaram AK, Panneerselvan L, Mukunthan K, Megharaj M. Effect of Pyroligneous Acid on the Microbial Community Composition and Plant Growth-Promoting Bacteria (PGPB) in Soils. Soil Systems. 2022; 6(1):10. https://doi.org/10.3390/soilsystems6010010
Chicago/Turabian StyleSivaram, Anithadevi Kenday, Logeshwaran Panneerselvan, Kannappar Mukunthan, and Mallavarapu Megharaj. 2022. "Effect of Pyroligneous Acid on the Microbial Community Composition and Plant Growth-Promoting Bacteria (PGPB) in Soils" Soil Systems 6, no. 1: 10. https://doi.org/10.3390/soilsystems6010010
APA StyleSivaram, A. K., Panneerselvan, L., Mukunthan, K., & Megharaj, M. (2022). Effect of Pyroligneous Acid on the Microbial Community Composition and Plant Growth-Promoting Bacteria (PGPB) in Soils. Soil Systems, 6(1), 10. https://doi.org/10.3390/soilsystems6010010







