Genetic Diversity and Plant Growth-Promoting Activities of Root-Nodulating Bacteria in Guar Plants Across Jazan Province
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Soil Samples
2.2. Plant Growth, Nodule Collection, and Bacteria Isolation
2.3. Phenotypic Characterization of Bacteria and Plant Nodulation Tests
2.4. Screening of PGPR for Multiple Plant Growth-Promoting Activities
2.5. Extraction of DNA and PCR Amplifications
2.6. 16S rRNA Gene Sequencing and Analysis
3. Results
3.1. Constitution of a Local Collection of Rhizobium and Soil Analysis
3.2. Phenotypic Characterization
3.3. Characterization for Plant Growth-Promoting Traits
3.4. Molecular Characterization
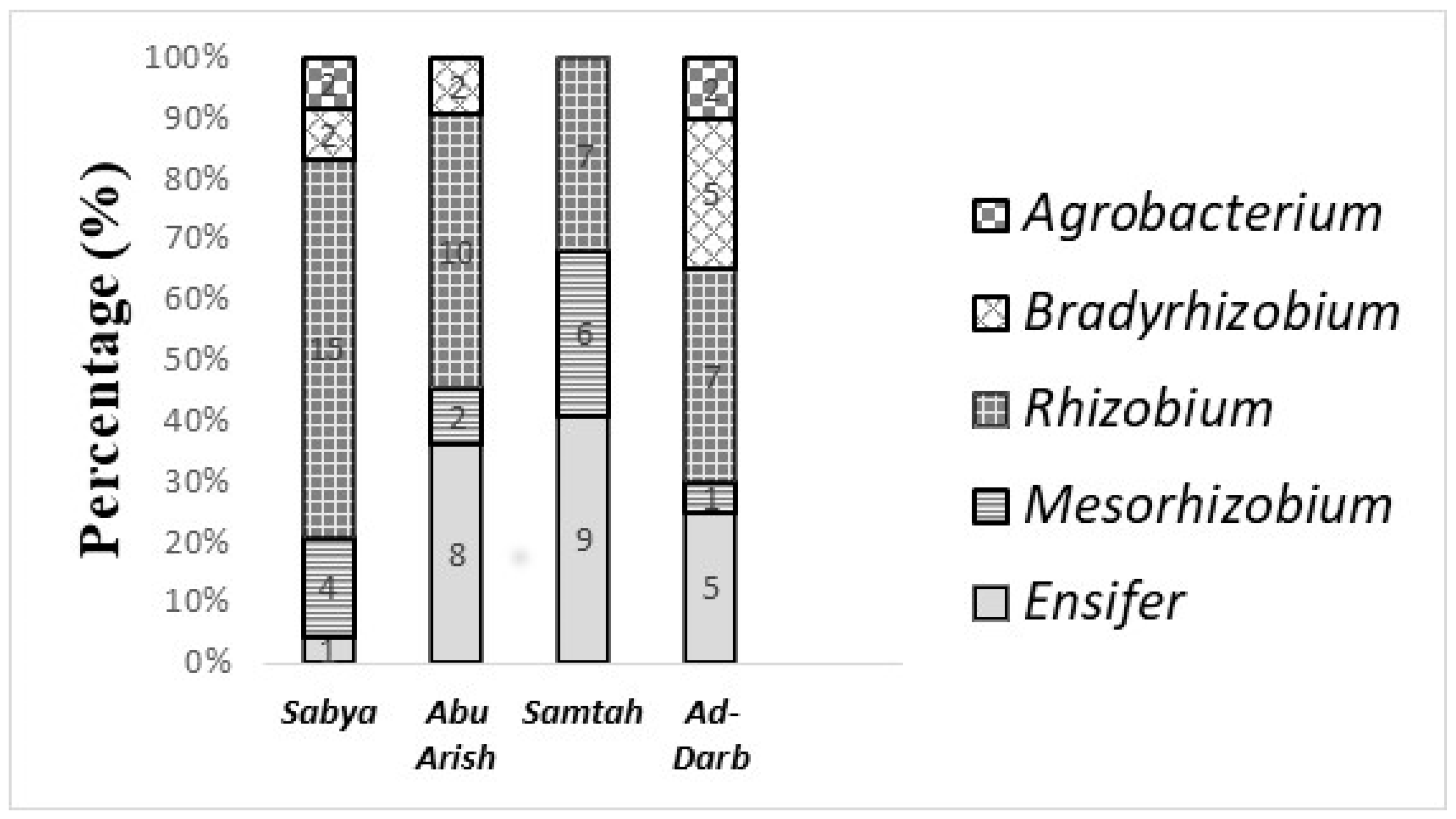
3.5. Taxonomic Affiliation of the Rhizobial Isolates and Their Distribution Across Sites of Origin
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). How to Feed the World in 2050; FAO: Rome, Italy, 2009; Available online: https://www.fao.org/publications (accessed on 20 February 2021).
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant breeding and drought in C-3 cereals: What should we breed for? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Zhao, C.X. Water-deficit stress-induced anatomical changes in higher plants. Comptes Rendus Biol. 2008, 331, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Patanè, C.; Saita, A.; Sortino, O. Eco-physiological response of a drought-tolerant and a drought-sensitive tomato genotype during water stress and recovery. Plant Biosyst. 2012, 146, 100–109. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt, and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Nitrogen fixation in perspective: An overview of research and extension needs. Field Crops Res. 2000, 65, 93–106. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Benhizia, Y.; Benhizia, H.; Benguedouar, A.; Muresu, R.; Giacomini, A.; Squartini, A. Gamma proteobacteria can nodulate legumes of the genus Hedysarum. Syst. Appl. Microbiol. 2004, 27, 462–468. [Google Scholar] [CrossRef]
- Muresu, R.; Polone, E.; Sulas, L.; Baldan, B.; Tondello, A.; Delogu, G.; Benguedouar, A. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 2008, 63, 383–400. [Google Scholar] [CrossRef]
- Mahdhi, M.; Fterich, A.; Rejili, M.; Rodriguez-Llorente, I.D.; Mars, M. Legume-Nodulating Bacteria (LNB) from three pasture legumes (Vicia sativa, Trigonella maritima and Hedysarum spinosissimum) in Tunisia. Ann. Microbiol. 2012, 62, 61–68. [Google Scholar] [CrossRef]
- Datta, C.; Basu, P.S. Indole acetic acid production by a Rhizobium species from root nodules of a leguminous shrub, Cajanus cajan. Microbiol Res. 2000, 155, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Lebrazi, S.; Fadil, M.; Chraibi, M.; Fikri-Benbrahim, K. Screening and optimization of indole-3-acetic acid production by Rhizobium sp. strain using response surface methodology. J. Genet. Eng. Biotechnol. 2020, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Sridevi, M.; Mallaiah, K.V. Phosphate solubilization by Rhizobium strains. Indian J. Microbiol. 2009, 49, 98–102. [Google Scholar] [CrossRef]
- Shome, S.; Barman, A.; Solaiman, Z.M. Rhizobium and Phosphate Solubilizing Bacteria Influence the Soil Nutrient Availability, Growth, Yield, and Quality of Soybean. Agriculture 2022, 12, 1136. [Google Scholar] [CrossRef]
- Manasa, K.R.; Subhash Reddy, S.; Triveni, B.; Kranthi, K.; Gowri Priya, N. Characterization of Rhizobium Isolates and their Potential PGPR Characteristics of different Rhizosphere Soils of Telangana Region. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2808–2813. [Google Scholar] [CrossRef]
- Saikia, S.P.; Bora, D.; Goswami, A.; Mudoi, K.D. Role of siderophores in chickpea (Cicer arietinum L.)—Rhizobium symbiosis. Microbiol. Res. 1998, 153, 47–53. [Google Scholar] [CrossRef]
- Deryło, M.; Choma, A.; Puchalski, B.; Suchanek, W. Siderophore activity in Rhizobium species isolated from different legumes. Acta Biochim. Pol. 1994, 41, 7–11. [Google Scholar] [CrossRef]
- Datta, B.; Chakrabartty, P.K. Siderophore biosynthesis genes of Rhizobium sp. isolated from Cicer arietinum L. 3 Biotech 2013, 4, 391–401. [Google Scholar] [CrossRef]
- Chandra, N.; Jack, K.J.; Wozniak, S.S.; Porter, M.L.; Friesen, M.L. Rhizobia protect their legume hosts against soil-borne microbial antagonists in a host-genotype-dependent manner. Rhizosphere 2019, 9, 47–55. [Google Scholar]
- Goyal, R.K.; Habtewold, J.Z. Evaluation of Legume–Rhizobial Symbiotic Interactions Beyond Nitrogen Fixation That Help the Host Survival and Diversification in Hostile Environments. Microorganisms 2023, 11, 1454. [Google Scholar] [CrossRef]
- Sijilmassi, B.; Filali-Maltouf, A.; Fahde, S.; Ennahli, Y.; Boughribil, S.; Kumar, S.; Amri, A. In-Vitro Plant Growth Promotion of Rhizobium Strains Isolated from Lentil Root Nodules under Abiotic Stresses. Agronomy 2020, 10, 1006. [Google Scholar] [CrossRef]
- Tulumello, J.; Chabert, N.; Rodriguez, J.; Long, J.; Nalin, R.; Achouak, W.; Heulin, T. Rhizobium alamii improves water stress tolerance in a non-legume. Sci. Total. Environ. 2021, 797, 148895. [Google Scholar] [CrossRef] [PubMed]
- Barquero, M.; Poveda, J.; Laureano-Marín, A.M.; Ortiz-Liébana, N.; Brañas, J.; González-Andrés, F. Mechanisms involved in drought stress tolerance triggered by rhizobia strains in wheat. Front. Plant Sci. 2022, 13, 1036973. [Google Scholar] [CrossRef] [PubMed]
- Alshameri, A.; Al-Qurainy, F.; Khan, S.; Nadeem, M.; Gaafar, A.R.; Tarroum, M.; Ashraf, M. Appraisal of guar [Cyamopsis tetragonoloba (L.) Taub.] accessions for forage purpose under the typical Saudi Arabian environmental conditions encompassing high temperature, salinity and drought. Pak. J. Bot. 2017, 49, 1405–1413. [Google Scholar]
- Gomaa, M.S.; Simons, C.; Brancale, A. Homology model of 1α, 25-dihydroxyvitamin D3 24-hydroxylase cytochrome P450 24A1 (CYP24A1): Active site architecture and ligand binding. J. Steroid Biochem. Mol. Biol. 2007, 104, 53–60. [Google Scholar] [CrossRef]
- Rao, N.K.; Shahid, M. Potential of cowpea [Vigna unguiculata (L.) Walp.] and guar [Cyamopsis tetragonoloba (L.) Taub.] as alternative forage legumes for the United Arab Emirates. Emir. J. Food Agric. 2011, 23, 147–156. [Google Scholar]
- Rasheed, M.J.Z.; Ahmad, K.; Qurainy, F.A.; Khan, S.; Athar, H.U.R. Screening of diverse local germplasm of guar [Cyamopsis tetragonoloba (L.) Taub.] for salt tolerance: A possible approach to utilize salt-affected soils. Pak. J. Bot. 2015, 47, 1721–1726. [Google Scholar]
- Mishra, B.K.; Yadav, V.; Vishal, M.K.; Kant, K. Physiological and molecular characterization of clusterbean (Cyamopsis tetragonoloba (L.) Taub) rhizobia isolated from different areas of Rajasthan, India. Legume Res. 2013, 36, 299–305. [Google Scholar]
- El Hussein, A.A. Characterization of Native Guar Rhizobia and Their Cross Inoculation Abilities Among Different Guar (Cyamopsis tetragonoloba (L.) Taub.) Lines. Master’s Thesis, University of Khartoum, Khartoum, Sudan, 2015. [Google Scholar]
- Subha, D.; Rajesh, G. Assessing Stress Tolerant Rhizobial Isolates of Clusterbean (Cymopsis tetragonoloba (L.) Taub.) Retrieved from Semi- Arid Regions of Haryana, India. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 744–753. [Google Scholar]
- Ibrahim, K.A.; Naeim, E.A.; El Naim, A.M.; Elsheikh, M.A. Response of guar (Cyamopsis teteragonolopa L.) to Bradyrhizobium inoculations in semi-arid Environment. Int. J. Agric. For. 2016, 6, 137–141. [Google Scholar]
- Yami, B. Phenotypic and Genetic Diversity of Root-Nodulating Bacteria Associated with Guar (Cyamopsis tetragonoloba L. Taub.) Growing in Different Regions of Jazan. Master’s Thesis, Jazan University Saudi Arabia, Jazan, Saudi Arabia, 2021; p. 18. [Google Scholar]
- Vincent, J.M. A Manual for the Practical Study of Root Nodule Bacteria; Blackwell Scientific Publications: Oxford, UK, 1970. [Google Scholar]
- Yelton, M.M.; Buckley, D.W.; Post, J.A. Growth and nitrogen fixation of Rhizobium japonicum and its mutant strains. Appl. Environ. Microbiol. 1983, 45, 602–605. [Google Scholar]
- Mohamed, M.A.H.; Harris, P.J.C.; Henderson, J. In vitro selection and characterisation of a drought tolerant clone of Tagetes minuta. Plant Sci. 2000, 159, 213–222. [Google Scholar] [CrossRef]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual, 4th ed.; Benjamin/Cummings: San Francisco, CA, USA, 1992. [Google Scholar]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiol. 1948, 17, 362–370. [Google Scholar]
- Mahdhi, M.; Tounekti, T.; Khemira, H. Phenotypic and Genotypic Characterization of Microsymbionts of Acacia Species Plants Grown in South-Western Saudi Arabia. J. Soil. Sci. Plant Nutr. 2019, 19, 631–638. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Jakobsen, I.B.; Nei, M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 2001, 17, 1244–1245. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Hernandez, B.S.; Focht, D.D. Invalidity of the concept of slow growth and alkali production in cowpea Rhizobia. Appl. Environ. Microbiol. 1984, 48, 206–210. [Google Scholar] [CrossRef]
- Sarma, R.K.; Saikia, R.; Thakur, D. Impact of plant growth-promoting endophytic bacteria on nutrient uptake and growth enhancement of aromatic rice and maize under low fertility condition. Microbiol. Res. 2021, 250, 126789. [Google Scholar]
- Alemayehu, W. The effect of indigenous Root Nodulating Bacteria on Nodulation and Growth of faba bean (Vicia faba) in low input agricultural systems of Tigray Highlands, Northern Ethopia. Momona Ethiop. J Sci. 2009, 1, 30–43. [Google Scholar]
- Al-mujahidy, S.J.; Hassan, M.; Rahman, M.; Mamun-Or-Rashid, A.N.M. Isolation and characterization of Rhizobium spp. and determination of their potency for growth factor production. Int. Res. J. Biotechnol. 2013, 4, 117–123. [Google Scholar]
- Jebara, M.; Mhamdi, R.; Aouani, M.E.; Ghrir, R.; Mars, M. Genetic diversity of Sinorhizobium populations recovered from different Medicago varieties cultivated in Tunisia soils. Can. J. Microbiol. 2001, 47, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Odee, D.W.; Haukka, K.; Sprent, J.I.; Sutherland, J.M.; Young, J.P.W. Genetic and symbiotic characterization of rhizobia isolated from tree and herbaceous legumes grown in soils from ecologically diverse sites in Kenya. Soil Biol. Biochem. 2002, 34, 801–811. [Google Scholar] [CrossRef]
- Mutumba, F.A.; Zagal, E.; Gerding, M.; Castillo-Rosales, D.; Paulino, L.; Schoebitz, M. Plant growth promoting rhizobacteria for improved water stress tolerance in wheat genotypes. J. Soil Sci. Plant Nutr. 2018, 18, 1080–1096. [Google Scholar] [CrossRef]
- Saghafi, D.; Ghorbanpour, M.; Lajayer, B.A. Efficiency of Rhizobium strains as plant growth promoting rhizobacteria on morphophysiological properties of Brassica napus L. under salinity stress. J. Soil Sci. Plant Nutr. 2018, 18, 253–268. [Google Scholar]
- Karanja, N.K.; Wood, M. Selecting Rhizobium phaseoli strains for use with beans (Phaseolus vulgaris L.) in Kenya: Tolerance of high temperature and antibiotic resistance. Plant Soil 1988, 112, 15–22. [Google Scholar] [CrossRef]
- Lebrazi, S.; Chraibi, M.; Fadil, M.; Barkai, H.; Fikri-Benbrahim, K. Phenotypic, Genotypic and Symbiotic Characterization of Rhizobial Isolates Nodulating Acacia sp. in Morocco. J. Pure Appl. Microbiol. 2018, 12, 249–263. [Google Scholar] [CrossRef]
- Coba de la Peña, T.; Fedorova, E.; Pueyo, J.J.; Lucas, M.M. The Symbiosome: Legume and Rhizobia Co-evolution toward a Nitrogen-Fixing Organelle? Front. Plant Sci. 2018, 8, 2229. [Google Scholar] [CrossRef]
- Shetta, D.; EL-Sayed, A.W.B.; Nasr, T.A.; Shaarawy, N.M. Influences of mineral fertilization with NPK, inoculation and methods of inoculation on seedling growth of two woody legume trees. World Appl. Sci. J. 2014, 29, 825–834. [Google Scholar]
- Jordan, D.C. Family III Rhizobiacae. In Bergey’s Manual of Systematic Bacteriology; krieg, N.R., Holt, J.G., Eds.; Williams & Wilkins: Baltimore, ML, USA, 1984; pp. 234–242. [Google Scholar]
- Graham, P.H. Stress tolerance in Rhizobium and Bradyrhizobium, and nodulation under adverse soil conditions. Can. J. Microbiol. 1992, 38, 475–484. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Sci. Hortic. 2012, 128, 963401. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.D.; Ghosh, A.K.; Kaur, S. Role of Rhizobium in Plant Growth Promotion and Nitrogen Fixation. Biotechnol. Lett. 2014, 36, 1399–1408. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Ghosh, P.; Maiti, T.K. Production and Metabolism of Indole Acetic Acid (IAA) by Root Nodule Bacteria (Rhizobium): A Review. Appl. Microbiol. 2011, 5, 523–540. [Google Scholar]
- Ramesh, B.D.; Subramanian, K.R.V. Siderophore production by Rhizobium and Bradyrhizobium species and its role in iron acquisition. Microbial. Ecol. 2014, 67, 667–676. [Google Scholar]
- Tahir, M.; Sarwar, M.A. Plant Growth Promoting Rhizobacteria (PGPR): A Budding Complement of Synthetic Fertilizers for Improving Crop Production. Group 2013, 19, 79–87. [Google Scholar]
- Igiehon, N.O.; Babalola, O.O.; Cheseto, X. Siderophores from rhizosphere bacteria as a potential biocontrol tool for managing plant pathogens. Environ. Sustain. 2019, 2, 57–71. [Google Scholar]
- Cubillos-Hinojosa, J.G.; de Sá, E.L.S.; Muniz, A.W.; Dick, D.P. Combined application of humic substances and PGPR inoculated and co-inoculated in plants of Phaseolus lunatus (L.) and Leucaena leucocephala (Lam.) de Wit. Rev. Colomb. Cienc. Hort. 2024, 18, e17621. [Google Scholar] [CrossRef]
- Ríos-Ruiz, W.F.; Castro-Tuanama, R.; Valdez-Nuñez, R.A.; Torres-Bernal, L.; Jave-Concepción, H.G.; Daza-Pérez, A.C.; Barrera-Lozano, M.; Archentti-Reátegui, F. Co-Inoculation of Phosphate-Solubilizing Bacteria and Rhizobia Increases Phosphorus Availability and Promotes the Development of Forage Legumes. Agronomy 2024, 14, 2493. [Google Scholar] [CrossRef]
- Damo, J.L.C.; Pedro, M.; Sison, M.L. Phosphate Solubilization and Plant Growth Promotion by Enterobacter sp. Isolate. Appl. Microbiol. 2024, 4, 1177–1192. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sathya, A.; Vijayabharathi, R.; Varshney, R.K.; Gowda, C.L.; Krishnamurthy, L. Plant growth promoting rhizobia: Challenges and opportunities. 3 Biotech 2015, 5, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Laslo, É.; Mara, G. Is PGPR an Alternative for NPK Fertilizers in Sustainable Agriculture? In Microbial Interventions in Agriculture and Environment; Singh, D.P., Gupta, V.K., Prabha, R., Eds.; Springer: Singapore, 2019; pp. 51–62. [Google Scholar]
- Glick, B.R.; de-Bashan, L.E. Enhanced Plant Growth by ACC Deaminase-Producing Bacteria. Plant Physiol. Biochem. 2017, 119, 57–65. [Google Scholar]
- Chibeba, A.M.; Kyei-Boahen, S.; de Fátima Guimarães, M.; Nogueira, M.A.; Hungria, M. Isolation and Screening of Elite Inoculant Strains of Bradyrhizobium spp. for Soybean Cultivation in Mozambique. Appl. Soil Ecol. 2017, 120, 55–67. [Google Scholar]
- Alikhani, H.A.; Saleh-Rastin, N.; Antoun, H. Phosphate Solubilization Activity of Rhizobia Native to Iranian Soils. Plant Soil 2006, 287, 35–41. [Google Scholar] [CrossRef]
- Fukaki, H.; Tasaka, M. Hormone Interactions During Lateral Root Formation. Plant Mol. Biol. 2020, 104, 73–83. [Google Scholar] [CrossRef]
- Atieno, M.; Herrmann, L.; Okalebo, R.W.; Lesueur, D. Improvement of Maize Growth and Nutrient Uptake by Co-inoculation of Nitrogen Fixing and Phosphate Solubilizing Rhizobacteria under Field Conditions in Kenya. Soil Sci. Plant Nutr. 2017, 63, 398–409. [Google Scholar]
- Zakhia, F.; Jeder, H.; Domergue, O.; Willems, A.; Cleyet-Marel, J.-C.; Gillis, M.; Dreyfus, B.; De Lajudie, P. Characterisation of Legume-Nodulating Bacteria (LNB) in arid regions of Tunisia. Syst. Appl. Microbiol. 2004, 27, 380–395. [Google Scholar] [CrossRef]
- Zakhia, F.; Jeder, H.; Willems, A.; Gillis, M.; Dreyfus, B.; de Lajudie, P. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb. Ecol. 2006, 51, 375–393. [Google Scholar] [CrossRef]
- Khalid, R.; Zhang, X.X.; Hayat, R.; Ahmed, M. Molecular Characteristics of Rhizobia Isolated from Arachis hypogaea Grown under Stress Environment. Sustainability 2020, 12, 6259. [Google Scholar] [CrossRef]
- Singh, R.P.; Manchanda, G.; Singh, R.N.; Srivastava, A.K.; Dubey, R.C. Selection of alkalotolerant and symbiotically efficient chickpea nodulating rhizobia from North-West Indo Gangetic Plains. J. Basic Microbiol. 2016, 56, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Richter, M.; Peplies, J.; Euzeby, J.; Amann, R.; Schleifer, K.-H.; Ludwig, W.; Glöckner, F.O.; Rosselló-Móra, R. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 2008, 31, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Zahran, H.H. Rhizobia from wild legumes: Diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J. Biotechnol. 2001, 91, 143–153. [Google Scholar] [CrossRef]
- Mahdhi, M.; Nzoué, A.; Gueye, F.; Merabet, C.; De Lajudie, P.; Mars, M. Phenotypic and genotypic diversity of Genista saharae microsymbionts from the infra-arid region of Tunisia. Lett. Appl. Microbiol. 2007, 45, 604–609. [Google Scholar] [CrossRef] [PubMed]
- de Lajudie, P.; Laurent-Fulele, E.; Willems, A.; Torck, U.; Coopman, R.; Collins, M.D.; Kersters, K.; Dreyfus, B.; Gillis, M. Allorhizobium undicola gen. nov., sp. nov., nitrogen-fixing bacteria that efficiently nodulate Neptunia natans in Senegal. Int. J. Syst. Evol. 1998, 4, 1277–1290. [Google Scholar] [CrossRef]
- Mhamdi, R.; Laguerre, G.; Aouani, M.E.; Mars, M.; Amarger, N. Different species and symbiotic genotypes of field rhizobia can nodulate Phaseolus vulgaris in Tunisian soils. FEMS Microbiol. Ecol. 2002, 41, 77–84. [Google Scholar] [CrossRef]
- Liu, J.; Wang, E.T.; Chen, W.X. Diverse rhizobia associated with woody legumes Wisteria sinensis, Cercis. Syst. Appl. Microbiol. 2005, 28, 465–477. [Google Scholar] [CrossRef]
- Ma, T.; Xue, H.; Piao, C.; Jiang, N.; Li, Y. Phylogenomic reappraisal of the family Rhizobiaceae at the genus and species levels, including the description of Ectorhizobium quercum gen. nov., sp. nov. Front. Microbiol. 2023, 14, 1207256. [Google Scholar] [CrossRef]

| Soil Texture | pH | EC 1 (dS/m) | Total N 2 (%) | |
|---|---|---|---|---|
| Sabya | silt | 7.3 ± 0.07 | 1.51 ± 0.04 | 0.13 ± 0.01 |
| Abu Arish | silt | 7.6 ± 0.06 | 1.42 ± 0.04 | 0.10 ± 0.01 |
| Al-Darb | sandy | 7.5 ± 0.06 | 1.39 ± 0.04 | 0.11 ± 0.01 |
| Samtah | sandy | 7.4 ± 0.10 | 1.53 ± 0.05 | 0.09 ± 0.01 |
| Isolate | Site of Origin | 16S rRNA Gene Type | Genus Level | Nodulation Test | Isolate | Site of Origin | 16S rRNA Gene Types | Genus Level | Nodulation Test |
|---|---|---|---|---|---|---|---|---|---|
| GD1 | Ad Darb | 14 | Bradyrhizobium | +(3) | GSA1 | Samtah | 4 | Mesorhizobium | +(8) |
| GD2 | Ad Darb | 3 | Ensifer | +(6) | GSA2 | Samtah | 4 | Mesorhizobium | +(2) |
| GD3 | Ad Darb | 3 | Ensifer | +(7) | GSA3 | Samtah | 2 | Ensifer | +(9) |
| GD4 | Ad Darb | 14 | Bradyrhizobium | +(5) | GSA4 | Samtah | 4 | Mesorhizobium | +(3) |
| GD5 | Ad Darb | 14 | Bradyrhizobium | +(4) | GSA5 | Samtah | 4 | Mesorhizobium | +(7) |
| GD6 | Ad Darb | 4 | Mesorhizobium | +(5) | GSA6 | Samtah | 5 | Mesorhizobium | +(2) |
| GD7 | Ad Darb | 11 | Rhizobium | - | GSA7 | Samtah | 3 | Ensifer | +(5) |
| GD8 | Ad Darb | 1 | Ensifer | +(4) | GSA8 | Samtah | 3 | Ensifer | +(2) |
| GD9 | Ad Darb | 2 | Ensifer | +(4) | GSA9 | Samtah | 3 | Ensifer | +(11) |
| GD10 | Ad Darb | 1 | Ensifer | +(3) | GSA10 | Samtah | 3 | Ensifer | +(2) |
| GD11 | Ad Darb | 15 | Bradyrhizobium | +(8) | GSA11 | Samtah | 3 | Ensifer | +(2) |
| GD12 | Ad Darb | 8 | Rhizobium | +(7) | GSA12 | Samtah | 7 | Rhizobium | +(4) |
| GD13 | Ad Darb | 8 | Rhizobium | +(2) | GSA13 | Samtah | 9 | Rhizobium | +(4) |
| GD14 | Ad Darb | 7 | Rhizobium | +(6) | GSA14 | Samtah | 9 | Rhizobium | +(6) |
| GD15 | Ad Darb | 9 | Rhizobium | +(4) | GSA15 | Samtah | 1 | Ensifer | +(7) |
| GD16 | Ad Darb | 9 | Rhizobium | +(7) | GSA16 | Samtah | 5 | Mesorhizobium | +(5) |
| GD17 | Ad Darb | 10 | Agrobacterium | - | GSA17 | Samtah | 12 | Rhizobium | +(9) |
| GD18 | Ad Darb | 14 | Bradyrhizobium | +(2) | GSA18 | Samtah | 12 | Rhizobium | +(2) |
| GD19 | Ad Darb | 10 | Agrobacterium | - | GSA19 | Samtah | 13 | Rhizobium | +(10) |
| GD20 | Ad Darb | 7 | Rhizobium | - | GSA20 | Samtah | 13 | Rhizobium | +(2) |
| GS1 | Sabya | 9 | Rhizobium | +(3) | GSA21 | Samtah | 3 | Ensifer | +(8) |
| GS2 | Sabya | 9 | Rhizobium | +(7) | GSA22 | Samtah | 3 | Ensifer | +(4 |
| GS3 | Sabya | 9 | Rhizobium | +(3) | GA1 | Abu Arish | 5 | Mesorhizobium | +(3) |
| GS4 | Sabya | 6 | Rhizobium | +(9) | GA2 | Abu Arish | 2 | Ensifer | +(7) |
| GS5 | Sabya | 12 | Rhizobium | +(3) | GA3 | Abu Arish | 1 | Ensifer | +(5) |
| GS6 | Sabya | 2 | Ensifer | +(5) | GA4 | Abu Arish | 1 | Ensifer | +(3) |
| GS7 | Sabya | 11 | Rhizobium | - | GA5 | Abu Arish | 1 | Ensifer | +(2) |
| GS8 | Sabya | 11 | Rhizobium | - | GA6 | Abu Arish | 2 | Ensifer | +(6) |
| GS9 | Sabya | 6 | Rhizobium | +(4) | GA7 | Abu Arish | 14 | Bradyrhizobium | +(2) |
| GS10 | Sabya | 6 | Rhizobium | +(4) | GA8 | Abu Arish | 14 | Bradyrhizobium | +(9) |
| GS11 | Sabya | 15 | Bradyrhizobium | +(7) | GA9 | Abu Arish | 8 | Rhizobium | +(2) |
| GS12 | Sabya | 15 | Bradyrhizobium | +(7) | GA10 | Abu Arish | 8 | Rhizobium | +(2) |
| GS13 | Sabya | 10 | Agrobacterium | - | GA11 | Abu Arish | 8 | Rhizobium | +(7) |
| GS14 | Sabya | 10 | Agrobacterium | - | GA12 | Abu Arish | 5 | Mesorhizobium | +(7) |
| GS15 | Sabya | 6 | Rhizobium | +(6) | GA13 | Abu Arish | 12 | Rhizobium | +(8) |
| GS16 | Sabya | 13 | Rhizobium | +(2) | GA14 | Abu Arish | 12 | Rhizobium | +(3) |
| GS17 | Sabya | 4 | Mesorhizobium | +(6) | GA15 | Abu Arish | 6 | Rhizobium | +(4) |
| GS18 | Sabya | 4 | Mesorhizobium | +(4) | GA16 | Abu Arish | 6 | Rhizobium | +(6 |
| GS19 | Sabya | 8 | Rhizobium | +(2) | GA17 | Abu Arish | 2 | Ensifer | +(9) |
| GS20 | Sabya | 6 | Rhizobium | +(3) | GA18 | Abu Arish | 3 | Ensifer | +(7) |
| GS21 | Sabya | 13 | Rhizobium | +(8) | GA19 | Abu Arish | 3 | Ensifer | +(5) |
| GS22 | Sabya | 5 | Mesorhizobium | +(7) | GA20 | Abu Arish | 7 | Rhizobium | +(5) |
| GS23 | Sabya | 9 | Rhizobium | +(9) | GA21 | Abu Arish | 7 | Rhizobium | +(4) |
| GS24 | Sabya | 5 | Mesorhizobium | +(7) | GA22 | Abu Arish | 13 | Rhizobium | +(2) |
| Isolate | IAA Production (µg/mL) | Siderophore Production | Ammonia Production | Phosphate Solubilization | Isolate | IAA Production (µg/mL) | Siderophore Production | Ammonia Production | Phosphate Solubilization |
|---|---|---|---|---|---|---|---|---|---|
| GD1 | - | + | + | - | GSA1 | - | - | + | - |
| GD2 | - | - | - | 1.33 | GSA2 | - | - | + | - |
| GD3 | - | - | - | 1.22 | GSA3 | - | - | - | - |
| GD4 | - | + | + | - | GSA4 | - | - | + | - |
| GD5 | - | + | + | - | GSA5 | - | - | + | - |
| GD6 | - | - | + | - | GSA6 | - | - | + | - |
| GD7 | - | - | - | - | GSA7 | - | - | + | 1.34 |
| GD8 | 55 | - | + | 2.02 | GSA8 | - | - | + | 1.37 |
| GD9 | - | - | + | - | GSA9 | - | - | + | 1.54 |
| GD10 | - | - | + | 2.11 | GSA10 | - | - | + | 1.66 |
| GD11 | - | - | + | - | GSA11 | - | - | - | - |
| GD12 | - | - | + | 1.4 | GSA12 | 36 | - | + | 2.19 |
| GD13 | - | - | + | 1.55 | GSA13 | 84 | - | + | - |
| GD14 | - | - | + | 1.61 | GSA14 | 93 | - | + | - |
| GD15 | 73 | + | + | 2.33 | GSA15 | - | - | + | - |
| GD16 | 39 | + | + | 2.-5 | GSA16 | - | - | + | - |
| GD17 | - | - | - | - | GSA17 | 15 | - | + | - |
| GD18 | - | + | - | - | GSA18 | 48 | - | + | - |
| GD19 | - | - | - | GSA19 | - | - | + | - | |
| GD20 | - | - | + | - | GSA20 | - | - | + | - |
| GS1 | 120 | + | + | 3.88 | GSA21 | - | - | + | - |
| GS2 | 45 | - | + | 2.22 | GSA22 | - | - | + | 1.04 |
| GS3 | 50 | - | + | 2.4 | GA1 | - | - | + | - |
| GS4 | 71 | - | + | 2.44 | GA2 | - | - | + | - |
| GS5 | - | - | + | - | GA3 | - | - | + | 1.22 |
| GS6 | - | - | - | - | GA4 | - | - | + | 1.44 |
| GS7 | - | - | - | - | GA5 | - | - | + | 1.88 |
| GS8 | - | - | - | - | GA6 | - | - | + | - |
| GS9 | 82 | + | + | 3.11 | GA7 | - | + | + | - |
| GS10 | - | + | 3.25 | GA8 | - | + | + | - | |
| GS11 | - | - | + | - | GA9 | 88 | - | + | - |
| GS12 | - | + | + | - | GA10 | 76 | - | + | 2.18 |
| GS13 | - | - | - | - | GA11 | 90 | - | + | 2.03 |
| GS14 | - | - | - | - | GA12 | - | - | + | - |
| GS15 | - | - | + | - | GA13 | 55 | - | + | 1.77 |
| GS16 | 92 | + | + | 3.44 | GA14 | 62 | - | + | - |
| GS17 | - | - | + | - | GA15 | 78 | - | + | 2.33 |
| GS18 | - | - | + | - | GA16 | 69 | - | + | 2.55 |
| GS19 | 53 | - | + | 2.92 | GA17 | - | - | + | - |
| GS20 | 70 | - | + | 2.77 | GA18 | 85 | - | + | 2.88 |
| GS21 | 67 | - | + | 2.03 | GA19 | - | - | + | 1.21 |
| GS22 | - | - | + | - | GA20 | 94 | - | + | 1.66 |
| GS23 | - | - | + | - | GA21 | 37 | - | + | 1.11 |
| GS24 | - | - | + | - | GA22 | - | - | + | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahdhi, M.; Yami, B.; Al Abboud, M.; Abada, E.; Khemira, H. Genetic Diversity and Plant Growth-Promoting Activities of Root-Nodulating Bacteria in Guar Plants Across Jazan Province. Soil Syst. 2025, 9, 39. https://doi.org/10.3390/soilsystems9020039
Mahdhi M, Yami B, Al Abboud M, Abada E, Khemira H. Genetic Diversity and Plant Growth-Promoting Activities of Root-Nodulating Bacteria in Guar Plants Across Jazan Province. Soil Systems. 2025; 9(2):39. https://doi.org/10.3390/soilsystems9020039
Chicago/Turabian StyleMahdhi, Mosbah, Boshra Yami, Mohamed Al Abboud, Emad Abada, and Habib Khemira. 2025. "Genetic Diversity and Plant Growth-Promoting Activities of Root-Nodulating Bacteria in Guar Plants Across Jazan Province" Soil Systems 9, no. 2: 39. https://doi.org/10.3390/soilsystems9020039
APA StyleMahdhi, M., Yami, B., Al Abboud, M., Abada, E., & Khemira, H. (2025). Genetic Diversity and Plant Growth-Promoting Activities of Root-Nodulating Bacteria in Guar Plants Across Jazan Province. Soil Systems, 9(2), 39. https://doi.org/10.3390/soilsystems9020039







