Simple Summary
Emerging evidence suggests that altered gut microbiota composition has the potential to promote hepatocellular carcinoma development and progression. This review outlines the role of gut microbiota in causing hepatocellular carcinoma, investigates its value in improving the its outcome after early diagnosis, and summarizes the latest progress regarding its potential for hepatocellular carcinoma therapy.
Abstract
In recent decades, gut microbiota have received emerging attention regarding their integral role in chronic liver disease progression, given the anatomic connection and the gut–liver axis. Emerging evidence has indicated a complex link between gut microbiota and hepatocellular carcinoma. This review explores the pathophysiological crosstalk between gut dysbiosis and hepatocarcinogenesis. The metabolic and immunologic effects mediated by gut-microbiota-derived metabolites, such as bile acids, short-chain fatty acids, and alcohol, could impact the aberrant biological behavior of hepatocellular carcinoma. This review also investigates the value of gut microbiota as novel non-invasive diagnostic biomarkers for the early detection of hepatocellular carcinoma, and summarizes the changes in the gut microbiota spectrum in patients with liver cancer. The current literature and studies on the role of the gut microbiota as adjuvant agents in liver cancer immunotherapy are reviewed.
1. Introduction
Primary liver cancer is the sixth most diagnosed cancer and the third leading cause of death among all types of cancer worldwide [1]. Globally, approximately 906,000 new cases of liver cancer were recorded and 830,000 deaths were attributed to it in 2020. Its incidence significantly increased, by 75%, between 1990 and 2015 [1,2]. Among the types of primary liver cancer, hepatocellular cancer (HCC) accounts for between 75% and 85% of cases [1]. The overall prognosis of HCC is poor, with a five-year survival rate below 20% [3,4]. For the best clinical outcome, curative treatment needs to be applied in the early stages of HCC. Treatment options include surgical resection, liver transplantation, and locoregional therapy, including ablation, chemoembolization, radioembolization, and systemic therapy for advanced HCC stages [5,6,7]. As early detection is believed to improve HCC survival rates, it is critical to understand the mechanisms of tumorigenesis in order to create novel approaches for HCC diagnosis and treatment.
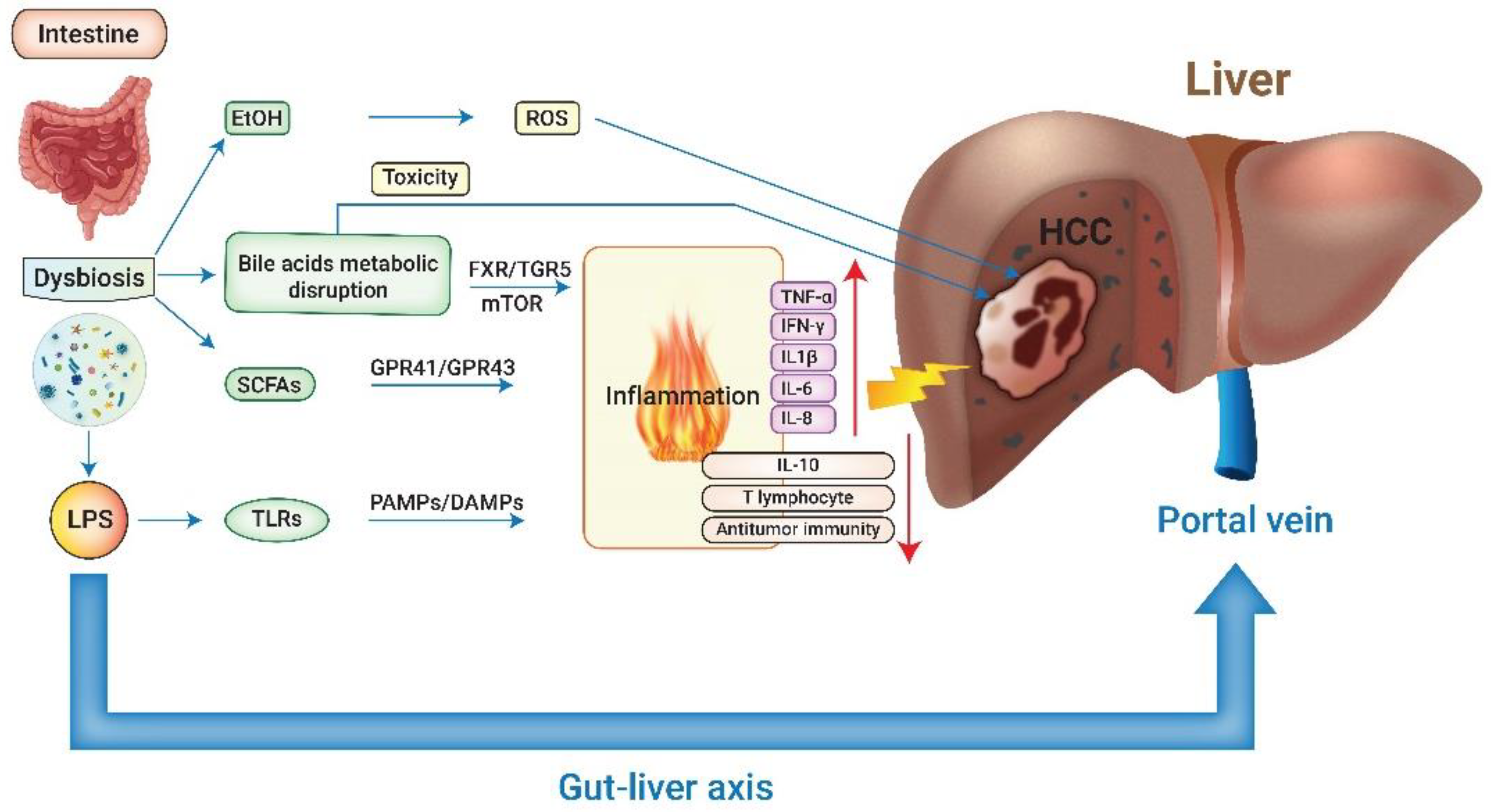
The main risk factors to promote HCC formation are chronic liver diseases related to hepatitis B virus (HBV) or hepatitis C virus (HCV), heavy alcohol intake, and non-alcoholic fatty liver disease (NAFLD) [1]. Recent emerging studies have elaborated on the integral role of gut microbiota in the progression of chronic liver disease via the modulation of inflammation, immunity, and metabolism [8,9,10,11,12]. There are more than 1000 bacteria species and 100 trillion bacteria colonizing the human intestinal tract. Their genes outnumber the genes of the human genome by 150-fold [13,14]. The composition and abundance of gut microbiota (GM) have considerable functional heterogeneity between individuals and are affected by many factors, including age, gender, diet, disease state, and medication. GM homeostasis boosts host health; in contrast, dysbiosis manifests as the imbalance between healthy and disease-promoting microorganisms and has been attributed to many pathological processes [11]. The gut–liver axis represents a complex bi-directional relationship between the gut, its microbiome, and the liver, created through a multitude of signals produced by genetic, dietary, and environmental factors (Figure 1).
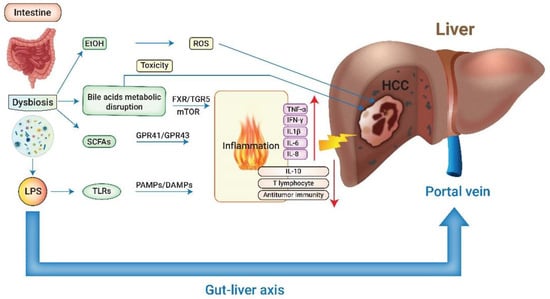
Figure 1.
The interplay between gut microbiota and hepatocellular carcinoma. LPS: lipopolysaccharide; EtOH: ethanol; SCFAs: short-chain fatty acids; TLRs: Toll-like receptors; ROS: reactive oxygen species; FXR: farnesoid X receptor; TGR5: transmembrane G protein-coupled receptor 5; GPR: G protein-coupled receptor; PAMPs: pathogen-associated molecular patterns; DAMPs: damage-associated molecular patterns; TNF-α: tumor necrosis factor-alfa; IFN-γ: interferon-gamma; IL-1β: interleukin 1 beta; IL-6: interleukin 6; IL-8: interleukin 8; IL-10: interleukin 10; HCC: hepatocellular carcinoma.
A growing number of studies continue to reveal the complex link between the gut microbiota and HCC. This review explores the pathophysiological connections between gut dysbiosis and hepatocarcinogenesis. Furthermore, it summarizes the microbiome-based predictive value of HCC diagnosis and its application in HCC treatment.
2. The Pathogenetic Links between Dysbiosis and Hepatocellular Carcinoma
The portal vein connects the gastrointestinal tract and the liver, not only supplying nutrition to the two organs, but also providing the anatomical basis for the gut–liver axis. The liver receives about 70% of its total blood supply from portal vein circulation. The portal vein blood contains nutritional macromolecules and GM-derived metabolites. An intact gut epithelium plays an essential role in modulating the gut–liver axis. The intestinal barrier functions as a mechanical and immunological barrier preventing hazardous substances from penetrating into the blood circulation [15]. However, dysbiosis can interrupt the tight junction of the gut epithelium, increase intestinal permeability, and result in consequential pathophysiological effects via a complex metabolic and immunologic network [11].
2.1. Interactions between Gut Microbiota and Host Immune System
Intestinal bacterial overgrowth weakens the intestinal integrity; results in the release of lipopolysaccharides (LPSs); and activates toll-like receptors (TLR), which are damage-associated molecular patterns (DAMP) and pattern recognition receptors that bind pathogen-associated molecular patterns (PAMPs) and further stimulate Kupffer cells. These TLRs release pro-inflammatory cytokines (tumor necrosis factor-alfa, interleukin 8, interleukin 1 beta) [11,16,17]. In NAFLD dysbiosis patients, both CD4-positive and CD8-positive cells are suppressed in the duodenal mucosa lamina propria. Meanwhile, pro-inflammatory cytokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-6, and interferon (IFN)-γ were increased [18].
Several pre-clinical animal studies strengthened the evidence of LPS-induced inflammatory effect on HCC formation. Yu et al. [19] showed that LPS was elevated in rats that developed HCC after diethylnitrosamine (DEN) exposure. However, once the LPS level was reduced by treatment with antibiotics or genetic knock-out Toll-like receptor 4 (TLR4), which mediates inflammatory cytokines, excessive tumor growth was prevented. The relationship between GM-related immune dysregulation and HCC is far beyond pro-inflammatory cascades. In a mouse model, intestinal dysbiosis suppressed antitumor immunity surveillance via the TLR-4-dependent expansion of hepatic monocytic myeloid-derived suppressor cells and suppression of T-cell abundance [20]. The LPS could induce hepatic stellate cells in the tumor microenvironment to develop a senescence-associated secretory phenotype (SASP) associated with the suppression of antitumor immunity [21,22].
2.2. Crosstalk between Gut Microbiota and Metabolites
2.2.1. Bile Acids
Bile acids (BAs) are synthesized from cholesterol, and the BA enterohepatic circulation serves a critical role in maintaining homeostasis of the gut–liver axis [23]. The GM is involved in multiple steps of BAs metabolism, including primary bile acid synthesis and the conjugation, reabsorption, deconjugation, and transformation to secondary bile acids. The disruption of key steps in bile acid metabolism could result in composition and abundance changes in BAs and impair homeostasis [11,23]. BAs directly affect liver cells when they accumulate at high concentrations [24]. The disruption of BA hemostasis is considered an adverse factor. The altered BA metabolism can promote tumorigenesis via inflammation, oxidative stress, fibrosis, and resistance to apoptosis [25,26].
The major signaling pathways by which bile acids regulate inflammation involve a few key regulators, such as farnesoid X receptor (FXR) and transmembrane G protein-coupled receptor 5 (TGR5) [27,28]. Via these pathways, BA hemostasis not only maintains intestinal barrier integrity, but also decreases hepatic inflammation and fibrosis [27,29]. Previous studies have indicated that bile acid analogs can activate FXR or TGR5 receptors to down-regulate pro-inflammatory gene expression while up-regulating monocyte and anti-inflammatory cytokine IL-10 production and alleviating the severity of hepatic inflammation, steatosis, and fibrosis [26,30]. Moreover, secondary BAs, via the mTOR signaling pathway, promote hepatic inflammation and carcinogenesis in NAFLD-related HCC [31]. In a pre-clinical mouse model, Shen et al. [32] found both in vitro and in vivo studies demonstrating that reducing gut bile salt hydrolase (BSH)-rich bacteria (i.e., Bifdobacteriales, Bacteroidales, Clostridiales, Lactobacillales) remarkably decreased serum conjugated deoxycholic acids (DCA), an effect which is thought to be associated with HCC growth. Glycodeoxycholic acid (GDCA) imitates conjugated DCA and inhibits the growth and migration of HCC both in vivo and in vitro. Knisely and colleagues [33] reported that canalicular bile salt export pump (BSEP)-deficient patients with cholestasis had a significant risk of developing HCC.
2.2.2. Short-Chain Fatty Acids
Short-chain fatty acids (SCFAs) are products fermented from dietary fiber by GM in the colon, and are primarily composed of acetate, propionate, and butyrate [11,34]. Like BAs, SCFAs also show metabolic and immunologic effects on tumorigenesis. SCFAs impact the genes related to regulated fatty acid β-oxidation, insulin sensitivity, lipogenesis, and ROS genesis via G-protein-coupled receptors GPR41 and GPR43 to alter the metabolism [11,35,36]. SCFAs play a complex role in immunomodulation. For example, butyrate can induce the differentiation of Treg cells, which are key players in suppressing inflammatory responses [37]. Furthermore, butyrate regulates pro-inflammatory responses by means of down-regulated TNF-alpha secretion via Prostaglandin E secretion and down-regulated NF-kappaB activation by lipopolysaccharides [38]. The anti-inflammatory effects are mediated by reducing the migration and proliferation of immune cells, reducing pro-inflammatory cytokines, and up-regulating anti-inflammatory cytokines [39,40], significantly affecting the tumor microenvironment and promoting tumorigenesis.
Singh et al. [41] recently published a study indicating that gut bacteria can ferment inulin into SCFAs and subsequently induce cholestatic HCC. They observed that HCC was microbiota-dependent in dysbiotic mice, but not in germ-free or antibiotics-treated mice. Dysbiosis and HCC occurred hand in hand in wild-type mice fed an inulin-enriched high-fat diet. Furthermore, intestinal SCFAs were markedly reduced by the pharmacologic inhibition of fermentation or eradication of fermenting bacteria by antibiotics such as metronidazole, and could potentially inhibit HCC’s development. Prolonged exposure to butyrate in the setting of dysbiosis caused hepatocyte injury, inflammation, and compensatory cell proliferation, and this synergetic effect eventually resulted in HCC.
2.2.3. Alcohol
Ethanol can be the product of glucose fermentation in the gut. The species that can produce endogenous ethanol include Candida species, Klebsiella pneumonia, and Saccharomyces cerevisiae [42]. NAFLD and obese patients have an increased abundance of ethanol-producing bacteria, and it has been found that serum ethanol was much higher in NAFLD and obese patients compared with their healthy counterparts, even without alcohol consumption [43,44,45]. Ethanol is a well-known carcinogen and acts synergistically with other risk factors in the liver. The elevated free radicals and reactive oxidative stress (ROS) derived from alcohol metabolism have been identified as essential mechanisms in hepatocarcinogenesis. ROS damages cellular macromolecules, impairs mitochondria, and forms lipid peroxides. It stimulates cytokine release, regulates immune processes, and promotes angiogenesis. ROS also affects DNA stability and impacts the processes of cell cycle arrest and apoptosis, which facilitate carcinogenesis [46].
2.3. GM Interplay with Chronic Liver Diseases and Cirrhosis Could Be Linked to Liver Cancer
Recent studies revealed that dysbiosis promotes chronic hepatitis B, alcohol-related liver disease (ALD), and NAFLD [47,48,49]. HCC develops as a result of chronic complex disease processes in the liver and persistent inflammation combined with immune dysregulation. About 80–90% of HCC cases occur in patients with cirrhosis. About one-third of patients with cirrhosis end up with HCC in their lifetime. Thus, chronic liver diseases should be considered as HCC precursors [15,50].
Advanced fibrosis and cirrhosis increase portal vein pressure. Portal hypertension with subsequent congestion of the intestinal venous circulation and increased exudation directly affects GM homeostasis, resulting in gut microbial dysbiosis [15]. Elevated endotoxin levels have been found in patients with cirrhosis, and there is a correlation between the severity of hepatic dysfunction and endotoxin concentrations in plasma [51]. Cirrhosis jeopardizes the capacity to clear LPS and its metabolites, and reciprocal dysbiosis impacts the development and progression of cirrhosis [52,53]. Tang et al. reported that IL2 and pro-inflammatory bacteria, such as phylum Proteobacteria, Fusobacterium, and Epsilonbacteraeota, were increased in patients with HBV cirrhosis, whereas Verrucomicrobia were decreased compared to the study’s healthy control group [54].
Complex network interactions between chronic liver inflammation and GM microbial dysbiosis may synergistically promote HCC in cirrhosis through the gut–liver axis [15]. Overall, the abundance changes in GM in chronic liver disease share many similarities with those in HCC.
2.3.1. NAFLD
The global prevalence of NAFLD is currently estimated to be about 25% of the population, with rapid growth globally driven by rising rates of obesity and type 2 diabetes [55]. NAFLD-related HCC showed increased levels of IL8, IL13, and chemokine ligands 3, 4, and 5, as well as activated circulating monocytes. The dysbiotic fingerprint in this cohort was very specific, characterized by a higher abundance of Enterobacteriaceae, Streptococcus, Bacteroides, and Ruminococcaceae and a reduced number of Akkermansia and Bifidobacterium. The abundance of Akkermansia and Bifidobacterium was inversely correlated with inflammation markers, such as calprotectin concentration. GM profile and systemic inflammation are significantly correlated, and can either promote or hinder hepatocarcinogenesis [56,57].
2.3.2. Alcohol-Related Liver Disease
It is well known that alcohol consumption jeopardizes intestinal epithelial integrity and permeability, which increases bacterial endotoxin and lipopolysaccharide penetration through the intestinal barrier and causes release into the portal vein. Previous studies have indicated a synergetic effect of GM and alcohol derivatives in hepatic inflammation, fibrosis, and tumorigenesis. Alcohol and its metabolic derivatives promote fibrogenesis and hepatocarcinogenesis via the interplay of the LPS/TLR4/MD-2/TNF-α/MAPK and TGF-β/Smad signaling pathways in ALD [58].
2.3.3. Chronic Viral Hepatitis
A pre-clinical model indicated that gut microbes contribute to immunity against HBV. The maturation of GM in adult mice stimulated liver immunity to reach rapid HBV clearance. On the other hand, the sterilization of GM using antibiotics also mitigated the capacity to rapidly clear HBV in adult mice [59]. A human study also concluded that a significantly altered GM environment in HBV-infected patients impacts disease progression [47]. Dysbiosis in patients with chronic HBV, specifically the abundance of Firmicutes, was lower, while Bacteroidetes was higher in patients with chronic HBV infection, liver cirrhosis, and HCC compared to the healthy controls. Meanwhile, an increase in glycan biosynthesis and metabolism-related genes was noted, as well as an increase in lipid metabolism-related genes in chronic HBV infection [60]. HCV- and HBV-infected patients had higher plasma levels of LPS, IL-6, sCD14 produced upon LPS activation of monocytes, and intestinal fatty acid binding protein, which was increased compared with the healthy controls. The severity of LPS-induced inflammation could predict the progression of cirrhosis [61]. Further investigations have suggested that dysbiosis impacted chronic HBV-infected patients by other mechanisms, such as the unmethylated CpG DNA-TLR9 pathway [62], the teichoic acid/peptidoglycan-TLR2 pathway, the flagellin-TLR5 pathway, and the MyD88-TRIF pathway [63,64,65,66].
3. The Gut Microbiome as a Non-Invasive Predictive Biomarker for Early Detection of HCC
As the overall prognosis of HCC remains poor, it is crucial to detect HCC in the early stages and start treatment in a timely manner to improve HCC survival rates. GM is thought to be a potential non-invasive diagnostic biomarker for HCC detection. Ni et al. [67] found an increase in pro-inflammatory gut bacteria and dysbiosis in patients with HCC compared with healthy controls, although the degree of dysbiosis was not associated with the stage of HCC. However, other investigators have reported associations between the stage of HCC and changes in GM and dysbiosis. A prospectively matched study on 30 patients with cirrhosis, published by Grat et al. [68] showed a higher level of fecal counts of Escherichia coli in patients with HCC, which predicted HCC presence based on Escherichia coli counts with a sensitivity of 66.7% and specificity of 73.3%. In patients with NAFLD cirrhosis, Ponziani et al. [57] concluded that the HCC group had a higher abundance of Bacteroides, Ruminococcaceae, Enterococcus, Phascolarctobacterium, and Oscillospira. Meanwhile, Bifidobacterium and Blautia were reduced compared to the controls.
Ren et al. [69] evaluated 486 fecal samples using a 16S rRNA sequencing MIseq platform, and showed that Actinobacteria was increased in early HCC compared to cirrhosis without HCC. Overall, 13 genera, including Gemmiger and Parabacteroides, were enriched in early HCC compared to cirrhosis without HCC. Compared with the healthy controls, 12 genera, including Alistipes, Phascolarctobacterium, and Ruminococcus, were reduced, while 6 genera, including Klebsiella and Haemophilus, were enriched in patients with early HCC. In addition, there was an increased abundance of LPS-producing bacteria and a reduced abundance of butyrate-producing bacteria in early HCC versus controls. Piñero et al. [70] studied 407 patients with a similar sequencing platform. The HCC group had a three-fold increase in the genus Erysipelotrichaceae and a fivefold decrease in the genus Leuconostocaceae compared to cirrhotic patients without HCC. There was a significant decrease in Fusobacterium and Dorea, of Lachnospiraceae, in patients with HCC, while Odoribacter and Butyricimonas were found to be more significantly enriched in HCC. The ratio of bactericides to prevotella was increased in HCC. The changes in bacterial abundance in association with HCC are summarized in Table 1.

Table 1.
“Favorable” and “less favorable” gut microbiota in HCC.
Komiyama and colleagues [71] reported that Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria serve as tumor-associated microbiota. In addition, Ruminococcus gnavus was found to be a signature taxon for patients with viral hepatitis-related HCC. Investigating metabolic syndrome-related HCC, Ni et al. [72] affirmed that there were differences in the GM between the subjects with and without HCC which could not be attributed to metabolic syndrome alone. They detected 49 bacterial genera and speculated that these might have a future role as an auxiliary tool for HCC diagnosis. Cho et al. [73] utilized circulating microbiome-based signatures with great AUC and accuracy of 0.875 and 79.8%, respectively, also speculating that they may serve as potential biomarkers for HCC detection.
The association between GM dysbiosis and HCC has received emerging attention, and it may potentially be interesting to utilize the gut microbiome profile as a predictive or diagnostic biomarker for HCC. However, many of the aforementioned studies were conducted on small sample sizes, and the study results might be significantly affected by ethnicity, country, living environment, and dietary preference. The results need to be validated using a large population with diverse backgrounds. The cost of 16S rDNA sequencing is another factor limiting the clinical application, and cost-effectiveness needs to be considered in clinical practice.
4. The Role of the Gut Microbiome in HCC Therapy and Prevention
The importance of the gut microbiome reaches far beyond its impact on the development and progression of chronic liver disease and HCC. There is mounting evidence that it also plays a role in the response to treatment of HCC, affecting the effectiveness of chemo- and immunotherapy. It is important to clearly understand the implications of pharmaco-microbiomics affecting cancer treatment agents’ ability to potentially modify the GM in order to improve treatment efficacy and reduce toxicity. In-depth insight into the prokaryotic metabolism of chemotherapeutic agents is required to understand the critical role of the GM in developing personalized cancer therapy strategies [74].
4.1. The Gut Microbiome May Influence the Efficacy of Surgery and Radiotherapy
There are emerging data on the influence of GM on the response of HCC to surgery or radiotherapy.
Li and colleagues [75] reported a study on patients receiving radiotherapy, speculating that disruption and modulation of the GM may impact the radiosensitivity of HCC. Dysbiosis prevents antigen presentation and inhibits T-cell activity through the cGAS–STING–IFN-I pathway. The stimulator of interferon genes (STING) acts as an important signal adaptor, and is responsible for cytosolic DNA-dependent innate immunity [76].
Wang and colleagues [77] prospectively explored the effect of the GM in 24 patients with HCC undergoing radiotherapy using 16S rRNA sequencing. They also suggested that resistance to radiotherapy could be affected by an altered cGAS–STING–IFN-I pathway, and identified cyclic-di-AMP of prominent gut bacteria as a strong STING agonist. They concluded that these pathways may represent a potential target to predict and potentially modulate the response to radiotherapy in HCC.
A small study on 30 patients with HBV-related HCC who underwent extended hepatectomy investigated the influence of GM on liver failure following surgery by sequencing 16S rRNA. This showed that the enrichment of bacterial taxa such as Allisonella, Bacteroides, Faecalibacterium, GCA-900066575, Helicobacter, Inquilinus, IS-44, Methylobacterium–Methylorubrum, Mycobacterium, Pantoea, and IS-44 was more prominent in patients who developed liver failure after surgery, while taxa such as Catabacter, Papillibacter, Scardovia, Senegalimassilia, and Turicibacter were significantly enriched in patients without postoperative liver failure. Interestingly, there was no difference between pre- and post-surgery GM composition, while markers of liver function (i.e., international normalised ratio INR, bilirubin, albumin, prealbumin) were strongly correlated with the composition of the gut microbiome [78]. The composition of the gut microbiome was distinct in patients of different age groups, which may play an additional role in decisions regarding surgery [78,79]. Furthermore, the data suggest that an abundance of Klebsiella can be a potential indicator of post-hepatectomy liver failure due to the altered levels of 3-methyl-2-oxobutanoic acid in the BCAA pathway. The investigators speculated that 3-methyl-2-oxobutanoic acid may have an additional value in targeted treatments for post-hepatectomy liver failure in patients with HCC [80].
Despite the fact that these data are preliminary, they nonetheless present potential avenues to further explore and use the GM as a predictor of the potential efficacy and outcomes of surgery and radiotherapy, as well as therapeutic alterations, to improve survival in patients with HCC.
4.2. Chemotherapy
Both in vivo and in vitro studies have documented that the gut flora are closely linked to the pharmacological effects of different chemotherapy agents, including 5-fluorouracil (5-FU), cyclophosphamide, irinotecan, oxaliplatin, gemcitabine, and methotrexate, as well as novel targeted immunotherapies, including anti-programmed cell death protein 1 (PD-1) and anti-CLTA-4 therapies. The GM modulates these agents via the so-called ‘TIMER’ mechanistic framework: translocation, immunomodulation, metabolism, enzymatic degradation, and reduced diversity, accompanied by ecological changes [74].
Yuan et al. [81] compared the size of colorectal cancer (CRC) tumors in mice treated with 5-FU depending on the analyzed gut microbiota, and found that antibiotic administration reduced the antitumor efficacy of 5-FU; the addition of probiotics did not significantly increase the efficacy of 5-FU treatment. Analysis showed that antibiotics significantly reduced the biodiversity and composition of the GM, with an increased abundance of the pathogenic bacteria Escherichia shigella and Enterobacter, as well as Blautia, Lachnospiracea_NK4 A136, Bacteroides, Odoribacter, and Mucispirillum, compared to the control group. Additionally, the functional capacity analysis of the gut microbiota showed that genes involved in amino acid metabolism, replication, and repair, as well as nucleotide metabolism, were expressed at a much lower rate in the group receiving antibiotics and 5-FU compared to the other groups. The authors concluded that these results strongly suggest that antibiotics affect the gut microbiota, which contributed to the reduction in the antitumor efficacy of 5-FU in pre-clinical studies.
Currently, there are only sparse data on the influence of the GM on the efficacy of chemotherapy in HCC. Wu et al. [82] reported that ZnCM-SD, a polyvinylpyrrolidone-based solid dispersion of Zn (II)-curcumin, may work as a safe novel chemosensitizer for doxorubicin in the treatment of HCC, as the zinc homeostasis was affected by dysbiosis and improved by FMT.
4.3. The Gut Microbiome and Immunotherapy
Tumor immune escape is an essential mechanism of tumorigenesis. It is well-documented that malignant cells are capable of escaping the immune system [83]. The most critical interactions in tumor immune escape are thought to be between the cancer cells, the cancer microenvironment, and the immune status of the host. GM composition and metabolites affect tumor immune escape mechanisms [84]. Tumor immunotherapy supports the restoration of the host’s antitumor immunity with the goal of controlling and eliminating cancer. Immunotherapy includes the application of cytokines, adoptive immune cells, and immune checkpoint inhibitors [85]. GM affects the function of the host’s immune cells, including the release of chemokines and cytokines. Its potential influence on the success or failure of immunotherapy seems obvious, but is not fully understood. The gut microbiome can be altered by antibiotic cocktails, probiotics, and fecal microbiota transplant (FMT), which may all be potential adjuvants to tumor immunotherapy [86,87].
Checkpoint immunotherapy has enabled breakthroughs in the treatment of solid malignancies. These drugs effectively inhibit tumor immune escape by targeting programmed cell death 1 (PD-1) and its ligand (PD-L1), lymphocyte-activating gene-3 (LAG3), and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) [88]. Overall, responses to checkpoint immune therapy are reported to be heterogeneous and not robust, with objective response rates of only 10–30% [88].
For years, sorafenib, an oral multi-kinase inhibitor, was the only systemic therapeutic option for advanced-stage HCC, with an objective response rate as low as 5%. The newer checkpoint blockade immunotherapy agents targeting PD-1 show promising efficacy for treating advanced-stage HCC. An objective response rate of about 20% using anti-PD-1 immunotherapy in sorafenib refractory HCC was reported in two multicenter studies. Atezolizumab combined with bevacizumab is now recommended as a first-line systemic therapy. The reported median progression-free survival (PFS) was 6.8 months, and the median overall survival (OS) was 19.2 months for patients receiving this combination [89,90]. Chemotherapy with gemcitabine and cisplatin is considered the primary treatment for unresectable cholangiocarcinoma; it resulted in a median PFS of 8.0 months and a median OS of 11.7 months. A subsequent line of treatment with lenvatinib combined with pembrolizumab could be considered after disease progression with chemotherapy [91].
As the gut microbiome promotes carcinogenesis and mediates the host immune response, one is compelled to assume that it would have a role in supporting or opposing the efficacy of immunotherapy agents. Inflammation, dysbiosis, DNA damage, and genotoxins are all influencers of oncogenesis. As documented in previous studies, the gut microbiome promotes or inhibits antitumor immunity through various host immune cells, such as macrophages, dendritic cells, CD8-T cells, B cells, Treg cells, TH1, TH17 cells, natural killer T cells, intraepithelial lymphocytes, and mucosal-associated invariant T cells [92], as well as through cytokine production, including IL-2,6,8,10,12,17, IFN-γ, and granzyme B [93,94]. The data suggest that the efficacy of immunotherapy is linked to GM composition. Studies have shown that the oral and intestinal microbiomes of individuals differ in diversity and composition between treatment responders versus non-responders. For example, a microbiome composition with abundant tumor-infiltrating lymphocytes combined with a low number of myeloid-derived suppressor cells seems to promote a response to immunotherapy [95]. There is strong evidence from pre-clinical tumor model studies [96,97,98] and patient cohorts [99,100] that an altered GM environment can increase or hamper response to immunotherapy and affect cancer-therapy-related toxicity. Certain bacterial taxa, such as Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia phyla, were found to be more prominent in responders to immunotherapy [93,95].
Routy et al. [99] showed that an environment enriched with Akkermansia muciniphila was linked to a favorable clinical response to anti-PD-1 immunotherapy in patients with different epithelial cancers, including non-small cell lung cancer, melanoma, and renal cell cancer. An abundance of Faecalibacterium and Ruminococcaceae [101], as well as Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium [102], could be seen in patients with advanced melanoma who showed a favorable response to anti-PD-1 immunotherapy. In one study, Gopalakrishnan et al. [93] showed that patients who responded to treatment had higher within-sample diversity in their intestinal microbiome, as well as a higher abundance of bacteria from the Ruminococcaceae family and the Faecalibacterium genus. By contrast, non-responders had a lower within-sample diversity of their intestinal microbiome and had a higher abundance of members of the Bacteroidales species.
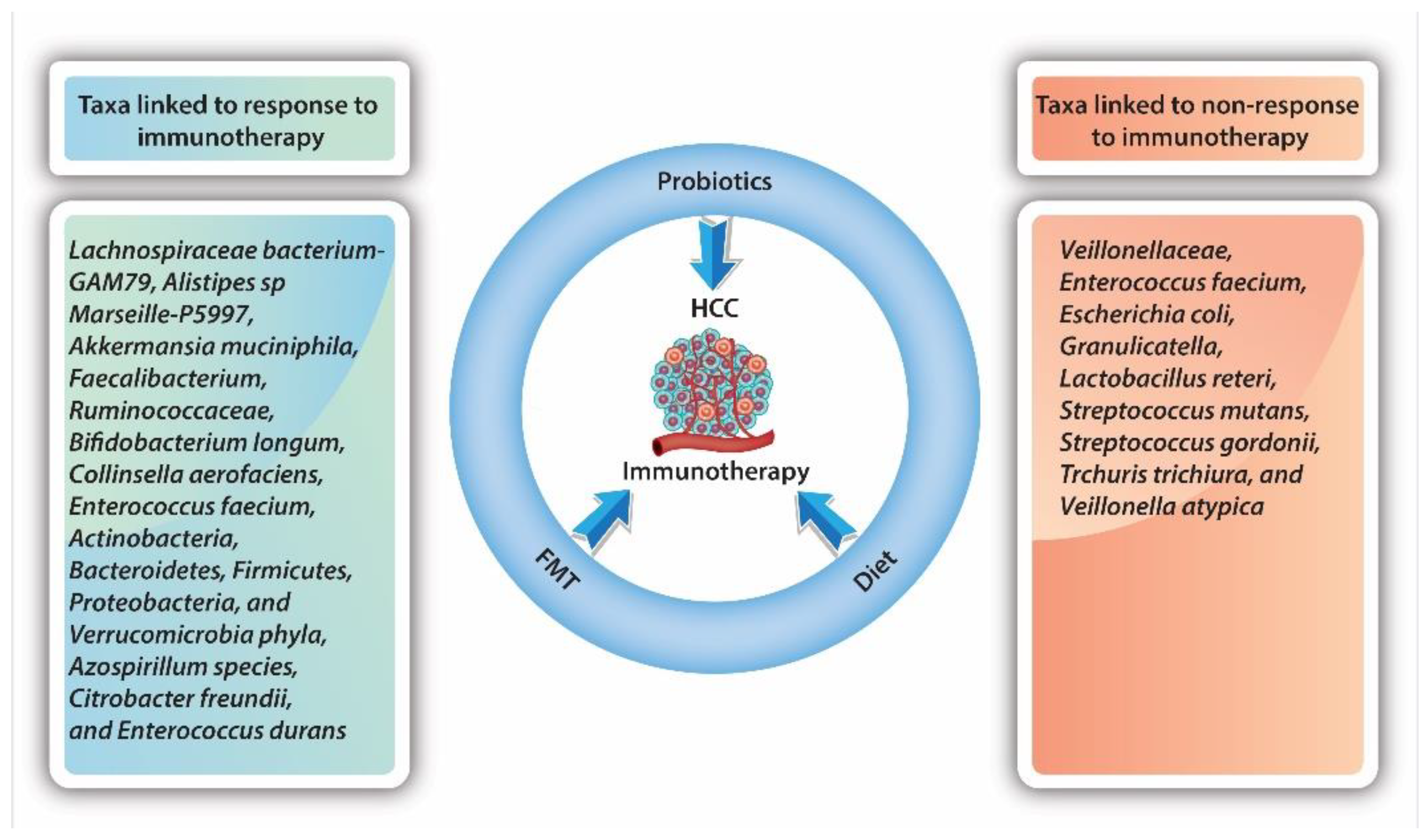
As the GM plays a vital role in the development and progression of HCC, it is not surprising that there is growing interest in identifying bacterial taxa which may promote or inhibit response to HCC therapy and specific immunotherapy (Figure 2). As only a small fraction of patients will benefit from immunotherapy, subsequently identifying those potential responders is crucial. At present, the data are diverse, but remain conflicting.
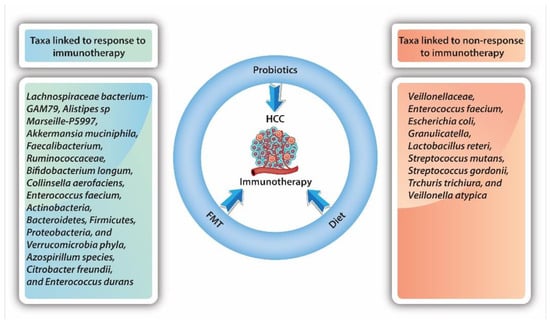
Figure 2.
Influence of bacterial taxa on response to immunotherapy.
Mao et al. [96] showed that, in patients with advanced hepatobiliary cancers receiving anti-PD-1 treatment, 74 unique taxa that are particularly associated with energy metabolism were significantly higher in responders to immunotherapy. In comparison, 40 unique taxa mainly associated with amino acid metabolism were significantly higher in non-responders. They also found that immunotherapy-related adverse events were influenced by GM diversity and the abundance or lack of certain taxa [96]. A higher abundance of Lachnospiraceae bacterium-GAM79 and Alistipes sp. Marseille-P5997 was associated with longer OS and PFS, while an abundance of Ruminococcus calidus and Erysipelotichaceae bacterium-GAM147 was associated with increased PFS only. On the other hand, taxa such as Veillonellaceae were found to be enriched in non-responders to immunotherapy.
Nivolumab has shown efficacy in the treatment of various cancers, such as melanoma and squamous cell carcinoma of the head and neck. In 2017, it was approved as a second-line agent for advanced HCC after sorafenib’s failure. The most recent evidence suggests that dysbiosis plays an important role as a prognostic marker for the response to nivolumab in the treatment of HCC. A small study published by Chung et al. [103] investigated the GM in fecal samples from eight patients with advanced HCC receiving nivolumab. In patients with HCC, progression data on nivolumab samples were collected at the time of progression. In patients who showed a good response to nivolumab, samples were obtained 5–7 months later. Metagenomic data from 16S ribosomal RNA sequencing were analyzed using CLC Genomics Workbench. The investigators found significant differences between the phylogenetic diversity and overall composition of the bacterial community in treatment responders and non-responders in general, but these were not associated with the immunotherapy drug being used. Specific taxa were linked to therapeutic responses: Dialister pneumosintes, Enterococcus faecium, Escherichia coli, Granulicatella, Lactobacillus reteri, Streptococcus mutans, Streptococcus gordonii, Trchuris trichiura, and Veillonella atypica were linked to non-response, while Azospirillum species, Citrobacter freundii, and Enterococcus durans were linked to response. In addition, a skewed Firmicutes-to-Bacteroidetes ratio of less than 0.5 or greater than 1.5 and a low Prevotella-to-Bacteroides ratio were good predictors of non-response, while Akkermansia species were predictive of a favorable treatment response [103]. Peng et al. [104] showed that bacteria with SCFA production, such as Eubacterium, Lactobacillus, and Streptococcus, had a positive association with the response to anti-PD-1 immunotherapy in patients with different GI cancers (i.e., CRC, stomach, esophagus). Akkermansia and Lactobacillus were, in previous studies, positively associated with response to anti-PD-1 immunotherapy in melanoma. Peng and colleagues found that in patients with GI cancers, a higher abundance of Akkermansia (p = 0.031), but not Lactobacillus (p = 0.56), was associated with a positive response to anti-PD-1 or PD-L1 immunotherapy [104].
Recent data have reported a link between different GI cancers and the Firmicutes-to-Bacteroidetes ratio, with the highest Firmicutes-to-Bacteroidetes ratio seen in early gastric cancer [105]. A substantially reduced Firmicutes-to-Bacteroidetes ratio was reported in cirrhosis and HCC [106]. The investigators also speculate that these changes in the GM are physiologic in nature and not related to cancer, as an increased Firmicutes-to-Bacteroidetes ratio is associated with older age and obesity [107].
In summary, these studies support the gut microbiome’s influence on the response or non-response to immunotherapy with anti-PD-1 and PD-L1 in at least a subset of patients with different cancers, including HCC. This should be considered as a potential marker for immune-checkpoint blockade response.
4.4. The Value of Probiotics and Fecal Microbiota Transplantation in HCC Treatment and Prevention
Accumulating evidence from pre-clinical studies suggests the intestinal microbiota–liver axis is a promising target, not only for preventing the progression of chronic liver disease, but also for developing HCC. In 1991, bacteria were first introduced as part of cancer therapy [108], and interest in this approach has steadily grown. At present, the GM is being used in three ways: as an oral probiotic, as a type of dietary intervention, and in fecal microbiota transplants (FMTs). Colorectal cancer (CRC) is the prime example in which probiotic supplements seem to have antitumor effects, as has been shown in pre-clinical and clinical studies. Clostridium butyricum and Bacillus subtilis have been proven to slow CRC progression in mice [109,110]. Supplementation with Lactobacillus johnsonii reduced the CRC recurrence rate after surgery. A prospective clinical study following patients with CRC over 12 years showed that patients regularly consuming yogurt containing Streptococcus thermophilus and Lactobacillus bulgaricus had significantly less advanced severe CRC [111]. However, it remains largely uncertain whether probiotic supplements enhance or hamper the response to cancer and immunotherapy, as the current data show conflicting results [112]. The negative effects of probiotics in response to immunotherapy have also been reported, as shown by Spencer et al., in patients with melanoma [113].
Suez et al. [114] reported that the use of probiotics after antibiotics led to delayed and incomplete GM reconstitution; FMT, however, induced a rapid and almost complete recovery within days. Subsequently, it was concluded that the benefits of post-antibiotic probiotics may be counter-acted by a delayed gut mucosal recovery. A future goal could be the development of personalized probiotic supplements, as well as FMT approaches, to achieve mucosa protection without compromising microbiome recolonization.
Dietary adjustments can rapidly affect the GM composition, and might be a simple yet safe approach to influence GM in patients receiving cancer treatment and immunotherapy [115,116]. Intake of a high-fiber diet was shown to be associated with significantly improved PFS in 128 patients with melanoma receiving immunotherapy, and interestingly, the largest benefit was seen in those not receiving probiotic supplements. The investigators also showed a reduced response to anti–PD-1 immunotherapy when mice were fed with a low-fiber diet or probiotics; they speculated that this was caused by the lack of interferon-γ-positive cytotoxic T cells [113].
The goal of fecal microbiota transplantation (FMT) is to restore the physiological gut microbiome in individuals with gut dysbiosis. FMT preparations can be administered orally with lyophilized pills or by endoscopy procedures (colonoscopy or gastroscopy). This procedure was initially used to treat clostridium difficile infection resistant to traditional therapy. The use of FMT has been widely reported in patients with many different diseases, including malignancies. An abundance of data have been reported in pre-clinical studies and clinical trials [99,101,102]. In mice fed with certain bacteria taxa or after the application of FMT, an increased response to immunotherapy was observed. Pre-clinical studies showed that FMT to germ-free mice from individuals that responded to anti-PD1 therapy led to increased antitumor immunity compared with mice who received FMT from non-responder donors. Increased levels of antitumor CD8+ T cells were found in mice that received FMT from responders, whereas higher levels of immunosuppressive CD4+ T cells were seen in those that received FMT from non-responders. The investigator concluded that increased CD8+ T cell activation and intra-tumor lymphocyte infiltration help to overcome resistance to anti-PD-1 therapy in melanoma [117,118].
To date, there have been promising results showing that resistance to immunotherapy can be overturned. Two clinical trials found that FMT from checkpoint immunotherapy responders combined with anti-PD-1 therapy overcame resistance to PD-1 blockade in patients with melanoma [117,118]. Another phase I clinical trial showed that (NCT03353402), in patients with metastatic melanoma who developed resistance to anti-PD-1 immunotherapy, application of FMT led to a favorable response in 3 of the 10 participants [117]. Another clinical trial (NCT03341143) in 15 patients with melanoma with non-response to anti-PD-1 therapy reported a partial response in 3 patients after the application of FMT combined with pembrolizumab, and stable disease in 3 patients for more than 12 months. Multiple clinical trials investigating the application of FMT to modulate the efficacy of, and adverse events related to, immunotherapy in various cancers (e.g., prostate, lung, renal cell, melanoma) are underway, although none are currently focusing on HCC.
As the gut microbiome modulates the response to cancer immunotherapy, it may become the next therapeutic target. The identification of bacterial taxa that directly or indirectly induce antitumor activities is critical to developing a microbiome-based treatment that may help to improve the overall response rate of immunotherapy. Differential bacteria taxa in different response groups can identify clinically relevant microbial species as potential biomarkers and, thus, help to guide the development of new therapies (Figure 2).
5. Conclusions and Future Directions
The crosstalk between GM dysbiosis and HCC has received increasing attention in recent decades, and emerging studies have dissected the complex immunological and metabolic network and elaborated on the signaling pathways and other pathophysiological effects of dysbiosis on HCC tumorigenesis. The more we understand the molecular mechanisms behind the development and proliferation of HCC affected by the gut microbiome, the more novel diagnostic and therapeutic strategies can be explored. Recent studies have indicated that compositional dysbiosis occurs in patients with HCC, and, therefore, GM has potential as a diagnostic biomarker for the early diagnosis of HCC. However, these results need to be validated in larger and more heterogeneous populations. Treatment strategies for HCC have come a long way, and further research on GM is a step forward in the quest to combat HCC and improve the outcomes for these patients. The synergistic role of GM in HCC immunotherapy promises therapeutic potential. Thus, future studies are warranted to validate microbial signatures in HCC and use them to our advantage.
Author Contributions
C.X.: original draft preparation, writing, reviewing, editing, and figure and tablet creation. C.P.: conceptualizing ideas, literature review, writing, reviewing, editing, and tablet creation. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [PubMed]
- Rattan, P.; Minacapelli, C.D.; Rustgi, V. The Microbiome and Hepatocellular Carcinoma. Liver Transplant. 2020, 26, 1316–1327. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Trends in Liver Cancer Mortality among Adults Aged 25 and over in the United States, 2000–2016. NCHS Data Brief 2018, 314, 1–8. [Google Scholar]
- Vogel, A.; Saborowski, A. Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat. Rev. 2020, 82, 101946. [Google Scholar] [CrossRef]
- Vitale, A.; Trevisani, F.; Farinati, F.; Cillo, U. Treatment of Hepatocellular Carcinoma in the Precision Medicine Era: From Treatment Stage Migration to Therapeutic Hierarchy. Hepatology 2020, 72, 2206–2218. [Google Scholar] [CrossRef]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef]
- Zafari, N.; Velayati, M.; Fahim, M.; Maftouh, M.; Pourali, G.; Khazaei, M.; Nassiri, M.; Hassanian, S.M.; Ghayour-Mobarhan, M.; Ferns, G.A.; et al. Role of gut bacterial and non-bacterial microbiota in alcohol-associated liver disease: Molecular mechanisms, biomarkers, and therapeutic prospective. Life Sci. 2022, 305, 120760. [Google Scholar] [CrossRef]
- Ciocan, D.; Spatz, M.; Trainel, N.; Hardonnière, K.; Domenichini, S.; Mercier-Nomé, F.; Desmons, A.; Humbert, L.; Durand, S.; Kroemer, G.; et al. Modulation of the Bile Acid Enterohepatic Cycle by Intestinal Microbiota Alleviates Alcohol Liver Disease. Cells 2022, 11, 968. [Google Scholar] [CrossRef]
- Kassa, Y.; Million, Y.; Gedefie, A.; Moges, F. Alteration of Gut Microbiota and Its Impact on Immune Response in Patients with Chronic HBV Infection: A Review. Infect. Drug Resist. 2021, 14, 2571–2578. [Google Scholar] [CrossRef]
- Xie, C.; Halegoua-DeMarzio, D. Role of Probiotics in Non-alcoholic Fatty Liver Disease: Does Gut Microbiota Matter? Nutrients 2019, 11, 2837. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M.; Knudsen, C.; Beaumont, M.; Rodriguez, J.; Neyrinck, A.M.; Bindels, L.B. Contribution of the gut microbiota to the regulation of host metabolism and energy balance: A focus on the gut–liver axis. Proc. Nutr. Soc. 2019, 78, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Núñez, G. Regulation of the Immune System by the Resident Intestinal Bacteria. Gastroenterology 2014, 146, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Etienne-Mesmin, L.; Gewirtz, A.T. Microbiota-liver axis in hepatic disease. Hepatology 2014, 59, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-W.; Chen, X.-H.; Ren, Z.-G.; Zheng, S.-S. Gut microbial dysbiosis associates hepatocellular carcinoma via the gut-liver axis. Hepatobiliary Pancreat. Dis. Int. 2018, 18, 19–27. [Google Scholar] [CrossRef]
- Miura, K.; Ohnishi, H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 7381–7391. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef]
- Yu, L.-X.; Yan, H.-X.; Liu, Q.; Yang, W.; Wu, H.-P.; Dong, W.; Tang, L.; Lin, Y.; He, Y.-Q.; Zou, S.-S.; et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 2010, 52, 1322–1333. [Google Scholar] [CrossRef]
- Schneider, K.M.; Mohs, A.; Gui, W.; Galvez, E.J.C.; Candels, L.S.; Hoenicke, L.; Muthukumarasamy, U.; Holland, C.H.; Elfers, C.; Kilic, K.; et al. Imbalanced gut microbiota fuels hepatocellular carcinoma development by shaping the hepatic inflammatory microenvironment. Nat. Commun. 2022, 13, 3964. [Google Scholar] [CrossRef]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE2-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Wu, L.; Feng, J.; Li, J.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. The gut microbiome-bile acid axis in hepatocarcinogenesis. Biomed. Pharmacother. 2021, 133, 111036. [Google Scholar] [CrossRef]
- Galle, P.R.; Theilmann, L.; Raedsch, R.; Otto, G.; Stiehl, A. Ursodeoxycholate reduces hepatotoxicity of bile salts in primary human hepatocytes. Hepatology 1990, 12, 486–491. [Google Scholar] [CrossRef]
- Jia, B.; Jeon, C.O. Promotion and induction of liver cancer by gut microbiome-mediated modulation of bile acids. PLoS Pathog. 2019, 15, e1007954. [Google Scholar] [CrossRef]
- Wang, X.; Fu, X.; Van Ness, C.; Meng, Z.; Ma, X.; Huang, W. Bile Acid Receptors and Liver Cancer. Curr. Pathobiol. Rep. 2013, 1, 29–35. [Google Scholar] [CrossRef]
- Chávez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 2017, 152, 1679–1694.e3. [Google Scholar] [CrossRef]
- Arab, J.P.; Karpen, S.J.; Dawson, P.A.; Arrese, M.; Trauner, M. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 2017, 65, 350–362. [Google Scholar] [CrossRef]
- Liu, H.; Pathak, P.; Boehme, S.; Chiang, J. Cholesterol 7α-hydroxylase protects the liver from inflammation and fibrosis by maintaining cholesterol homeostasis. J. Lipid Res. 2016, 57, 1831–1844. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Liu, Q.; Harnish, D.C. Farnesoid X receptor agonist WAY-362450 attenuates liver inflammation and fibrosis in murine model of non-alcoholic steatohepatitis. J. Hepatol. 2009, 51, 380–388. [Google Scholar] [CrossRef]
- Yamada, S.; Takashina, Y.; Watanabe, M.; Nagamine, R.; Saito, Y.; Kamada, N.; Saito, H. Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice. Oncotarget 2018, 9, 9925–9939. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Ke, L.; Li, Q.; Dang, X.; Shen, S.; Shen, J.; Li, S.; Liang, L.; Peng, B.; Kuang, M.; et al. Abnormal bile acid-microbiota crosstalk promotes the development of hepatocellular carcinoma. Hepatol. Int. 2022, 16, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Knisely, A.S.; Strautnieks, S.S.; Meier, Y.; Stieger, B.; Byrne, J.A.; Portmann, B.C.; Bull, L.N.; Pawlikowska, L.; Bilezikçi, B.; Özçay, F.; et al. Hepatocellular carcinoma in ten children under five years of age with bile salt export pump deficiency. Hepatology 2006, 44, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Hara, E. Gut-liver axis-mediated mechanism of liver cancer: A special focus on the role of gut microbiota. Cancer Sci. 2021, 112, 4433–4443. [Google Scholar] [CrossRef]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef]
- Kimura, I.; Inoue, D.; Maeda, T.; Hara, T.; Ichimura, A.; Miyauchi, S.; Kobayashi, M.; Hirasawa, A.; Tsujimoto, G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl. Acad. Sci. USA 2011, 108, 8030–8035. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Usami, M.; Kishimoto, K.; Ohata, A.; Miyoshi, M.; Aoyama, M.; Fueda, Y.; Kotani, J. Butyrate and trichostatin A attenuate nuclear factor kappaB activation and tumor necrosis factor alpha secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr. Res. 2008, 28, 321–328. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Singh, V.; San Yeoh, B.; Chassaing, B.; Xiao, X.; Saha, P.; Olvera, R.A.; Lapek, J.D., Jr.; Zhang, L.; Wang, W.B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694.e22. [Google Scholar] [CrossRef]
- Bayoumy, A.B.; Mulder, C.J.J.; Mol, J.J.; Tushuizen, M.E. Gut fermentation syndrome: A systematic review of case reports. United Eur. Gastroenterol. J. 2021, 9, 332–342. [Google Scholar] [CrossRef]
- Satapathy, S.K.M.; Banerjee, P.; Pierre, J.F.; Higgins, D.; Dutta, S.; Heda, R.B.; Khan, S.D.B.; Mupparaju, V.K.; Mas, V.; Nair, S.; et al. Characterization of Gut Microbiome in Liver Transplant Recipients with Nonalcoholic Steatohepatitis. Transplant. Direct 2020, 6, e625. [Google Scholar] [CrossRef]
- Nair, S.; Cope, K.; Terence, R.H.; Diehl, A.M. Obesity and female gender increase breath ethanol concentration: Potential implications for the pathogenesis of nonalcoholic steatohepatitis. Am. J. Gastroenterol. 2001, 96, 1200–1204. [Google Scholar] [CrossRef]
- Volynets, V.; Küper, M.A.; Strahl, S.; Maier, I.B.; Spruss, A.; Wagnerberger, S.; Königsrainer, A.; Bischoff, S.C.; Bergheim, I. Nutrition, Intestinal Permeability, and Blood Ethanol Levels Are Altered in Patients with Nonalcoholic Fatty Liver Disease (NAFLD). Dig. Dis. Sci. 2012, 57, 1932–1941. [Google Scholar] [CrossRef]
- Pocha, C.; Xie, C. Hepatocellular carcinoma in alcoholic and non-alcoholic fatty liver disease-one of a kind or two different enemies? Transl. Gastroenterol. Hepatol. 2019, 4, 72. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Zhang, X.; Liu, J.; Zhang, Q.; Zhao, Y.; Peng, J.; Feng, Q.; Dai, J.; Sun, S.; et al. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front. Microbiol. 2017, 8, 2222. [Google Scholar] [CrossRef]
- Shen, F.; Zheng, R.-D.; Sun, X.-Q.; Ding, W.-J.; Wang, X.-Y.; Fan, J.-G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2017, 16, 375–381. [Google Scholar] [CrossRef]
- Mutlu, E.A.; Gillevet, P.M.; Rangwala, H.; Sikaroodi, M.; Naqvi, A.; Engen, P.A.; Kwasny, M.; Lau, C.K.; Keshavarzian, A. Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G966–G978. [Google Scholar] [CrossRef]
- El-Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef]
- Lin, R.S.; Lee, F.Y.; Lee, S.D.; Tsai, Y.T.; Lin, H.C.; Lu, R.H.; Hsu, W.C.; Huang, C.C.; Wang, S.S.; Lo, K.J. Endotoxemia in patients with chronic liver diseases: Relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J. Hepatol. 1995, 22, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhou, H.; Xiang, Y.; Cui, F. The diagnostic potential of gut microbiome for early hepatitis B virus-related hepatocellular carcinoma. Eur. J. Gastroenterol. Hepatol. 2021, 33, e167–e175. [Google Scholar] [CrossRef]
- Younossi, Z.; Aggarwal, P.; Shrestha, I.; Fernandes, J.; Johansen, P.; Augusto, M.; Nair, S. The burden of non-alcoholic steatohepatitis: A systematic review of health-related quality of life and patient-reported outcomes. JHEP Rep. 2022, 4, 100525. [Google Scholar] [CrossRef]
- Cassano, M.; Dufour, J.F. Inflammation and Microbiota Fingerprint: Delphi’s Oracle for Nonalcoholic Fatty Liver Disease-Related Hepatocellular Carcinoma? Hepatology 2019, 69, 12–15. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Nicoletti, A.; Gasbarrini, A.; Pompili, M. Diagnostic and therapeutic potential of the gut microbiota in patients with early hepatocellular carcinoma. Ther. Adv. Med. Oncol. 2019, 11, 1758835919848184. [Google Scholar] [CrossRef]
- Boye, A.; Zou, Y.H.; Yang, Y. Metabolic derivatives of alcohol and the molecular culprits of fibro-hepatocarcinogenesis: Allies or enemies? World J. Gastroenterol. 2016, 22, 50–71. [Google Scholar] [CrossRef]
- Chou, H.-H.; Chien, W.-H.; Wu, L.-L.; Cheng, C.-H.; Chung, C.-H.; Horng, J.-H.; Ni, Y.-H.; Tseng, H.-T.; Wu, D.; Lu, X.; et al. Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc. Natl. Acad. Sci. USA 2015, 112, 2175–2180. [Google Scholar] [CrossRef]
- Zeng, Y.; Chen, S.; Fu, Y.; Wu, W.; Chen, T.; Chen, J.; Yang, B.; Ou, Q. Gut microbiota dysbiosis in patients with hepatitis B virus-induced chronic liver disease covering chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. J. Viral Hepat. 2020, 27, 143–155. [Google Scholar] [CrossRef]
- Sandler, N.G.; Koh, C.; Roque, A.; Eccleston, J.L.; Siegel, R.B.; Demino, M.; Kleiner, D.; Deeks, S.G.; Liang, T.J.; Heller, T.; et al. Host Response to Translocated Microbial Products Predicts Outcomes of Patients with HBV or HCV Infection. Gastroenterology 2011, 141, 1220–1230.e3. [Google Scholar] [CrossRef]
- Gao, K.; Liu, L.; Wang, H. Advances in immunomodulation of microbial unmethylated CpG DNA on animal intestinal tract—A review. Acta Microbiol. Sin. 2015, 55, 543–550. [Google Scholar]
- Cinar, M.U.; Islam, M.A.; Pröll, M.; Kocamis, H.; Tholen, E.; Tesfaye, D.; Looft, C.; Schellander, K.; Uddin, M.J. Evaluation of suitable reference genes for gene expression studies in porcine PBMCs in response to LPS and LTA. BMC Res. Notes 2013, 6, 56. [Google Scholar] [CrossRef]
- Lemaitre, B.; Girardin, S.E. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat. Rev. Genet. 2013, 11, 365–369. [Google Scholar] [CrossRef]
- Yang, R.; Xu, Y.; Dai, Z.; Lin, X.; Wang, H. The Immunologic Role of Gut Microbiota in Patients with Chronic HBV Infection. J. Immunol. Res. 2018, 2018, 2361963. [Google Scholar] [CrossRef]
- Ignacio, A.; Morales, C.I.; Câmara, N.O.S.; Almeida, R.R. Innate Sensing of the Gut Microbiota: Modulation of Inflammatory and Autoimmune Diseases. Front. Immunol. 2016, 7, 54. [Google Scholar] [CrossRef]
- Ni, J.; Huang, R.; Zhou, H.; Xu, X.; Li, Y.; Cao, P.; Zhong, K.; Ge, M.; Chen, X.; Hou, B.; et al. Analysis of the Relationship Between the Degree of Dysbiosis in Gut Microbiota and Prognosis at Different Stages of Primary Hepatocellular Carcinoma. Front. Microbiol. 2019, 10, 1458. [Google Scholar] [CrossRef]
- Grąt, M.; Wronka, K.M.; Krasnodębski, M.; Masior, L.; Lewandowski, Z.; Kosińska, I.; Grąt, K.; Stypułkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of Gut Microbiota Associated with the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2018, 68, 1014–1023. [Google Scholar] [CrossRef]
- Piñero, F.; Vazquez, M.; Baré, P.; Rohr, C.; Mendizabal, M.; Sciara, M.; Alonso, C.; Fay, F.; Silva, M. A different gut microbiome linked to inflammation found in cirrhotic patients with and without hepatocellular carcinoma. Ann. Hepatol. 2019, 18, 480–487. [Google Scholar] [CrossRef]
- Komiyama, S.; Yamada, T.; Takemura, N.; Kokudo, N.; Hase, K.; Kawamura, Y.I. Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci. Rep. 2021, 11, 10589. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Fu, C.; Huang, R.; Li, Z.; Li, S.; Cao, P.; Zhong, K.; Ge, M.; Gao, Y. Metabolic syndrome cannot mask the changes of faecal microbiota compositions caused by primary hepatocellular carcinoma. Lett. Appl. Microbiol. 2021, 73, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Leem, S.; Kim, S.A.; Yang, J.; Bin Lee, Y.; Kim, S.S.; Cheong, J.Y.; Cho, S.W.; Kim, J.W.; Kim, S.-M.; et al. Circulating Microbiota-Based Metagenomic Signature for Detection of Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 7536. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Hong, W.; Wang, B.; Chen, Y.; Yang, P.; Zhou, J.; Fan, J.; Zeng, Z.; Du, S. Gut microbiota modulate radiotherapy-associated antitumor immune responses against hepatocellular carcinoma via STING signaling. Gut Microbes 2022, 14, 2119055. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Li, Z.; Ke, X.; Zuo, D.; Wang, Z.; Fang, F.; Bo, L. New Insights into the Relationship between Gut Microbiota and Radiotherapy for Cancer. Nutrients 2022, 15, 48. [Google Scholar] [CrossRef]
- Peng, Y.-C.; Xu, J.-X.; Zeng, C.-F.; Zhao, X.-H.; Li, L.-Q.; Qi, L.-N. Gut microbiome dysbiosis in patients with hepatitis B virus-related hepatocellular carcinoma after extended hepatectomy liver failure. Ann. Transl. Med. 2022, 10, 549. [Google Scholar] [CrossRef]
- Peng, Y.-C.; Xu, J.-X.; Zeng, C.-F.; Zhao, X.-H.; You, X.-M.; Xu, P.-P.; Li, L.-Q.; Qi, L.-N. Operable hepatitis B virus-related hepatocellular carcinoma: Gut microbiota profile of patients at different ages. Ann. Transl. Med. 2022, 10, 477. [Google Scholar] [CrossRef]
- Peng, Y.-C.; Zhao, X.-H.; Zeng, C.-F.; Xu, J.-X.; Qi, L.-N.; Li, L.-Q. Integrated omics analysis: The relationship between significantly increased Klebsiella post-hepatectomy and decreased hub-metabolite 3-methyl-2-oxobutanoic acid is associated with induced liver failure. J. Gastrointest. Oncol. 2022, 13, 326–343. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, S.; Li, H.; Yang, F.; Mushtaq, N.; Ullah, S.; Shi, Y.; An, C.; Xu, J. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed. Pharmacother. 2018, 108, 184–193. [Google Scholar] [CrossRef]
- Wu, R.; Mei, X.; Ye, Y.; Xue, T.; Wang, J.; Sun, W.; Lin, C.; Xue, R.; Zhang, J.; Xu, D. Zn(II)-curcumin solid dispersion impairs hepatocellular carcinoma growth and enhances chemotherapy by modulating gut microbiota-mediated zinc homeostasis. Pharmacol. Res. 2019, 150, 104454. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef]
- Houot, R.; Schultz, L.M.; Marabelle, A.; Kohrt, H. T-cell–based Immunotherapy: Adoptive Cell Transfer and Checkpoint Inhibition. Cancer Immunol. Res. 2015, 3, 1115–1122. [Google Scholar] [CrossRef]
- Arias-Borrego, A.; Selma-Royo, M.; Collado, M.C.; Abril, N.; García-Barrera, T. Impact of “chemical cocktails” exposure in shaping mice gut microbiota and the role of selenium supplementation combining metallomics, metabolomics, and metataxonomics. J. Hazard. Mater. 2022, 438, 129444. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, X.; Wang, M.; He, Z.; Li, H.; Wang, J.; Li, Q. Gut microbiota influence immunotherapy responses: Mechanisms and therapeutic strategies. J. Hematol. Oncol. 2022, 15, 47. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Lim, H.Y.; Kudo, M.; Breder, V.; Merle, P.; et al. Updated efficacy and safety data from IMbrave150: Atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J. Hepatol. 2022, 76, 862–873. [Google Scholar] [CrossRef]
- NCCN. Clinical Practice Guidelines in Oncology. Hepatobiliary cancers. Version 2. JNCCN 2021, 19, 541–565. [Google Scholar] [CrossRef]
- Skelly, A.; Sato, Y.; Kearney, S.; Honda, K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 2019, 19, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Chervin, C.S.; Gajewski, T.F. Cancer and the Microbiome-Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology 2021, 160, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Mao, J.; Wang, D.; Long, J.; Yang, X.; Lin, J.; Song, Y.; Xie, F.; Xun, Z.; Wang, Y.; Wang, Y.; et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J. Immunother. Cancer 2021, 9, e003334. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- Routy, B.; Gopalakrishnan, V.; Daillère, R.; Zitvogel, L.; Wargo, J.A.; Kroemer, G. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 2018, 15, 382–396. [Google Scholar] [CrossRef]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Chung, M.-W.; Kim, M.-J.; Won, E.J.; Lee, Y.J.; Yun, Y.-W.; Cho, S.B.; Joo, Y.-E.; Hwang, J.-E.; Bae, W.K.; Chung, I.-J.; et al. Gut microbiome composition can predict the response to nivolumab in advanced hepatocellular carcinoma patients. World J. Gastroenterol. 2021, 27, 7340–7349. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, S.; Kou, Y.; Wang, Z.; Jin, R.; Hu, H.; Zhang, X.; Gong, J.F.; Li, J.; Lu, M.; et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020, 8, 1251–1261. [Google Scholar] [CrossRef]
- Yu, C.; Su, Z.; Li, Y.; Li, Y.; Liu, K.; Chu, F.; Liu, T.; Chen, R.; Ding, X. Dysbiosis of gut microbiota is associated with gastric carcinogenesis in rats. Biomed. Pharmacother. 2020, 126, 110036. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Y.-N.; Chen, T.; Ren, C.-H.; Li, X.; Liu, G.-X. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 149–157. [Google Scholar] [CrossRef]
- Payne, A.N.; Chassard, C.; Zimmermann, M.; Müller, P.; Stinca, S.; Lacroix, C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr. Diabetes 2011, 1, e12. [Google Scholar] [CrossRef]
- Coley, W.B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. 1893. Clin. Orthop. Relat. Res. 1991, 262, 3–11. [Google Scholar]
- Chen, Z.-F.; Ai, L.-Y.; Wang, J.-L.; Ren, L.-L.; Yu, Y.-N.; Xu, J.; Chen, H.-Y.; Yu, J.; Li, M.; Qin, W.-X.; et al. Probiotics Clostridium butyricum and Bacillus subtilis ameliorate intestinal tumorigenesis. Future Microbiol. 2015, 10, 1433–1445. [Google Scholar] [CrossRef]
- Gianotti, L.; Morelli, L.; Galbiati, F.; Rocchetti, S.; Coppola, S.; Beneduce, A.; Gilardini, C.; Zonenschain, D.; Nespoli, A.; Braga, M. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J. Gastroenterol. 2010, 16, 167–175. [Google Scholar] [CrossRef]
- Pala, V.; Sieri, S.; Berrino, F.; Vineis, P.; Sacerdote, C.; Palli, D.; Masala, G.; Panico, S.; Mattiello, A.; Tumino, R.; et al. Yogurt consumption and risk of colorectal cancer in the Italian European prospective investigation into cancer and nutrition cohort. Int. J. Cancer 2011, 129, 2712–2719. [Google Scholar] [CrossRef] [PubMed]
- Janket, S.J.; Ackerson, L.K.; Diamandis, E.P. Gut microbiotas and immune checkpoint inhibitor therapy response: A causal or coincidental relationship? Clin. Chem. Lab. Med. 2019, 58, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Khan, A.W.; Zhang, X.; White, M.G.; Peterson, C.B.; et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Zmora, N.; Zilberman-Schapira, G.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; Federici, S.; et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell 2018, 174, 1406–1423.e16. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Lee, K.A.; Shaw, H.M.; Bataille, V.; Nathan, P.; Spector, T.D. Role of the gut microbiome for cancer patients receiving immunotherapy: Dietary and treatment implications. Eur. J. Cancer 2020, 138, 149–155. [Google Scholar] [CrossRef]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.-M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).