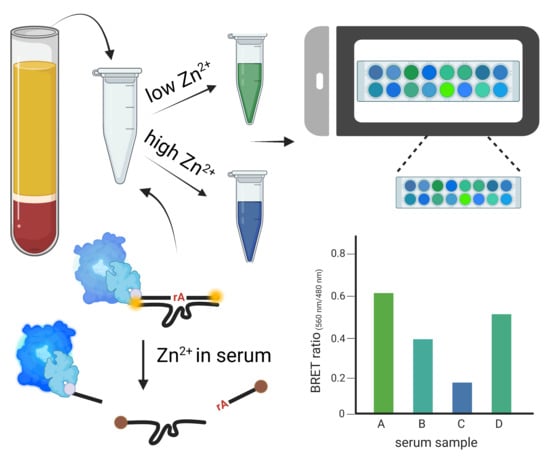
Ratiometric Detection of Zn2+ Using DNAzyme-Based Bioluminescence Resonance Energy Transfer Sensors
Abstract
1. Introduction
2. Materials and Methods
2.1. Instrumentation
2.2. Chemicals
2.3. DNA Sequences
2.4. BRET Assays
2.5. BRET POC Imaging
2.6. ICP-MS
2.7. Statistical Analysis
3. Results and Discussion
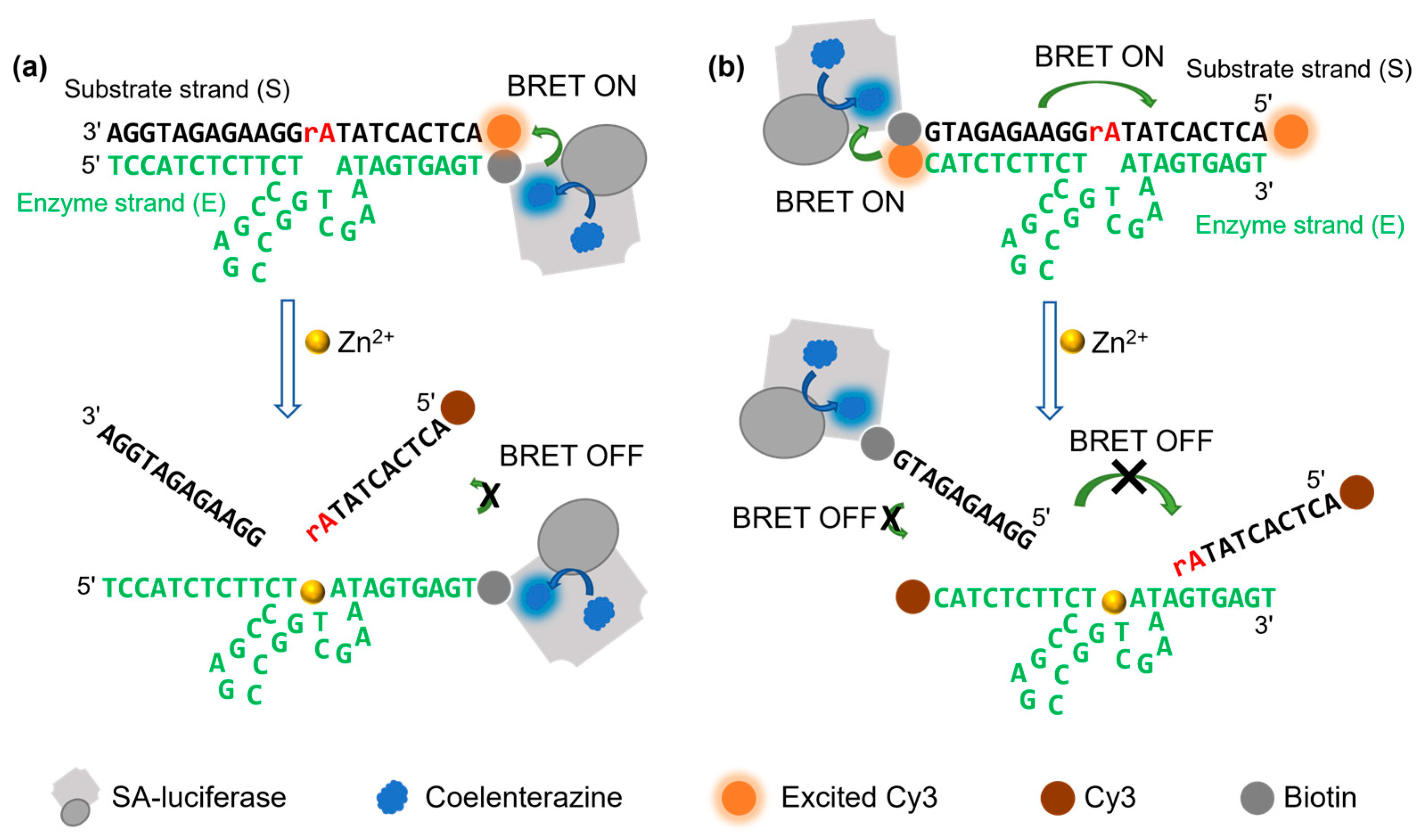
3.1. Design of a DNAzyme–BRET Sensor
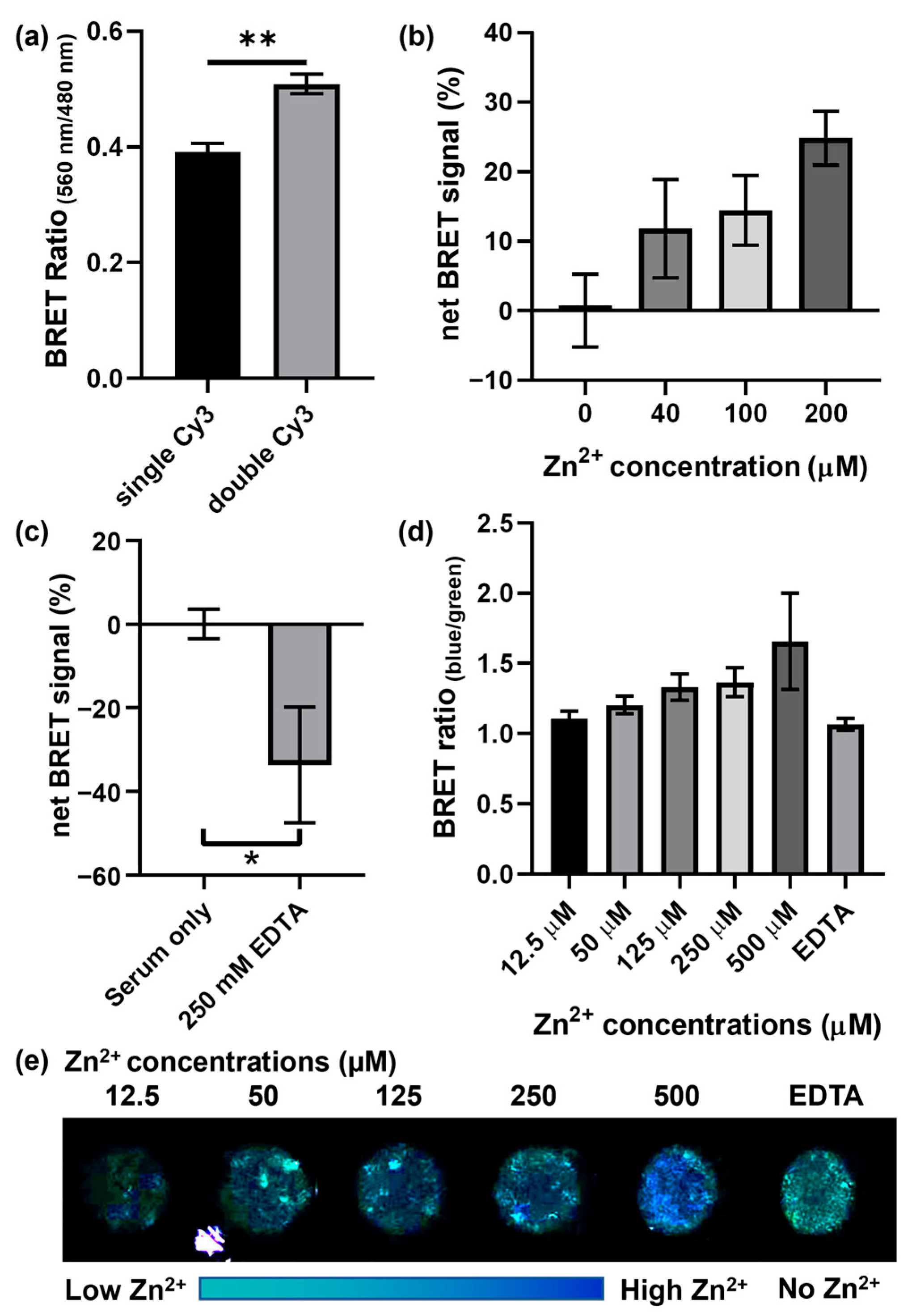
3.2. Optimization and Demonstration of the BRET-DNAzyme Sensor
3.3. Zn2+ Detection in Human Serum with Smartphone
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Que, E.L.; Domaille, D.W.; Chang, C.J. Metals in Neurobiology: Probing Their Chemistry and Biology with Molecular Imaging. Chem. Rev. 2008, 108, 1517–1549. [Google Scholar] [CrossRef] [PubMed]
- Sigel, A.; Sigel, H.; Sigel, R.K.O. (Eds.) Interrelations between Essential Metal Ions and Human Diseases; Metal Ions in Life Sciences; Springer: Dordrecht, The Netherlands, 2013; Volume 13, ISBN 978-94-007-7499-5. [Google Scholar]
- Moustakas, M. The Role of Metal Ions in Biology, Biochemistry and Medicine. Materials 2021, 14, 549. [Google Scholar] [CrossRef] [PubMed]
- Cvijanovich, N.Z.; King, J.C.; Flori, H.R.; Gildengorin, G.; Wong, H.R. Zinc Homeostasis in Pediatric Critical Illness. Pediatr. Crit. Care Med. J. Soc. Crit. Care Med. World Fed. Pediatr. Intensive Crit. Care Soc. 2009, 10, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S. Discovery of Human Zinc Deficiency: Its Impact on Human Health and Disease. Adv. Nutr. 2013, 4, 176–190. [Google Scholar] [CrossRef]
- Roohani, N.; Hurrell, R.; Kelishadi, R.; Schulin, R. Zinc and Its Importance for Human Health: An Integrative Review. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2013, 18, 144–157. [Google Scholar]
- Hussein, M.; Fathy, W.; Hassan, A.; Elkareem, R.A.; Marzouk, S.; Kamal, Y.S. Zinc Deficiency Correlates with Severity of Diabetic Polyneuropathy. Brain Behav. 2021, 11, e2349. [Google Scholar] [CrossRef]
- Nakanishi, K.; Toyoshima, M.; Ichikawa, G.; Suzuki, S. Zinc Deficiency Is Associated with Gynecologic Cancer Recurrence. Front. Oncol. 2022, 12, 1025060. [Google Scholar] [CrossRef]
- Maxfield, L.; Shukla, S.; Crane, J.S. Zinc Deficiency. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Wang, Y.; Sun, Z.; Li, A.; Zhang, Y. Association between Serum Zinc Levels and Lung Cancer: A Meta-Analysis of Observational Studies. World J. Surg. Oncol. 2019, 17, 78. [Google Scholar] [CrossRef]
- Farooq, D.M.; Alamri, A.F.; Alwhahabi, B.K.; Metwally, A.M.; Kareem, K.A. The Status of Zinc in Type 2 Diabetic Patients and Its Association with Glycemic Control. J. Fam. Community Med. 2020, 27, 29–36. [Google Scholar] [CrossRef]
- Knez, M.; Glibetic, M. Zinc as a Biomarker of Cardiovascular Health. Front. Nutr. 2021, 8, 686078. [Google Scholar] [CrossRef]
- Knez, M.; Boy, E. Existing Knowledge on Zn Status Biomarkers (1963–2021) with a Particular Focus on FADS1 and FADS2 Diagnostic Performance and Recommendations for Further Research. Front. Nutr. 2023, 9, 1057156. [Google Scholar] [CrossRef]
- Ruel, M.T.; Rivera, J.A.; Santizo, M.C.; Lönnerdal, B.; Brown, K.H. Impact of Zinc Supplementation on Morbidity from Diarrhea and Respiratory Infections among Rural Guatemalan Children. Pediatrics 1997, 99, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, Z.A.; Nizami, S.Q.; Isani, Z. Zinc Supplementation in Malnourished Children with Persistent Diarrhea in Pakistan. Pediatrics 1999, 103, e42. [Google Scholar] [CrossRef] [PubMed]
- Sazawal, S.; Black, R.E.; Ramsan, M.; Chwaya, H.M.; Dutta, A.; Dhingra, U.; Stoltzfus, R.J.; Othman, M.K.; Kabole, F.M. Effect of Zinc Supplementation on Mortality in Children Aged 1–48 Months: A Community-Based Randomised Placebo-Controlled Trial. Lancet. 2007, 369, 927–934. [Google Scholar] [CrossRef]
- Prasad, A.S.; Beck, F.W.J.; Snell, D.C.; Kucuk, O. Zinc in Cancer Prevention. Nutr. Cancer 2009, 61, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Dib, R.E.; Gameiro, O.L.; Ogata, M.S.; Módolo, N.S.; Braz, L.G.; Jorge, E.C.; Junior, P.D.N.; Beletate, V. Zinc Supplementation for the Prevention of Type 2 Diabetes Mellitus in Adults with Insulin Resistance. Cochrane Database Syst. Rev. 2015, 2015, CD005525. [Google Scholar] [CrossRef]
- Shakoor, H.; Feehan, J.; Al Dhaheri, A.S.; Ali, H.I.; Platat, C.; Ismail, L.C.; Apostolopoulos, V.; Stojanovska, L. Immune-Boosting Role of Vitamins D, C, E, Zinc, Selenium and Omega-3 Fatty Acids: Could They Help against COVID-19? Maturitas 2021, 143, 1–9. [Google Scholar] [CrossRef]
- Fosmire, G.J. Zinc Toxicity. Am. J. Clin. Nutr. 1990, 51, 225–227. [Google Scholar] [CrossRef]
- Duan, M.; Li, T.; Liu, B.; Yin, S.; Zang, J.; Lv, C.; Zhao, G.; Zhang, T. Zinc Nutrition and Dietary Zinc Supplements. Crit. Rev. Food Sci. Nutr. 2023, 63, 1277–1292. [Google Scholar] [CrossRef]
- de Blas, O.J.; Rodriguez, R.S.; Mendez, J.H.; Tomero, J.A.; Gomez, B.D.L.; Gonzalez, S.V. Determination of Zinc in Serum, Blood, and Ultrafiltrate Fluid from Patients on Hemofiltration by Graphite Furnace/Atomic Absorption Spectroscopy or Flow Injection Analysis/Atomic Absorption Spectroscopy. J. AOAC Int. 1994, 77, 722–727. [Google Scholar] [CrossRef]
- Zhang, L.; Shao, J.; Tan, S.-W.; Ye, H.-P.; Shan, X.-Y. Association between Serum Copper/Zinc Ratio and Lung Cancer: A Systematic Review with Meta-Analysis. J. Trace Elem. Med. Biol. 2022, 74, 127061. [Google Scholar] [CrossRef]
- Zerhusen, B.; de Silva, G.; Pei, X.; Papautsky, I.; Beyette, F.R. Point-of-Care Device for Quantification of Zinc in Serum. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2013, 2013, 4171–4174. [Google Scholar] [CrossRef] [PubMed]
- McNerney, M.P.; Zhang, Y.; Steppe, P.; Silverman, A.D.; Jewett, M.C.; Styczynski, M.P. Point-of-Care Biomarker Quantification Enabled by Sample-Specific Calibration. Sci. Adv. 2019, 5, eaax4473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, Y. Advancing Point-of-Care Diagnostics of Metabolites Through Engineering Semisynthetic Proteins. Clin. Chem. 2019, 65, 507–509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lan, T.; Lu, Y. Translating In Vitro Diagnostics from Centralized Laboratories to Point-of-Care Locations Using Commercially-Available Handheld Meters. TrAC Trends Anal. Chem. 2020, 124, 115782. [Google Scholar] [CrossRef]
- Bravo, I.D.B.; Castro, R.S.; Riquelme, N.L.; Díaz, C.T.; Goyenaga, D.A. Optimization of the Trace Element Determination by ICP-MS in Human Blood Serum. J. Trace Elem. Med. Biol. 2007, 21, 14–17. [Google Scholar] [CrossRef]
- Li, J.; Cen, D.; Huang, D.; Li, X.; Xu, J.; Fu, S.; Cai, R.; Wu, X.; Tang, M.; Sun, Y.; et al. Detection and Analysis of 12 Heavy Metals in Blood and Hair Sample from a General Population of Pearl River Delta Area. Cell Biochem. Biophys. 2014, 70, 1663–1669. [Google Scholar] [CrossRef]
- Hall, A.G.; King, J.C.; McDonald, C.M. Comparison of Serum, Plasma, and Liver Zinc Measurements by AAS, ICP-OES, and ICP-MS in Diverse Laboratory Settings. Biol. Trace Elem. Res. 2022, 200, 2606–2613. [Google Scholar] [CrossRef]
- World Leader in AA, ICP-OES and ICP-MS. Available online: https://resources.perkinelmer.com/corporate/content/relatedmaterials/brochures/bro_worldleaderaaicpmsicpms.pdf (accessed on 27 July 2023).
- Wilschefski, S.C.; Baxter, M.R. Inductively Coupled Plasma Mass Spectrometry: Introduction to Analytical Aspects. Clin. Biochem. Rev. 2019, 40, 115–133. [Google Scholar] [CrossRef]
- Nolan, E.M.; Lippard, S.J. Small-Molecule Fluorescent Sensors for Investigating Zinc Metalloneurochemistry. Acc. Chem. Res. 2009, 42, 193–203. [Google Scholar] [CrossRef]
- Woo, H.; You, Y.; Kim, T.; Jhon, G.-J.; Nam, W. Fluorescence Ratiometric Zinc Sensors Based on Controlled Energy Transfer. J. Mater. Chem. 2012, 22, 17100–17112. [Google Scholar] [CrossRef]
- Ali, S.; Cuajungco, M.P. Protocol for Quantifying Zinc Flux in Cultured Cells Using Fluorescent Indicators. STAR Protoc. 2020, 1, 100050. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.S.; Zakaria, N.H.; Daud, N.; Tan, L.L.; Ta, G.C.; Heng, L.Y.; Hassan, N.I. The Role of 8-Amidoquinoline Derivatives as Fluorescent Probes for Zinc Ion Determination. Sensors 2021, 21, 311. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, H.; Li, J.; Zhou, C.; Li, L.; Wu, X.; Lei, C. A near Infrared Fluorescent Probe for Detection and Bioimaging of Zinc Ions and Hypochlorous Acid. Anal. Chim. Acta 2022, 1206, 339750. [Google Scholar] [CrossRef] [PubMed]
- Salonia, J.A.; Wuilloud, R.G.; Gásquez, J.A.; Olsina, R.A.; Martinez, L.D. On-Line Complexation of Zinc with 5-Br-PADAP and Preconcentration Using a Knotted Reactor for Inductively Coupled Plasma Atomic Emission Spectrometric Determination in River Water Samples. Fresenius J. Anal. Chem. 2000, 367, 653–657. [Google Scholar] [CrossRef]
- Menek, N.; Topçu, S.; Uçar, M. Voltammetric and Spectrophotometric Studies of 2-(5-Bromo-2-Pyridylazo)-5-Diethylamino)Phenol Copper(Ii) Complex. Anal. Lett. 2001, 34, 1733–1740. [Google Scholar] [CrossRef]
- Ratnani, S.; Agnihotri, N.K. An Eco-Friendly, Sensitive and Selective Derivative Spectrophotometric Method for Simultaneous Determination of Copper and Zinc in Synthetic Mixtures, Alloys and Environmental Samples. Int. J. Environ. Anal. Chem. 2021, 1–14. [Google Scholar] [CrossRef]
- Enbanathan, S.; Munusamy, S.; Jothi, D.; Manojkumar, S.; Manickam, S.; KulathuIyer, S. Zinc Ion Detection Using a Benzothiazole-Based Highly Selective Fluorescence “Turn-on” Chemosensor and Its Real-Time Application. RSC Adv. 2022, 12, 27839–27845. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y. A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J. Am. Chem. Soc. 2000, 122, 10466–10467. [Google Scholar] [CrossRef]
- Li, J.; Zheng, W.; Kwon, A.H.; Lu, Y. In Vitro Selection and Characterization of a Highly Efficient Zn(II)-Dependent RNA-Cleaving Deoxyribozyme. Nucleic Acids Res. 2000, 28, 481–488. [Google Scholar] [CrossRef]
- Xiao, Y.; Rowe, A.A.; Plaxco, K.W. Electrochemical Detection of Parts-Per-Billion Lead via an Electrode-Bound DNAzyme Assembly. J. Am. Chem. Soc. 2007, 129, 262–263. [Google Scholar] [CrossRef]
- Liu, J.; Brown, A.K.; Meng, X.; Cropek, D.M.; Istok, J.D.; Watson, D.B.; Lu, Y. A Catalytic Beacon Sensor for Uranium with Parts-per-Trillion Sensitivity and Millionfold Selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 2056–2061. [Google Scholar] [CrossRef]
- Torabi, S.-F.; Wu, P.; McGhee, C.E.; Chen, L.; Hwang, K.; Zheng, N.; Cheng, J.; Lu, Y. In Vitro Selection of a Sodium-Specific DNAzyme and Its Application in Intracellular Sensing. Proc. Natl. Acad. Sci. USA 2015, 112, 5903–5908. [Google Scholar] [CrossRef]
- Huang, P.-J.J.; Liu, J. Rational Evolution of Cd2+-Specific DNAzymes with Phosphorothioate Modified Cleavage Junction and Cd2+ Sensing. Nucleic Acids Res. 2015, 43, 6125–6133. [Google Scholar] [CrossRef]
- Saran, R.; Liu, J. A Silver DNAzyme. Anal. Chem. 2016, 88, 4014–4020. [Google Scholar] [CrossRef]
- Zhou, W.; Vazin, M.; Yu, T.; Ding, J.; Liu, J. In Vitro Selection of Chromium-Dependent DNAzymes for Sensing Chromium(III) and Chromium(VI). Chem.-Eur. J. 2016, 22, 9835–9840. [Google Scholar] [CrossRef] [PubMed]
- McGhee, C.E.; Yang, Z.; Guo, W.; Wu, Y.; Lyu, M.; DeLong, C.J.; Hong, S.; Ma, Y.; McInnis, M.G.; O’Shea, K.S.; et al. DNAzyme-Based Lithium-Selective Imaging Reveals Higher Lithium Accumulation in Bipolar Disorder Patient-Derived Neurons. ACS Cent. Sci. 2021, 7, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Torabi, S.-F.; Lake, R.J.; Hong, S.; Yu, Z.; Wu, P.; Yang, Z.; Nelson, K.; Guo, W.; Pawel, G.T.; et al. Simultaneous Fe2+/Fe3+ Imaging Shows Fe3+ over Fe2+ Enrichment in Alzheimer’s Disease Mouse Brain. Sci. Adv. 2023, 9, eade7622. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; McGhee, C.E.; Lake, R.J.; Yang, Z.; Guo, Z.; Zhang, X.-B.; Lu, Y. A Highly Selective Mn(II)-Specific DNAzyme and Its Application in Intracellular Sensing. JACS Au 2023, 3, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Joyce, G.F. A DNA Enzyme That Cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. A General Purpose RNA-Cleaving DNA Enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef]
- Li, Y.; Breaker, R.R. Deoxyribozymes: New Players in the Ancient Game of Biocatalysis. Curr. Opin. Struct. Biol. 1999, 9, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Ihms, H.E.; Lu, Y. In Vitro Selection of Metal Ion-Selective DNAzymes. In Ribozymes. Methods in Molecular Biology; Hartig, J.S., Ed.; Humana Press: Totowa, NJ, USA, 2012; Volume 848, pp. 297–316. ISBN 978-1-61779-545-9. [Google Scholar]
- Liu, J.; Lu, Y. Improving Fluorescent DNAzyme Biosensors by Combining Inter- and Intramolecular Quenchers. Anal. Chem. 2003, 75, 6666–6672. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.H.J.; Liu, Z.; Brennan, J.D.; Li, Y. An Efficient RNA-Cleaving DNA Enzyme That Synchronizes Catalysis with Fluorescence Signaling. J. Am. Chem. Soc. 2003, 125, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Navani, N.K.; Li, Y. Nucleic Acid Aptamers and Enzymes as Sensors. Curr. Opin. Chem. Biol. 2006, 10, 272–281. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. A DNAzyme Catalytic Beacon Sensor for Paramagnetic Cu2+ Ions in Aqueous Solution with High Sensitivity and Selectivity. J. Am. Chem. Soc. 2007, 129, 9838–9839. [Google Scholar] [CrossRef]
- Huang, P.-J.J.; Liu, J. An Ultrasensitive Light-up Cu2+ Biosensor Using a New DNAzyme Cleaving a Phosphorothioate-Modified Substrate. Anal. Chem. 2016, 88, 3341–3347. [Google Scholar] [CrossRef]
- Lake, R.J.; Yang, Z.; Zhang, J.; Lu, Y. DNAzymes as Activity-Based Sensors for Metal Ions: Recent Applications, Demonstrated Advantages, Current Challenges, and Future Directions. Acc. Chem. Res. 2019, 52, 3275–3286. [Google Scholar] [CrossRef]
- Zhou, W.; Saran, R.; Liu, J. Metal Sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar] [CrossRef]
- McConnell, E.M.; Cozma, I.; Mou, Q.; Brennan, J.D.; Lu, Y.; Li, Y. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954–8994. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, X.; Wu, Z.; Jiang, J.-H. Recent Advances on DNAzyme-Based Sensing. Chem.-Asian J. 2022, 17, e202101414. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Ou, Y.; Wang, Z.; Fu, F.; Guo, L. DNAzyme-Based Biosensor for Cu2+ Ion by Combining Hybridization Chain Reaction with Fluorescence Resonance Energy Transfer Technique. Talanta 2016, 155, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ding, J.; Liu, J. Splitting a DNAzyme Enables a Na+-Dependent FRET Signal from the Embedded Aptamer. Org. Biomol. Chem. 2017, 15, 6959–6966. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Mou, Q.; Lake, R.J.; Xiong, M.; Holland, B.; Lu, Y. Metal-Dependent DNAzymes for the Quantitative Detection of Metal Ions in Living Cells: Recent Progress, Current Challenges, and Latest Results on FRET Ratiometric Sensors. Inorg. Chem. 2019, 58, 13696–13708. [Google Scholar] [CrossRef] [PubMed]
- Moon, W.J.; Yang, Y.; Liu, J. Zn2+-Dependent DNAzymes: From Solution Chemistry to Analytical, Materials and Therapeutic Applications. ChemBioChem 2021, 22, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Lin, Y.; Cai, L.; Basa, P.N.; Shigemoto, A.K.; Zheng, C.; Zhang, F.; Burdette, S.C.; Lu, Y. Detection and Quantification of Tightly Bound Zn2+ in Blood Serum Using a Photocaged Chelator and a DNAzyme Fluorescent Sensor. Anal. Chem. 2021, 93, 5856–5861. [Google Scholar] [CrossRef]
- Hwang, K.; Wu, P.; Kim, T.; Lei, L.; Tian, S.; Wang, Y.; Lu, Y. Photocaged DNAzymes as a General Method for Sensing Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2014, 53, 13798–13802. [Google Scholar] [CrossRef]
- Wang, W.; Satyavolu, N.S.R.; Wu, Z.; Zhang, J.-R.; Zhu, J.-J.; Lu, Y. Near-Infrared Photothermally Activated DNAzyme–Gold Nanoshells for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2017, 56, 6798–6802. [Google Scholar] [CrossRef]
- Lin, Y.; Yang, Z.; Lake, R.J.; Zheng, C.; Lu, Y. Enzyme-Mediated Endogenous and Bioorthogonal Control of a DNAzyme Fluorescent Sensor for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2019, 58, 17061–17067. [Google Scholar] [CrossRef]
- Xiong, M.; Yang, Z.; Lake, R.J.; Li, J.; Hong, S.; Fan, H.; Zhang, X.-B.; Lu, Y. DNAzyme-Mediated Genetically Encoded Sensors for Ratiometric Imaging of Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2020, 59, 1891–1896. [Google Scholar] [CrossRef]
- Yi, D.; Zhao, J.; Li, L. An Enzyme-Activatable Engineered DNAzyme Sensor for Cell-Selective Imaging of Metal Ions. Angew. Chem. Int. Ed. 2021, 60, 6300–6304. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Loh, K.Y.; Chu, Y.T.; Feng, R.; Satyavolu, N.S.R.; Xiong, M.; Nakamata Huynh, S.M.; Hwang, K.; Li, L.; Xing, H.; et al. Optical Control of Metal Ion Probes in Cells and Zebrafish Using Highly Selective DNAzymes Conjugated to Upconversion Nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kim, G.; Chu, J.L.; Song, T.; Yang, Z.; Guo, W.; Shao, X.; Oelze, M.L.; Li, K.C.; Lu, Y. Noninvasive and Spatiotemporal Control of DNAzyme-Based Imaging of Metal Ions In Vivo Using High-Intensity Focused Ultrasound. J. Am. Chem. Soc. 2022, 144, 5812–5819. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Pavlov, V.; Gill, R.; Bourenko, T.; Willner, I. Lighting up Biochemiluminescence by the Surface Self-Assembly of DNA-Hemin Complexes. ChemBioChem 2004, 5, 374–379. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. Lead(II)-Induced Allosteric G-Quadruplex DNAzyme as a Colorimetric and Chemiluminescence Sensor for Highly Sensitive and Selective Pb2+ Detection. Anal. Chem. 2010, 82, 1515–1520. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. Parallel G-Quadruplex-Specific Fluorescent Probe for Monitoring DNA Structural Changes and Label-Free Detection of Potassium Ion. Anal. Chem. 2010, 82, 7576–7580. [Google Scholar] [CrossRef]
- Shimron, S.; Wang, F.; Orbach, R.; Willner, I. Amplified Detection of DNA through the Enzyme-Free Autonomous Assembly of Hemin/G-Quadruplex DNAzyme Nanowires. Anal. Chem. 2012, 84, 1042–1048. [Google Scholar] [CrossRef]
- Li, T.; Dong, S.; Wang, E. Label-Free Colorimetric Detection of Aqueous Mercury Ion (Hg2+) Using Hg2+-Modulated G-Quadruplex-Based DNAzymes. Anal. Chem. 2009, 81, 2144–2149. [Google Scholar] [CrossRef]
- Du, Y.; Li, B.; Guo, S.; Zhou, Z.; Zhou, M.; Wang, E.; Dong, S. G-Quadruplex -Based DNAzyme for Colorimetric Detection of Cocaine: Using Magnetic Nanoparticles as the Separation and Amplification Element. Analyst 2011, 136, 493–497. [Google Scholar] [CrossRef]
- Sun, H.; Chen, H.; Zhang, X.; Liu, Y.; Guan, A.; Li, Q.; Yang, Q.; Shi, Y.; Xu, S.; Tang, Y. Colorimetric Detection of Sodium Ion in Serum Based on the G-Quadruplex Conformation Related DNAzyme Activity. Anal. Chim. Acta 2016, 912, 133–138. [Google Scholar] [CrossRef]
- Zheng, J.; Wai, J.L.; Lake, R.J.; New, S.Y.; He, Z.; Lu, Y. DNAzyme Sensor Uses Chemiluminescence Resonance Energy Transfer for Rapid, Portable, and Ratiometric Detection of Metal Ions. Anal. Chem. 2021, 93, 10834–10840. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, K.D.G.; Eidne, K.A. Illuminating Insights into Protein-Protein Interactions Using Bioluminescence Resonance Energy Transfer (BRET). Nat. Methods 2006, 3, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Mie, M.; Hirashima, R.; Mashimo, Y.; Kobatake, E. Construction of an Enzymatically-Conjugated DNA Aptamer–Protein Hybrid Molecule for Use as a BRET-Based Biosensor. Appl. Sci. 2020, 10, 7646. [Google Scholar] [CrossRef]
- Aper, S.J.A.; Dierickx, P.; Merkx, M. Dual Readout BRET/FRET Sensors for Measuring Intracellular Zinc. ACS Chem. Biol. 2016, 11, 2854–2864. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, E.G.; Stoddard, A.K.; Zeng, H.-H.; Franke, G.; Bourne, L.; Fierke, C.A.; Thompson, R.B. Ratiometric Zinc Biosensor Based on Bioluminescence Resonance Energy Transfer: Trace Metal Ion Determination with Tunable Response. Int. J. Mol. Sci. 2022, 23, 14936. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.B.; Maliwal, B.P.; Fierke, C.A. Selectivity and Sensitivity of Fluorescence Lifetime-Based Metal Ion Biosensing Using a Carbonic Anhydrase Transducer. Anal. Biochem. 1999, 267, 185–195. [Google Scholar] [CrossRef]
- Hessels, A.M.; Chabosseau, P.; Bakker, M.H.; Engelen, W.; Rutter, G.A.; Taylor, K.M.; Merkx, M. EZinCh-2: A Versatile, Genetically Encoded FRET Sensor for Cytosolic and Intraorganelle Zn2+ Imaging. ACS Chem. Biol. 2015, 10, 2126–2134. [Google Scholar] [CrossRef]
- Michielsen, C.M.S.; van Aalen, E.A.; Merkx, M. Ratiometric Bioluminescent Zinc Sensor Proteins to Quantify Serum and Intracellular Free Zn2+. ACS Chem. Biol. 2022, 17, 1567–1576. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Dacres, H.; Michie, M.; Wang, J.; Pfleger, K.D.G.; Trowell, S.C. Effect of Enhanced Renilla Luciferase and Fluorescent Protein Variants on the Förster Distance of Bioluminescence Resonance Energy Transfer (BRET). Biochem. Biophys. Res. Commun. 2012, 425, 625–629. [Google Scholar] [CrossRef]
- Borroto-Escuela, D.O.; Flajolet, M.; Agnati, L.F.; Greengard, P.; Fuxe, K. Bioluminiscence resonance energy transfer (bret) methods to study g protein-coupled receptor-receptor tyrosine kinase heteroreceptor complexes. Methods Cell Biol. 2013, 117, 141–164. [Google Scholar] [CrossRef]
- Jenne, A.; Famulok, M. Disruption of the Streptavidin Interaction with Biotinylated Nucleic Acid Probes by 2-Mercaptoethanol. BioTechniques 1999, 26, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Long, G.L.; Winefordner, J.D. Limit of Detection: A Closer Look at the IUPAC Definition. Anal. Chem. 1983, 55, 712A–724A. [Google Scholar] [CrossRef]
- Moon, W.J.; Huang, P.-J.J.; Liu, J. Probing Metal-Dependent Phosphate Binding for the Catalysis of the 17E DNAzyme. Biochemistry 2021, 60, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- FluoZinTM-3, Tetrapotassium Salt, Cell Impermeant. Available online: https://www.thermofisher.com/order/catalog/product/F24194 (accessed on 27 July 2023).
- Landero-Figueroa, J.A.; Vignesh, K.S.; Deepe, G.; Caruso, J. Selectivity and specificity of small molecule fluorescent dyes/probes used for the detection of Zn2+ and Ca2+ in cellS. Met. Integr. Biometal Sci. 2014, 6, 301–315. [Google Scholar] [CrossRef]
- Brown, N.E.; Blumer, J.B.; Hepler, J.R. Bioluminescence Resonance Energy Transfer to Detect Protein-Protein Interactions in Live Cells. In Protein-Protein Interactions: Methods and Applications; Meyerkord, C.L., Fu, H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 457–465. ISBN 978-1-4939-2425-7. [Google Scholar]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Biggins, S.W.; Kim, W.R.; Terrault, N.A.; Saab, S.; Balan, V.; Schiano, T.; Benson, J.; Therneau, T.; Kremers, W.; Wiesner, R.; et al. Evidence-Based Incorporation of Serum Sodium Concentration Into MELD. Gastroenterology 2006, 130, 1652–1660. [Google Scholar] [CrossRef]
- Costello, R.B.; Elin, R.J.; Rosanoff, A.; Wallace, T.C.; Guerrero-Romero, F.; Hruby, A.; Lutsey, P.L.; Nielsen, F.H.; Rodriguez-Moran, M.; Song, Y.; et al. Perspective: The Case for an Evidence-Based Reference Interval for Serum Magnesium: The Time Has Come. Adv. Nutr. 2016, 7, 977–993. [Google Scholar] [CrossRef]
- Calomme, M.R.; Vanden Berghe, D.A. Supplementation of Calves with Stabilized Orthosilicic Acid. Biol. Trace Elem. Res. 1997, 56, 153–165. [Google Scholar] [CrossRef]
- Serum Iron Test Information|Mount Sinai—New York. Available online: https://www.mountsinai.org/health-library/tests/serum-iron-test (accessed on 27 July 2023).
- Persijn, J.P.; van der Slik, W.; Riethorst, A. Determination of Serum Iron and Latent Iron-Binding Capacity (LIBC). Clin. Chim. Acta 1971, 35, 91–98. [Google Scholar] [CrossRef]
- Bárány, E.; Bergdahl, I.A.; Bratteby, L.E.; Lundh, T.; Samuelson, G.; Schütz, A.; Skerfving, S.; Oskarsson, A. Trace Element Levels in Whole Blood and Serum from Swedish Adolescents. Sci. Total Environ. 2002, 286, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Nemeth, E. Iron Metabolism: Interactions with Normal and Disordered Erythropoiesis. Cold Spring Harb. Perspect. Med. 2012, 2, a011668. [Google Scholar] [CrossRef] [PubMed]
- Kapaki, E.; Segditsa, J.; Papageorgiou, C. Zinc, Copper and Magnesium Concentration in Serum and CSF of Patients with Neurological Disorders. Acta Neurol. Scand. 1989, 79, 373–378. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Lewis, W.; Wai, J.L.; Xiong, M.; Zheng, J.; Yang, Z.; Gordon, C.; Lu, Y.; New, S.Y.; Zhang, X.-B.; et al. Ratiometric Detection of Zn2+ Using DNAzyme-Based Bioluminescence Resonance Energy Transfer Sensors. Chemistry 2023, 5, 1745-1759. https://doi.org/10.3390/chemistry5030119
Wu Y, Lewis W, Wai JL, Xiong M, Zheng J, Yang Z, Gordon C, Lu Y, New SY, Zhang X-B, et al. Ratiometric Detection of Zn2+ Using DNAzyme-Based Bioluminescence Resonance Energy Transfer Sensors. Chemistry. 2023; 5(3):1745-1759. https://doi.org/10.3390/chemistry5030119
Chicago/Turabian StyleWu, Yuting, Whitney Lewis, Jing Luen Wai, Mengyi Xiong, Jiao Zheng, Zhenglin Yang, Chloe Gordon, Ying Lu, Siu Yee New, Xiao-Bing Zhang, and et al. 2023. "Ratiometric Detection of Zn2+ Using DNAzyme-Based Bioluminescence Resonance Energy Transfer Sensors" Chemistry 5, no. 3: 1745-1759. https://doi.org/10.3390/chemistry5030119
APA StyleWu, Y., Lewis, W., Wai, J. L., Xiong, M., Zheng, J., Yang, Z., Gordon, C., Lu, Y., New, S. Y., Zhang, X.-B., & Lu, Y. (2023). Ratiometric Detection of Zn2+ Using DNAzyme-Based Bioluminescence Resonance Energy Transfer Sensors. Chemistry, 5(3), 1745-1759. https://doi.org/10.3390/chemistry5030119