Abstract
Apurinic/apyrimidinic endonuclease 1 (APE1), also known as redox factor-1 (Ref-1), is a multifunctional protein that exists widely in living organisms. It can specifically recognize and cleave the DNA in apurinic/apyrimidinic (AP) sites in the base excision repair (BER) pathway, as well as regulate the expression of genes to activate some transcription factors. The abnormal expression and disruptions in the biological functions of APE1 are linked to a number of diseases, including inflammation, immunodeficiency, and cancer. Hence, it is extremely desired to monitor the activity of APE1, acquiring a thorough understanding of the healing process of damaged DNA and making clinical diagnoses. Thanks to the advent of DNA nanotechnology, some nanodevices are used to image the activity of APE1 with great sensitivity and simplicity. In this review, we will summarize developments in DNA-nanotechnology-empowered fluorescence imaging in recent years for APE1 activity according to different types of DNA probes, which are classified into linear DNA probes, composite DNA nanomaterials, and three-dimensional (3D) DNA nanostructures. We also highlight the future research directions in the field of APE1 activity imaging.
1. Introduction
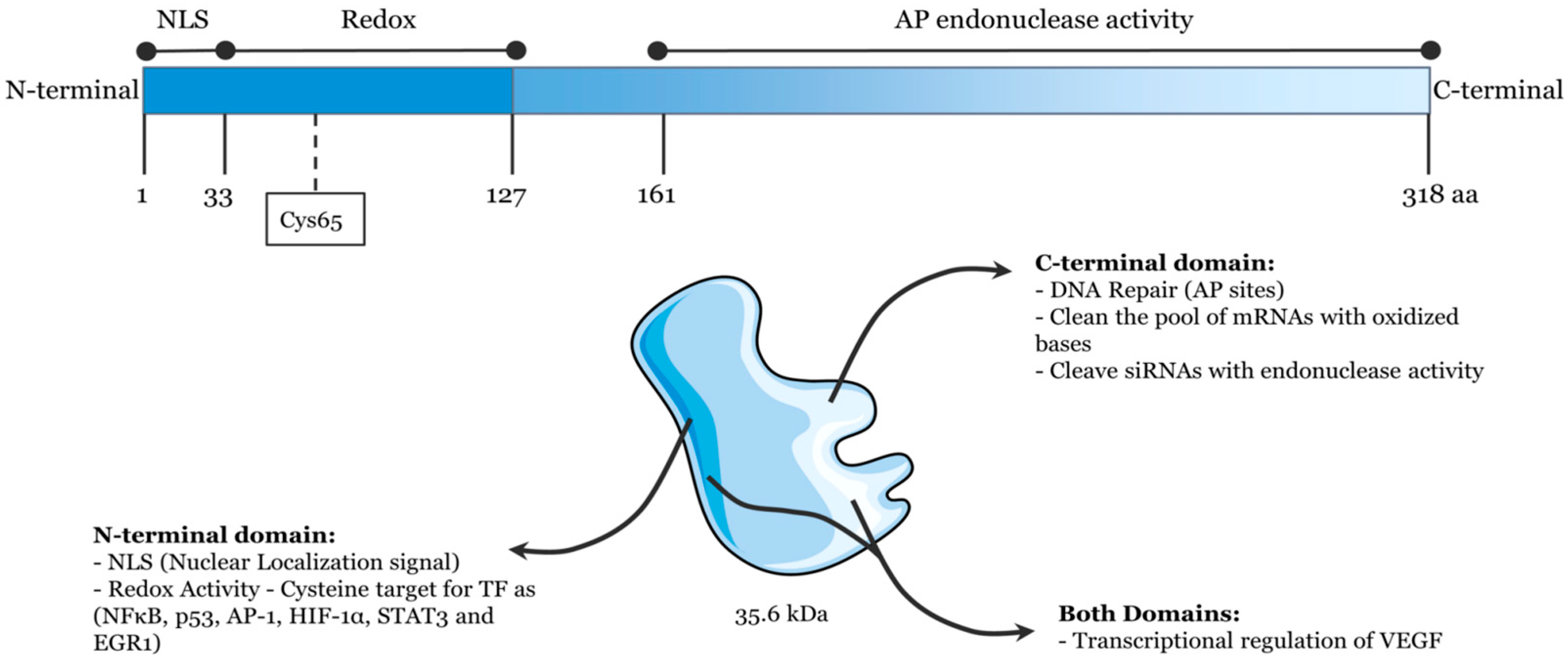
Human APE1, which displays homology to Escherichia coli exonuclease III, consists of a globular domain with 318 amino acid residues [1,2,3]. It has been discovered to be a 35.6 kDa cellular multifunctional protein that contains a redox function and AP endonuclease activity, and both functions are independent in their actions (Figure 1) [4,5,6,7]. The redox function of APE1, which is in its N-terminal domain, plays an important role in regulating the expression of genes to activate some transcription factors, such as p53, Egr-1, and NF-kB [8,9]. According to multiple studies, APE1 redox function is linked to the occurrence of various cancers, such as cutaneous squamous cell, pancreatic cancer, and esophageal adenocarcinoma [10,11,12,13]. Inhibiting APE1 redox function can decrease cell proliferation, prevent angiogenesis, and inhibit endothelial precursor cell differentiation [14,15,16]. On the other hand, as a central enzyme in the BER pathway, APE1 contains AP endonuclease activity in its C-terminal domain [17,18,19,20]. After APE1 recognizes and cleaves the AP site, it results in a single-strand break that may be processed further by the coordinated actions of DNA polymerase, deoxyribose phosphatase, and DNA ligase [21,22,23,24]. APE1 is mainly present in the nucleus and mitochondria. In addition, it can also be found in the cytoplasm or be secreted from cells. The state of disease dynamically regulates APE1 localization [7,25,26]. So far, APE1 has been found to be vital in modern life science, and it is engaged in a variety of biological functions such as apoptosis and cell cycle regulation [20,27]. Misfunctioning or the disturbed expression of APE1 can lead to a series of diseases [28], such as immunodeficiency [29], cancer [30,31], and cellular senescence [32]. Therefore, monitoring APE1 activity is extremely desired in order to acquire a complete understanding of the repair process of damaged DNA and make clinical diagnoses.
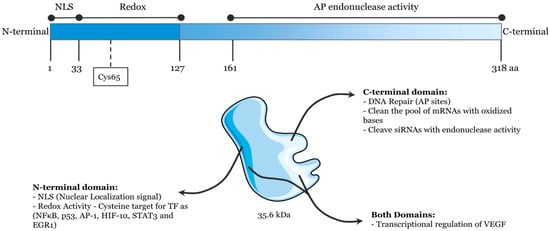
Figure 1.
Schematic illustration of the functional domains of APE1/Ref-1. (Reprinted with permission from Ref. [4]. Copyright 2022 Frontiers).
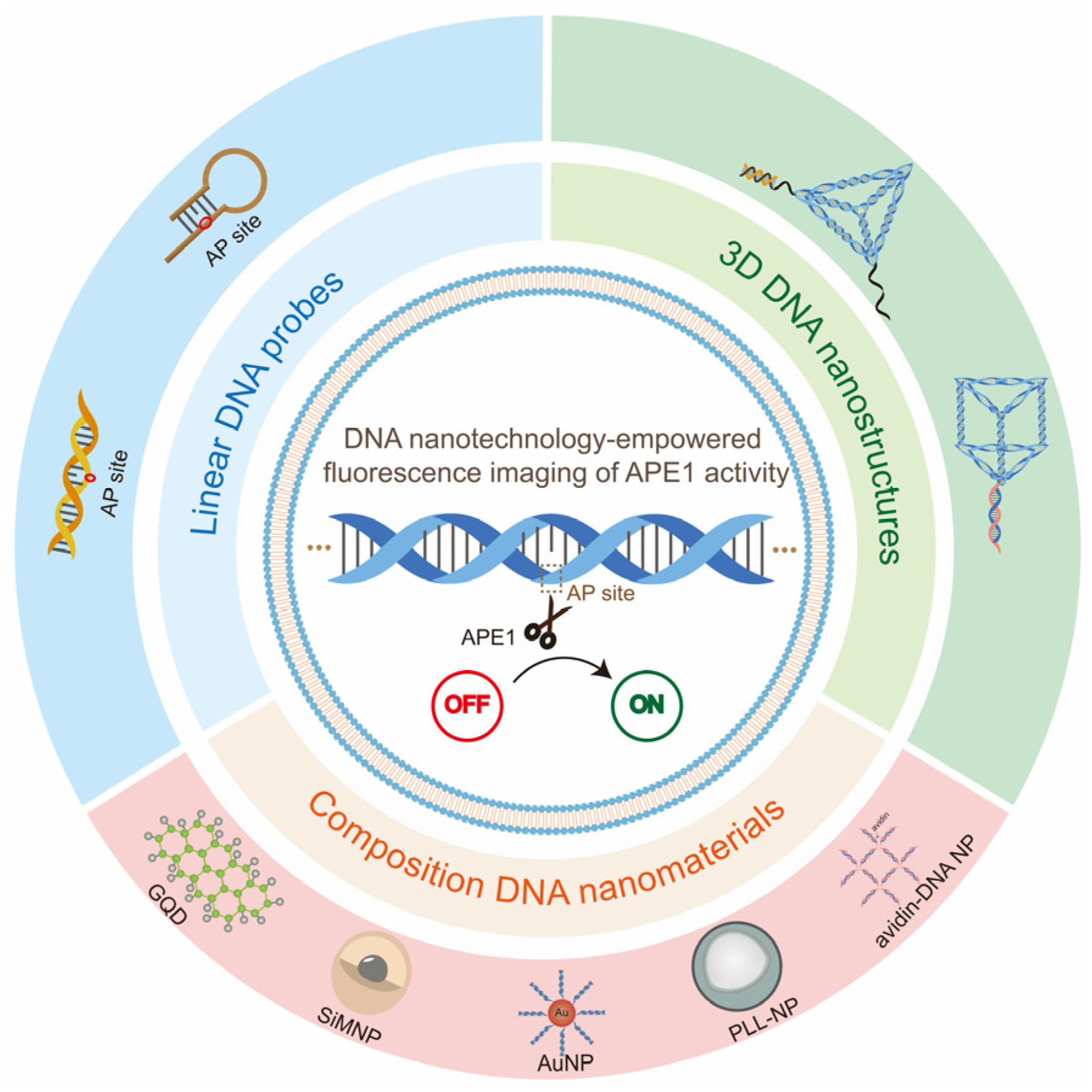
Because of its great sensitivity and real-time capabilities, fluorescence molecular imaging has been intensively pursued for the investigation of enzyme expression and distribution [33]. However, small molecular fluorescent probes are difficult to manage in cells, limiting their applicability in subcellular detection. In addition, complex genetic fusion mechanisms frequently restrict protein-based fluorescent sensors [34]. Alternatively, DNA-nanotechnology-based fluorescence probes are being developed for the sensing and imaging of biomarkers in living cells due to their great programmability, simplicity in synthesis, and biocompatibility [35]. Our group designed a series of DNA nanostructures for biomarker fluorescence imaging and cancer therapy, such as (1) a DNA tetrahedron-based nanocarrier with ATP aptamer for ATP detection [36]; (2) a DNA nanotriangle linked with split aptamer probes and loaded doxorubicin (Dox) as a platform for diagnosis and treatment [37]; (3) a Y-shaped DNA nanostructure with split i-motif fragments and loaded Dox for enhancing cancer imaging and therapy [38]; (4) a three-arm aptamer nanoclaw for enhancing imaging and tumor growth inhibition [39]; and (5) a hand-in-hand DNA tile assembly loaded with TK1 mRNA and Dox for the imaging of TK1 mRNA and chemotherapy. In addition, Willner’s group proposed that the formation of self-assembled nanostructures provided a basis for the formation of signal-amplifying sensor systems as well as for the bottom-up construction of functional scaffolds [40]. Since APE1 has a specific effect on the AP site, the DNA probes that modified the AP site, such as double-stranded DNA probes [41,42], gold–DNA nanomaterial complexes [43,44], and DNA prisms [45], etc., can be used for the imaging of APE1 activity simply. The different types of DNA probes used for APE1 activity fluorescence imaging can be classified into linear DNA probes, composite DNA nanomaterials, and 3D DNA nanostructures. Although many studies have focused on APE1 imaging, there are a few reviews on the fluorescence imaging of APE1 based on DNA nanotechnology. In this review, we will specifically summarize the DNA-nanotechnology-based fluorescence imaging of APE1 activity (Figure 2). First, we briefly describe the origin of AP sites and the mechanism of cleavage of AP site by APE1. Then, the most recent achievements of DNA nanotechnology as an excellent tool for APE1 activity fluorescence imaging are described by typical examples. Finally, we summarize the driving effect of the DNA-nanotechnology-empowered fluorescence imaging of APE1 activity and the directions and opportunities for future development. We hope this review will provide new insights into the design and development of new strategies for APE1 activity imaging, acquiring a complete understanding of the repair process of damaged DNA and aiding clinical diagnoses.
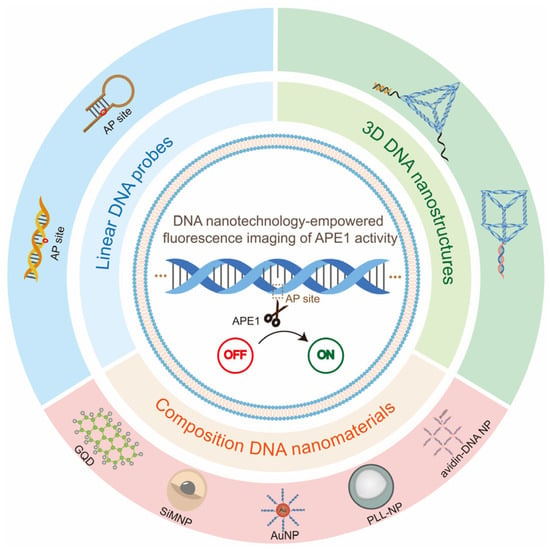
Figure 2.
Diagram of the content of this review. The DNA probes for APE1 activity fluorescence imaging can be mainly classified into three main parts according to different probe types: linear DNA probes, composite DNA nanomaterials, and 3D DNA nanostructures.
2. Origin of AP Sites and Mechanism of Cleavage of AP Site by APE1
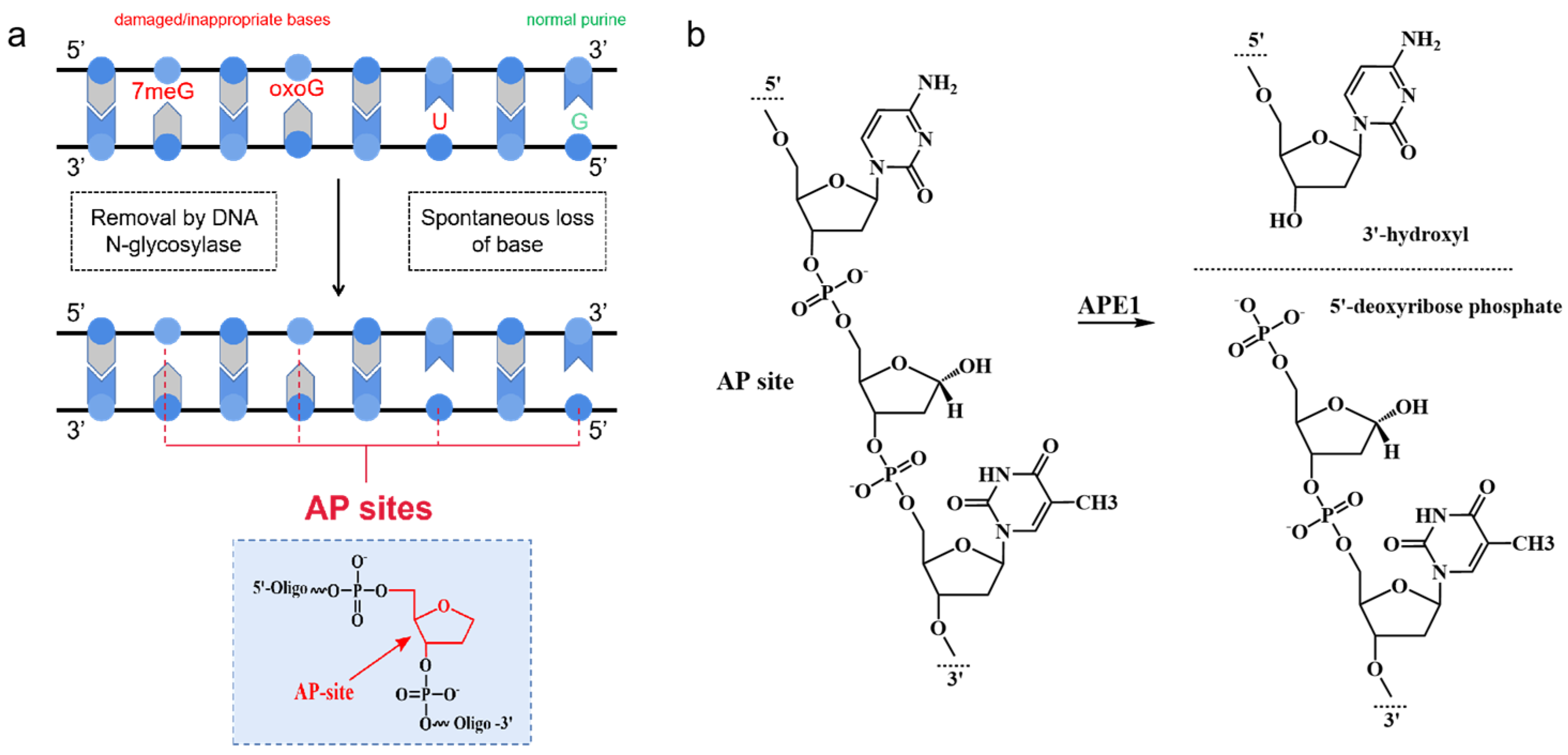
As one of the common lesions in genomic DNA, AP sites can block DNA replication and transcription [46]. The term “AP sites” refers to DNA damage products that are created when a nucleobase is lost, but the deoxyribose is retained, along with the conserved 3′- and 5′-phosphodiester linkages [47]. As shown in Figure 3a, there are two main sources of AP sites: (1) the spontaneous loss of a base and (2) removal by DNA N-glycosylase [48,49]. As the major repair enzyme of the BER pathway, APE1 recognizes the AP site and cleaves it on the 5′ side, producing a single-strand break with a 3′-hydroxyl end and a 5′-deoxyribose phosphate end (shown in Figure 3b) [50,51]. Studies have shown that increased APE1 levels lead to migration, the drug resistance of tumor cells, and reduced patient survival [15,30,52]. Differences in APE1 expression patterns between normal cells and cancer cells or inflammatory cells could serve as endogenous triggers. In DNA nanotechnology, the majority of APE1-responsive DNA systems rely on the design of AP sites that can be specifically recognized and bound by APE1. With this concept, a variety of DNA nanostructures containing AP sites have been rationally designed to be substrates for the imaging of APE1 activity.
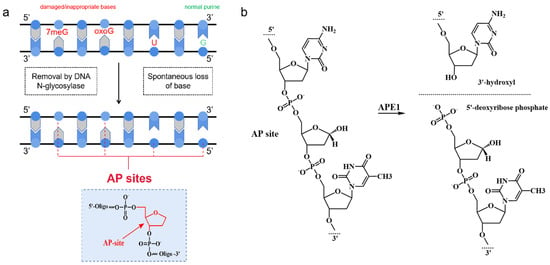
Figure 3.
(a) Origin of AP sites; (b)mechanism of cleavage of AP site by APE1.
3. DNA-Nanotechnology-Empowered Fluorescence Imaging of APE1 Activity
3.1. Linear DNA-Empowered Fluorescence Imaging of APE1 Activity
The basic DNA-based probes for intracellular biomarkers detection are linear DNA structures [53]. As it is simple to design and produce, linear DNA is frequently used as the fundamental unit for arranging different nanomaterials. According to their structural characteristics, linear DNA-based probes used for APE1 imaging can be classified as hairpin type and double-stranded linear type [54].
3.1.1. Hairpin-DNA-Based Nucleic Acid Probes
Hairpin DNA is formed by single-stranded DNA (ssDNA) molecules, which bend back to bring complementary base pairs close to each other [55,56]. It is widely used in biosensing assays, such as for the detection of proteins [57,58], metal ions [59], small molecules [60,61,62], and RNA [63,64]. Hairpin DNA probes used for APE1 imaging are typically designed with an AP site at the stem and tagged with fluorophores and quenched groups on both sides of the AP site. APE1 has the ability to cut the 5′ end of the AP site, causing the short strand to separate from the rest of the probe and making fluorescence recovery. Furthermore, APE1 can cleave the AP site to form a trigger sequence and combine with DNA signal amplification technology, such as Hybridization Chain Reaction (HCR), Catalytic Hairpin Assembly (CHA), and Rolling Circle Amplification (RCA) to achieve the signal amplification detection of APE1 activity.
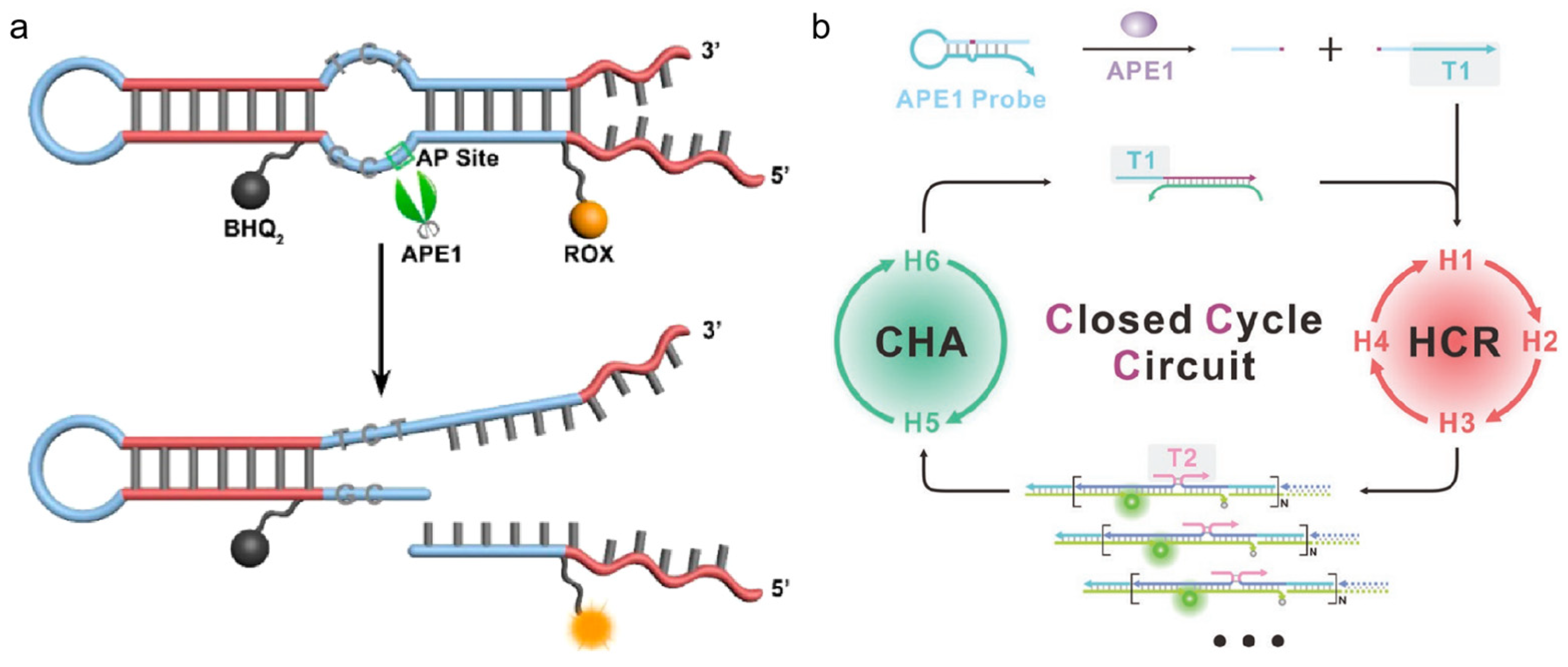
Signal reporters using hairpin DNA, based on Föster resonance energy transfer (FRET), have shown robustness for the imaging of APE1 activity [65,66,67]. Lu et al. designed a hairpin DNA probe with an AP site on the toehold section and a blunt end [68]. The presence of APE1 could expose the toehold part of the hairpin by cleaving the AP site. Then, miRNA-21 triggers the CHA, causing fluorescence recovery. With ease of use and low cost, this fluorescent biosensor would support the rapid diagnosis of APE1. To avoid severe interference by other nonspecific endonucleases and distinguish APE1 from similar nucleases, Fang et al. designed a fluorescent probe with a double loop (shown in Figure 4a) [69]. As this structure contained an AP site with a 3′ internal loop, which enabled effective differentiation between APE1 and nonspecific endonuclease, it was a practical tool for measuring APE1 activity in several biological samples with high throughput. Furthermore, DNA signal amplification techniques are one of the most effective approaches to improve detection effect, and they were in combination with hairpin DNA for APE1 imaging. Especially, the cascade integration of different amplification modes can improve amplification depth and strengthen anti-interference capacity. As shown in Figure 4b, Chai et al. employed a probe containing an AP site and triggering T1, which was complementary to the 5′-toehold of H1 [70]. APE1 could cleave the AP site and release T1. Then, T1 activated the cascade cycle of the HCR and CHA to provide the amplified signal for quantitative analysis.
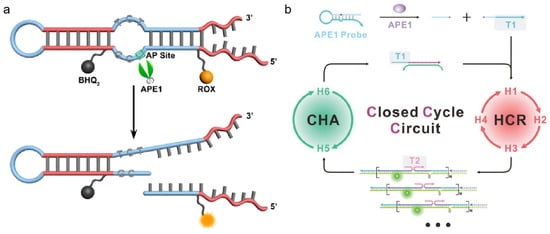
Figure 4.
Hairpin-DNA-based probe design strategies for APE1 activity imaging. (a) Schematic illustration of the fluorescent probe with a double loop for imaging of APE1 activity (reprinted with permission from Ref. [69]. Copyright 2015 American Chemical Society); (b) schematic illustration of the closed cyclic-based DNA machine for APE1 detection (reprinted with permission from Ref. [70]. Copyright 2023 Wiley-VCH GmbH).
However, problems such as the degradation of nucleases in cells and the nonspecific separation of fluorescent groups from quenched groups often result in false positive signals. In comparison to the FRET strategy, the label-free strategy preserves many benefits: (1) great specificity; (2) low time consumption; and (3) stability of the sample activity for a significantly longer period of time. Huang and co-workers used a hairpin-structured probe to detect the activity of APE1 sensitively [42]. After incision by APE1, with the help of Klenow fragment (KF) polymerase, the products could initiate polymerization. Then, it might make the hairpin structure open, resulting in double-stranded DNA (dsDNA) with an Nt.BbvCI endonuclease recognition site. By means of polymerase-catalyzed strand displacement and binding with N-methylmesoporphyrin IX (NMM), a G-rich segment was released to produce a powerful fluorescence emission. In a subsequent work, Liu et al. reported a hairpin-structured probe (HP) that contained an AP site and a circular DNA probe (CDT) as the template of RCA [41]. The primer sequence was released by APE1. Then, with the assistance of DNA polymerase and dNTPs, it quickly initiated RCA to produce long-chain DNA. As the production contained a number of repetitive G-quadruplex sequences that could combine with Thioflavin T (ThT), it generated strong fluorescent signals. This experiment successfully developed a simple, label-free, and highly sensitive fluorescence method for APE1 detection.
3.1.2. Double-Stranded Linear-DNA-Based Nucleic Acid Probes
In addition to hairpin DNA probes, double-stranded linear DNA probes are among the most widely used linear DNA probes for APE1 analysis. Simeonov et al. used a duplex probe to identify APE1 inhibitions [71,72]. Subsequently, Liu and co-workers designed a natural substrate of dsDNA probes for APE1 assay [73]. As this probe contained one uracil residue, the probes were pretreated with uracil DNA glycosylase (UDG) and then reduced with NaBH4 to create an AP site. When APE1 cleaved DNA that was labeled with fluorophore (TAMRA), it quickly dissociated, restoring fluorescence. This strategy featured rapidity and high simplicity. They used UDG rather than a directly designed AP site as substrate, which could be easily adaptable to alternative DNA repair enzymes by changing the damaged base. Conventional dsDNA-based probes are susceptible to nonspecific cleavage by other nucleases. Lu et al. developed a DNA/RNA hybrid-based fluorescent probe. This hybrid probe contained a 3′-base mismatch that could effectively enhance the activity of APE1 endonuclease. It showed good stability against other nucleases, and it made APE1 detection in complex matrices possible [74]. The hybrid probe, which allowed for the one-step detection of APE1 subcellular activity, provided a practical and potential tool for the high-throughput measurement of APE1 subcellular activity. The label-free method can also be used with dsDNA for assays of APE1 activity. Li et al. used the AP-site-binding fluorophore 2-Amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND) for APE1 activity imaging [75]. As a C base was in the opposite location, the fluorescence of ATMND was quenched. After the AP site was cleaved by APE1, the ATMND was released, and the fluorescence recovered. This label-free technique provided good sensitivity and selectivity for APE1 activity detection.
Since APE1 can specifically recognize and cleave the AP site at the duplex DNA, Li and co-workers developed a DNA system that contained a motor chain, a track strand, and four stator strands to achieve autonomous movement with high controllability and processivity [54]. As the motor-stator hybrid probe contained an AP site, it could be incised by APE1, while the ssDNA could not cleave. The cleaved short fragment products can activate the toehold domain for motor strand movement. As a proof of concept for developing gene regulators for creating cellular diagnostic tools, this DNA machine showed great potential. Although APE1 is applicable to a variety of DNA substrates containing AP sites, the relative activity of APE1 varied with the sequence. Wu’s group constructed a probe with 2-Aminopurine for APE1 activity imaging [76]. 2-Aminopurine can produce different fluorescence in different DNA microenvironments (mononucleotide > ssDNA > dsDNA). The 2-Aminopurine fluorescence at dsDNA is completely extinguished. However, it could be cut by APE1 and release a DNA fragment that contained 2-Aminopurine, in which case the fragment was digested by Exo I and released 2-Aminopurine mononucleotide to make fluorescence recovery. This strategy eliminated the influence from the little exonuclease activity of APE1 in comparison to the standard dual-terminal labeling. It was shown that the relative endonuclease activity of APE1 varied with the close or opposition base sequence. This study provided a reference for sequence design for imaging APE1 activity or APE1-mediated biosensors.
Linear DNA probes are widely used in APE1 imaging. Additionally, they are common in many DNA substrates for signal amplification. However, as a hydrophilic molecule with a negative charge, linear DNA is difficult to enter living cells in the absence of a transfection agent and vector because the cell membrane is negatively charged. In the complex cellular environment, linear DNA probes are also easily cleaved by other intracellular enzymes, causing false positives [77,78]. With the development of nanotechnology, some intelligent and multifunctional nanomaterials have become good nucleic acid probes carriers. Selected functional groups can be modified to efficiently bring large numbers of nucleic acid probes into living cells.
3.2. Composite DNA-Nanomaterials-Empowered Fluorescence Imaging of APE1 Activity
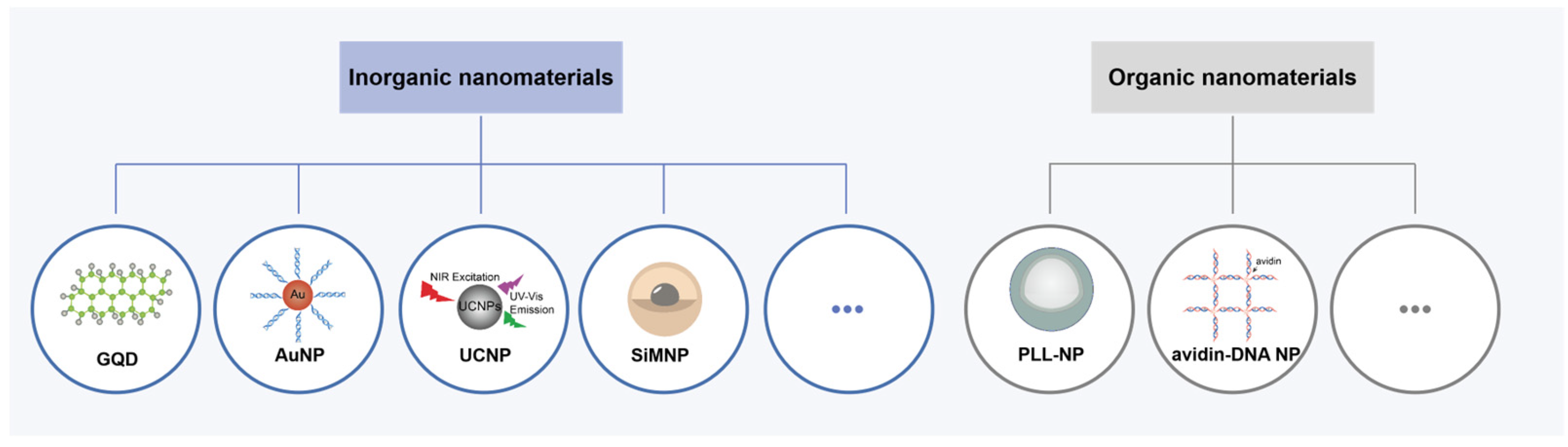
Nanomaterials have attracted much attention as carriers for APE1 probe delivery. According to Willner’s group, combining biomolecules with nanomaterials can produce new hybrid nanostructures with distinctive traits that mix the characteristics of the biomolecules and the nanomaterials [79]. DNA containing an AP site is usually linked to nanomaterials by adsorption, covalent coupling, and encapsulation to improve the stability of the probe in the plasma and further facilitate their cellular uptake [77,78,80]. According to the properties, nanomaterials are categorized into inorganic nanomaterials and organic nanomaterials (Figure 5). Inorganic nanomaterials like graphene quantum dots (GQDs), gold nanoparticles (AuNPs), lanthanide-doped upconversion nanoparticles (UCNPs), or those of other metals are desired as participants in these hybrid materials for APE1 activity imaging [81,82]. In addition, organic nanomaterials such as cationic polymer-coated nanoparticles (PLL-NPs) and proteins can also be used for the same role as they can provide predefined and biocompatible cargo-carrying and targeting capabilities [83,84].
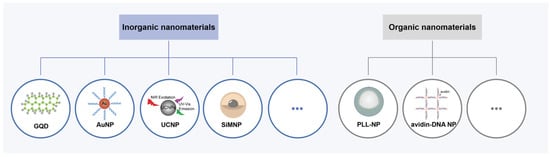
Figure 5.
Schematic illustration of inorganic nanomaterials and organic nanomaterials for APE1 activity assays.
3.2.1. DNA/Inorganic Nanomaterial Composites
GQDs are zero-dimensional graphene nanomaterials with excellent properties that are used in various applications [73,85]. Nanocomposites constructed of DNA and fine-sized GQDs can be effectively used to detect intracellular APE1 activity. Zhang et al. constructed a biocompatible functional nanocomposite, which was designed as a combination of single-molecule DNA and fine-sized GQDs for cellular APE1 imaging [86]. The GQD moiety of this nanocomposite was designed to be a quench agent that turned off the fluoresce of Cy3 in the DNA before the cleavage by APE1. In addition, it also facilitated the entry of functional nanocomposites into the cytoplasm by endocytosis. This study showed that the sensitive, selective, and rapid detection of APE1 activity in living cells was possible by utilizing GQD-based nanocomposites.
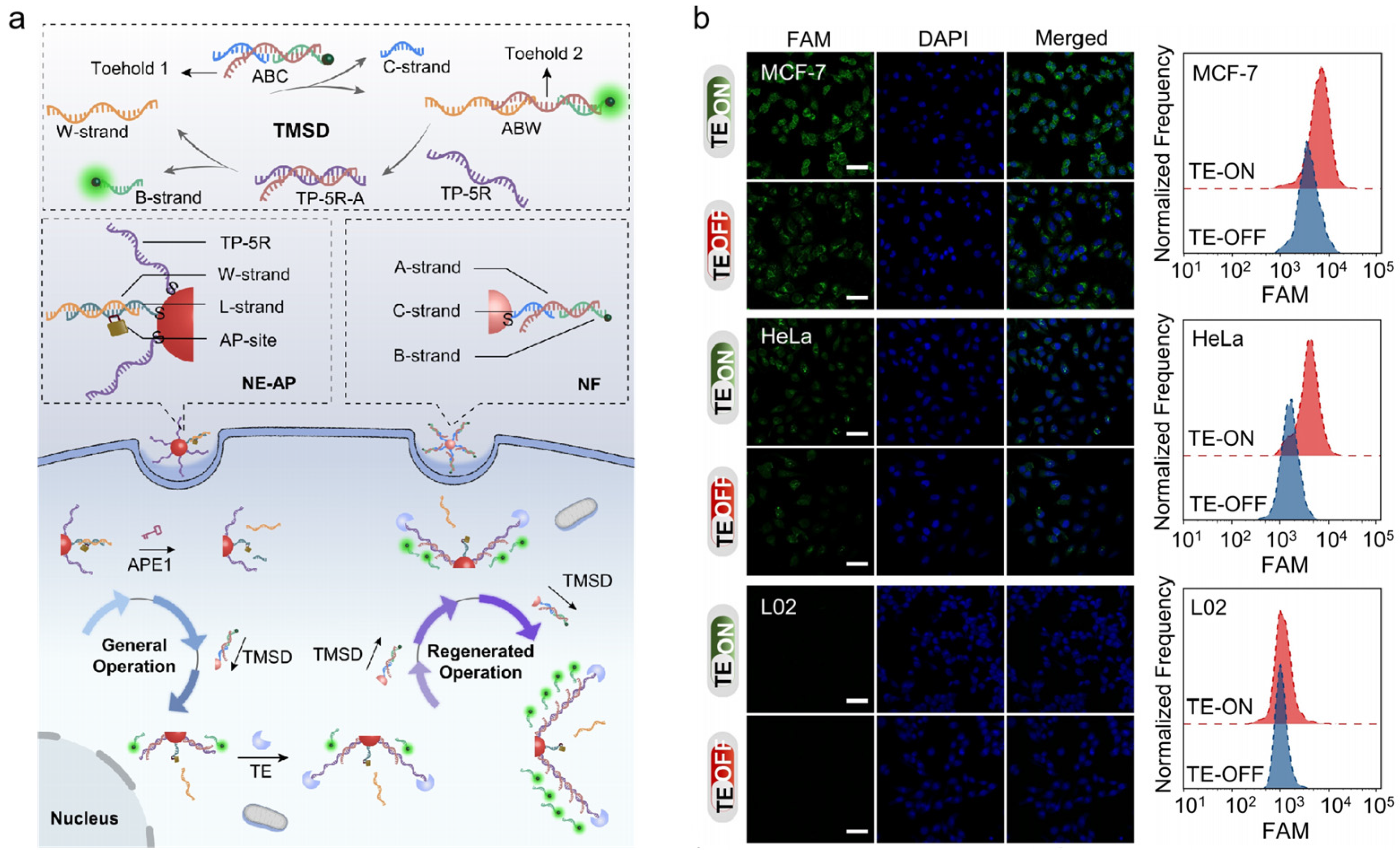
AuNPs are also widely used in the intracellular imaging of APE1 activity due to their unique properties and ease of molecular functionalization [43,44]. Cai and co-workers reported a DNA nanomachine to detect miRNA-21 and APE1 concurrently [87]. The detecting device had two operating modes: OR and AND. The existence of any target could result in fluorescence in the OR mode, which was suited for sensitive detection. In the AND mode, fluorescence could be produced by the presence of all targets, which was more suited for precise detection. It had potential in medical application and provided reference for further biomarker detection. In another study, as shown in Figure 6a, Zhang et al. presented a DNA nanomachine for the signal amplification imaging of APE1 in live cells [88]. APE1 served as a switch in this DNA nanomachine to get it moving, and telomerase served as an engineer to continually expand new tracks. They constructed a nanoeffector (NE-AP) and a nanofueler (NF). Since the NE-AP contained double-stranded DNA containing an AP site, a walker strand (W-strand) could be released to make a toehold-mediated strand displacement (TMSD) reaction occur with the presence of APE1. The W-strand could be recycled between the NE-AP and NF for signal amplification. At the same time, the track strand TP-5R could be continually extended by TE, significantly creating magnified fluorescence signals under the NF. The fluorophore-labeled DNA strand was continuously released while the walker walked, enhancing fluorescence signals for APE1 imaging in different cell types (shown in Figure 6b). Compared to traditional nanomachines, this nanomachine performs regenerative motion and shows higher specificity and sensitivity.
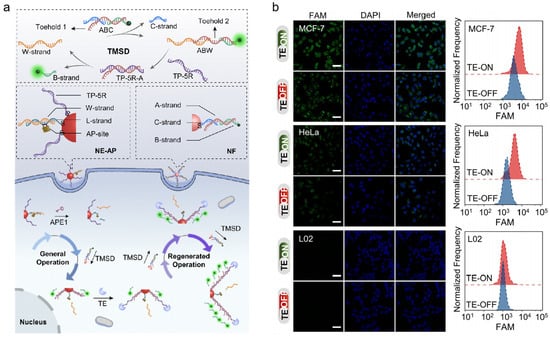
Figure 6.
(a) Schematic illustration of the TE-based DNA nanomachine (NE-AP/NF) for imaging of intracellular APE1 activity; (b) effect of NE-AP/NF probes on imaging of APE1 in different cells. (Reprinted with permission from Ref. [88]. Copyright 2022 Wiley-VCH GmbH).
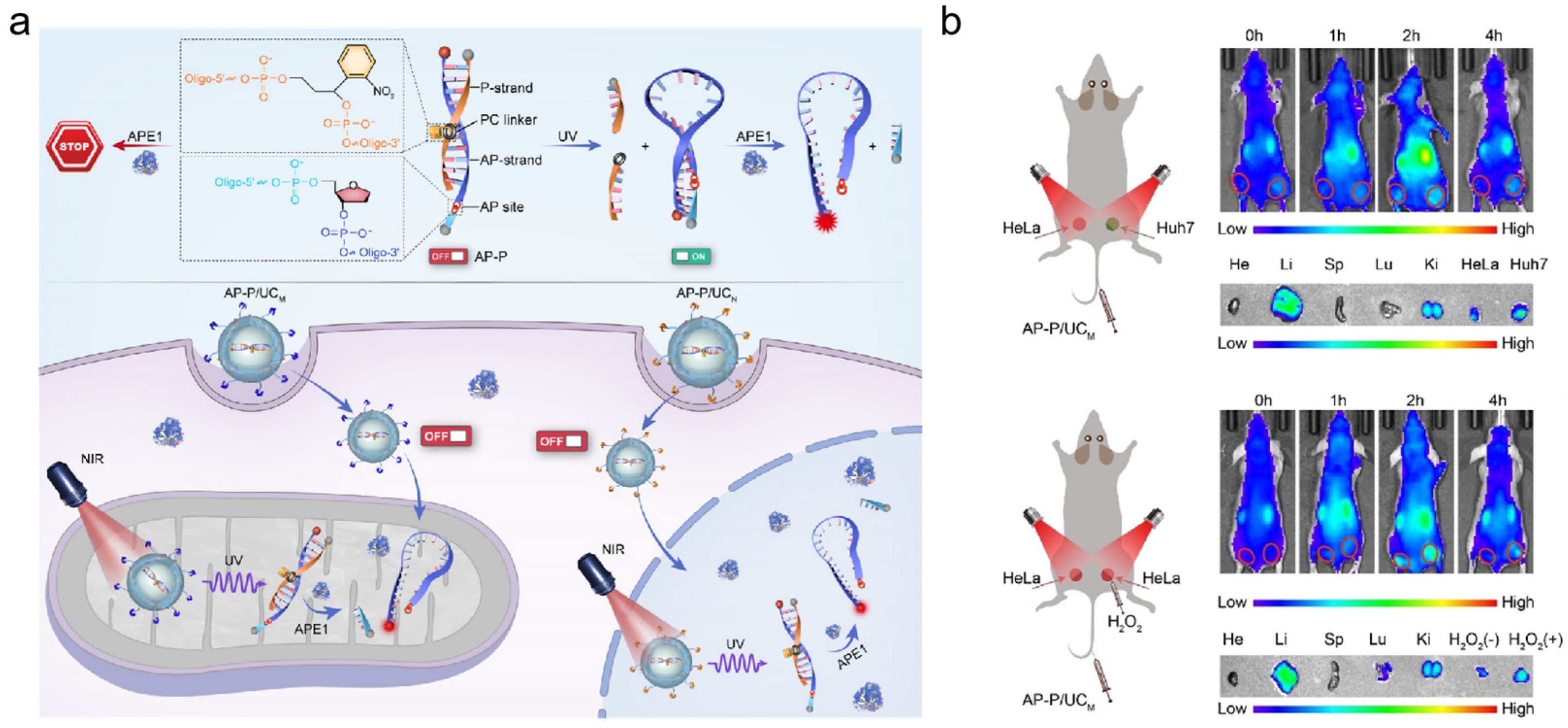
In recent years, UCNPs have received a lot of attention to offer spatial and temporal control of APE1 activity detection. They can be activated in the biological window of the near-infrared (650–1700 nm) for upconversion luminescence (UCL), which results in deeper penetration and reduced bioabsorption [89,90]. As the cytoplasmic/nuclear distribution of APE1 is crucial for tumor initiation and metastasis [91], it is important to visualize APE1 at high subcellular resolution. Li’s group introduced a sensor technology that allowed in situ localization and photoactivation imaging of APE1 in the nucleus as well as mitochondria (shown in Figure 7a) [92]. The P-strand contained a photocleavable (PC) linker, and it was used to hybridize the AP-strand to block the fluorescent sequence of the target APE1. The UCNPs were used as carriers of a DNA probe (AP-P) to improve the capability to penetrate deep tissue. This detection probe, the AP-P, could effectively eliminate potential false-positive signals. They also evaluated the potential of the photoactivatable imaging of subcellular APE1 in vivo (shown in Figure 7b). They established a bilateral tumor model. As the mitochondrial APE1 of Huh7 cells was higher than that of HeLa cells, the Huh7 tumor showed higher fluorescence intensity than the HeLa tumor. Additionally, to demonstrate the effectiveness of the nanosensor in monitoring the re-localization of APE1, the tumors pretreated with H2O2 exhibited a significantly enhanced fluorescence intensity. Then, Li’s group presented a nanoplatform (UR-HAPT) for the dynamic imaging of mitochondrial APE1 [93]. It was constructed by the combination of a UCNP, a rose bengal (RB)-modified UCNP, triphenyllphosphonium (TPP) ligands, and a DNA probe (HAP). Upon the TPP targeting, the nanostructure entered the mitochondria. During photodynamic therapy (PDT), the NIR light irradiation induced subcellular ROS production by the energy transfer from the UCNPs to the photosensitizer’s RB-modified UCNP, which increased the mitochondrial APE1 expression. Then, APE1 cleaved the AP site in HAP and led to a recovery of fluorescence signal. Since PDT could increase the content of APE1 in mitochondria, this system could be used as an index to evaluate the efficacy of PDT in real time. Additionally, this nanoplatform possessed an in vivo biosensing capability for monitoring mitochondrial APE1 dynamics.
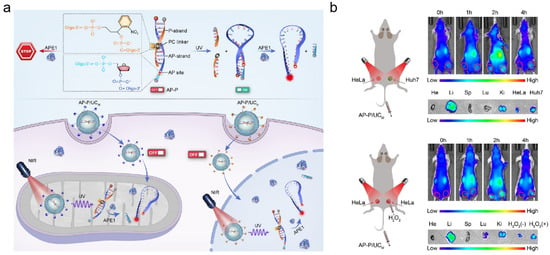
Figure 7.
DNA/UCNPs design strategies for APE1 activity imaging. (a) Schematic illustration of the design of the DNA nanosensor for photoactivation imaging of APE1 in the nucleus as well as mitochondria; (b) photoactivated imaging of mitochondrial APE1 in vivo. (Reprinted with permission from Ref. [92]. Copyright 2021 Wiley-VCH GmbH).
For intracellular diagnosis or genetic treatment, silica-coated magnetic core-shell NPs (SiMNPs) have also been employed to carry APE1-DNA probes into different types of cells [94,95]. As SiMNPs completely inhibit the interaction between DNA and nucleases, including APE1 [96], it is not feasible to use SiMNPs only for APE1 activity imaging. Excitingly, interactions with avidin allow it to attract APE1 to DNA on nanoparticles. This allowed avidin-modified SiMNPs to be used for APE1 imaging. Zhai et al. demonstrated that APE1 might precisely and effectively cleave the AP site in DNA, which links to the avidin-modified surface of SiMNPs. It could, thus, resist nonspecific cleavage by other enzymes [97]. This study demonstrated the potential application of avidin in the detection of nuclease activity.
3.2.2. DNA/Organic Nanomaterial Composites
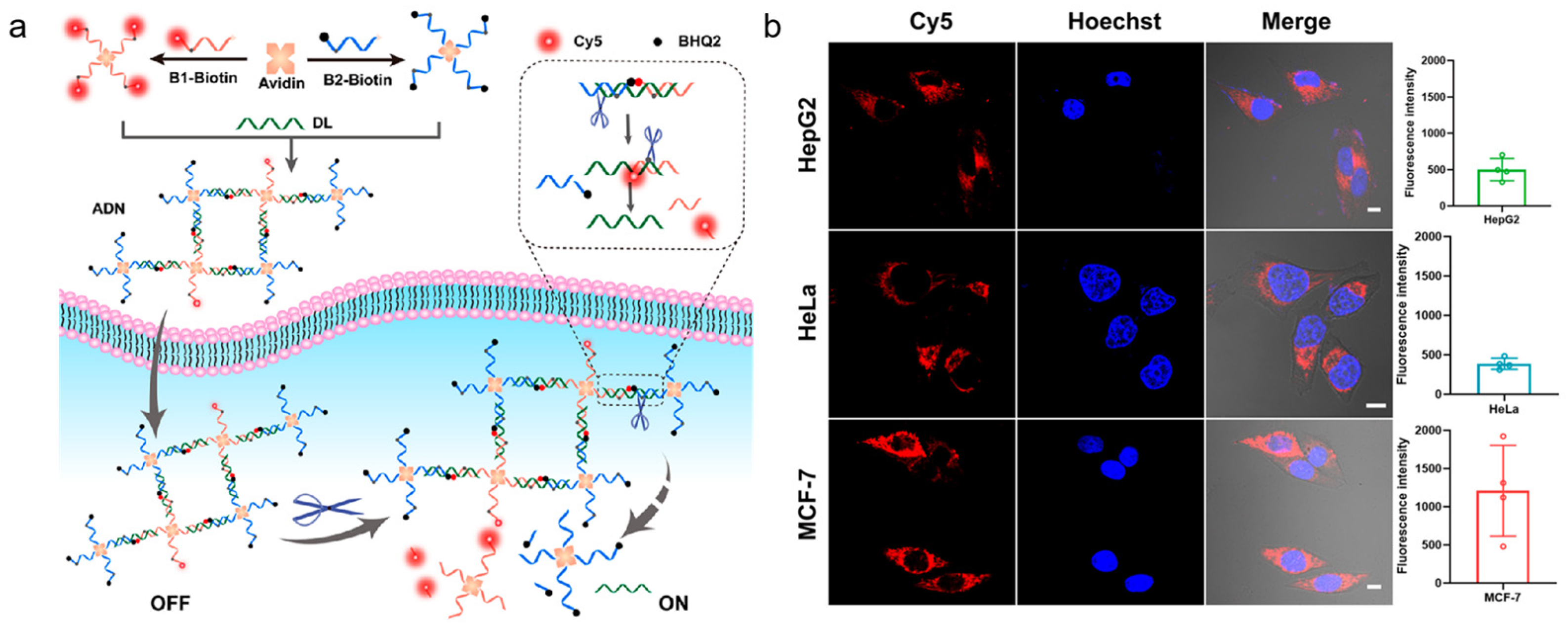
It is difficult for naked DNA probes to penetrate the cell membrane because both DNA and biological cell membranes are negatively charged. However, cationic polymer-coated NPs can be employed for the delivery of DNA probes as well as for stability improvement. Li and co-workers designed activatable DNA nanoparticles for the imaging of cytochrome c and APE1 [98]. The nanoparticles coated with cationic polymer were used for DNA probe delivery. After the AP site was cleaved by APE1, it led to the restoration of the aptamer recognition ability of cytochrome c. This method allowed for the imaging of cytochrome c and APE1 with a DNA probe, which provided a basis for the detection of biological functions of related proteins. Inspired by the strong interaction between avidin and APE1, Li et al. proposed a DNA nanostructure, which consisted of a protein (avidin) as a connecting structure, for imaging the activity of APE1 in different live cells (shown in Figure 8) [99]. The avidin-scaffolded DNA nanostructure, which was prepared based on the interaction between avidin and biotin, had a relatively loose structure with little steric hindrance. It favored APE1 entry into the AP sites. APE1 cleaved the nanostructure effectively and precisely, resulting in activated fluorescence for APE1 detection. By assembling an avidin-scaffolded nanostructure, it could effectively improve the specificity of APE1 detection and reduce nonspecific cleavage by other nucleases.
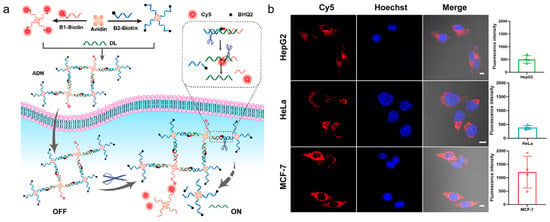
Figure 8.
DNA/organic nanomaterial composite design strategies for imaging of APE1 activity. (a) Schematic illustration of the avidin-scaffolded DNA nanostructure for live-cell APE1 imaging; (b) confocal fluorescence images for different cells treated with avidin-scaffolded DNA nanostructure. (Reprinted with permission from Ref. [99]. Copyright 2023 American Chemical Society).
Organic and inorganic nanomaterials can be used to effectively improve the stability of APE1 probes in the plasma and further facilitate their cellular uptake. On the other hand, composite DNA nanomaterials combine APE1-responsive DNA systems with extra functions of nanomaterials to improve the performance in APE1 imaging [78,100]. Despite the rapid development in composite DNA nanomaterials, the fabrication process of nanomaterials is complex and time-consuming. Differently from these nanomaterials, DNA possesses many unique properties, including biocompatibility, programmability, ease of synthesis, and controllability on the nanoscale [101,102,103]. Because of the self-assembly ability of DNA, three-dimensional (3D) nanostructures, such as a DNA tetrahedron [104], DNA polyhedron [105], 3D DNA origami [106], and DNA nanoflowers [107,108], can be created for APE1 activity imaging [109].
3.3. 3D DNA-Nanostructure-Empowered Fluorescence Imaging of APE1 Activity
A strong hydrogen bond holds the 3D DNA nanostructure together. It has great qualities such as (1) biocompatibility and nontoxicity; (2) precise size and shape management; and (3) facile target-decorating in numerous places [110]. Currently, 3D DNA nanostructures for APE1 activity imaging include DNA prisms and DNA tetrahedrons [45]. Herein, we concentrate on the usage of a DNA tetrahedron for APE1 activity imaging since tetrahedrons are extensively employed because of their high productivity and rigid structures [111].
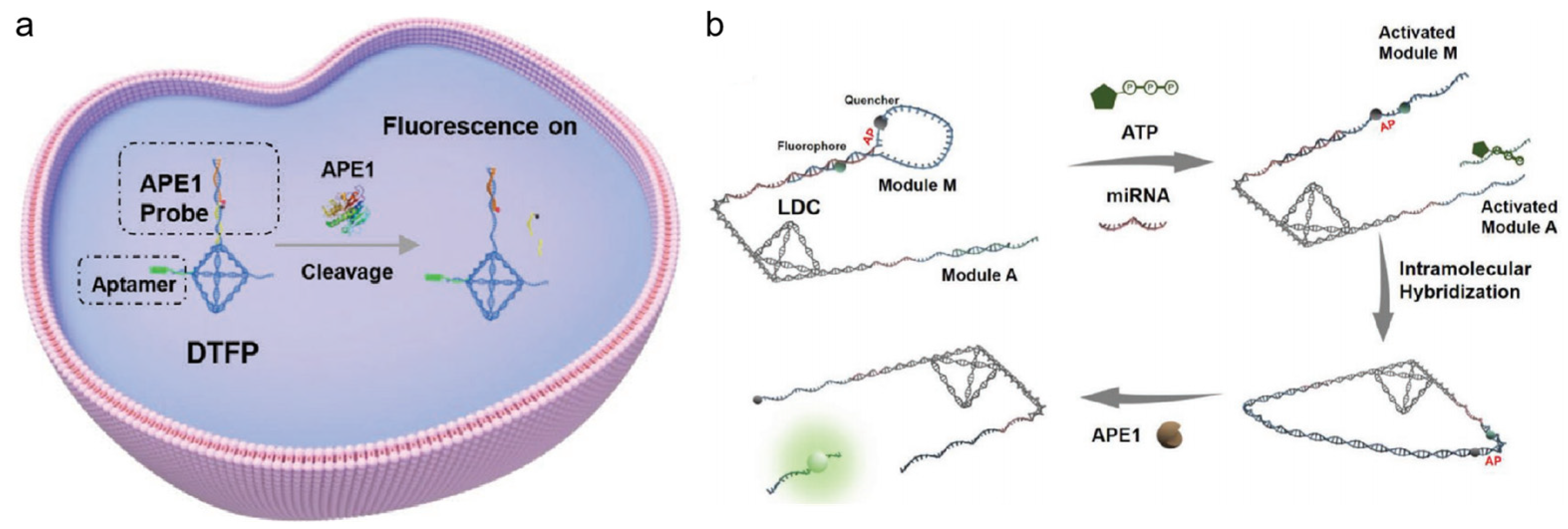
A DNA tetrahedron is typically formed by four ssDNA strands. The four strands can be arranged to create a tetrahedral frame structure with six double-helix edges. Due to their superior mechanical rigidity and structural stability, DNA tetrahedrons make excellent molecular scaffolds [112,113]. Moreover, DNA tetrahedrons have multiple sites for targeting decoration [114,115], and functional nucleic acids can be designed and constructed in DNA tetrahedrons with superior properties for APE1 imaging. Su’s group reported a tumor-aptamer-modified and AP-site-contained DNA tetrahedrons probe (DTFP) for fluorescent imaging of APE1 activity (shown in Figure 9a) [116]. The AP site of the DNA could be cleaved by APE1, and short chains containing fluorophores could be released. As platinum-resistant cell lines with down-regulated APE1 expression were highly sensitive to platinum treatment, the potential to overcome this drug-resistance was demonstrated. Compared to traditional single-molecule biomarker assays, the use of multi-target detection can effectively enhance targeting accuracy. Moreover, the four overhangs of the tetrahedral structure make the design more flexible and programmable. Additionally, the compact tetrahedral structure can link a large number of recognition chains with a large local concentration, thus greatly increasing the reaction rate. In Su’s group, Kou et al. created the intramolecular hybridization of localized DNA circuits (LDCs) with excellent sensitivity and quick reaction times, which could concurrently detect intracellular miRNAs, APE1, and ATP (shown in Figure 9b) [117]. To carry out an AND logic gate operation, three response elements were positioned on the DNA tetrahedron nanostructure. The release of a fluorescence signal was dependent on the coexistence of miRNA and APE1 with a significant amount of ATP. This discovery offered a potent tool for accurate diagnostics and substantially enhanced the performance of DNA-based nanodevices.
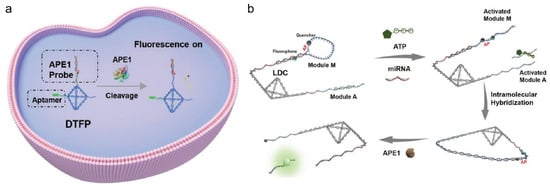
Figure 9.
(a) Schematic illustration of aptamer-modified DNA tetrahedron-based nanostructure (DTFP) for APE1 activity detection in cancer cells. (Reprinted with permission from Ref. [116]. Copyright 2022 Wiley-VCH GmbH.); (b) Schematic illustration of LDC working principle (reprinted with permission from Ref. [117]. Copyright 2022 Wiley-VCH GmbH).
Additionally, the high-efficiency amplification technology provides a robust tool for the sensitive detection of APE1, which can improve accurate diagnoses. Meanwhile, as a disease biomarker, APE1 can be used to release small interfering RNAs (siRNAs) for precise treatment. Two kinds of functionalized tetrahedral DNA nanostructures (f-TND1 and f-TND2) were constructed to form a DNA nanostructure by Zhou and co-workers [118]. f-TND1 contained tetrahedral DNA and two hairpin DNA probes, H1 and H2, while f-TND2 contained tetrahedral DNA, H1, and H3. As there were aptamer Sgc8 and AP sites in H1, Cy5, BHQ2, and siRNA in H2, APE1 could cause the AP sites to split, releasing S1 to start CHA reactions on two tetrahedral DNA nanostructures. It then created a massive network of nanostructures that significantly increased the production of fluorescent signals, enabling sensitive detection and intracellular imaging of APE1. To fulfill the goal of targeted tumor therapy, siRNA produced during the CHA response may inhibit cell survival. This work offered a superb and efficient nanoplatform for the accurate gene treatment of cancers, sensitive biomarker detection, and disease-specific diagnostics.
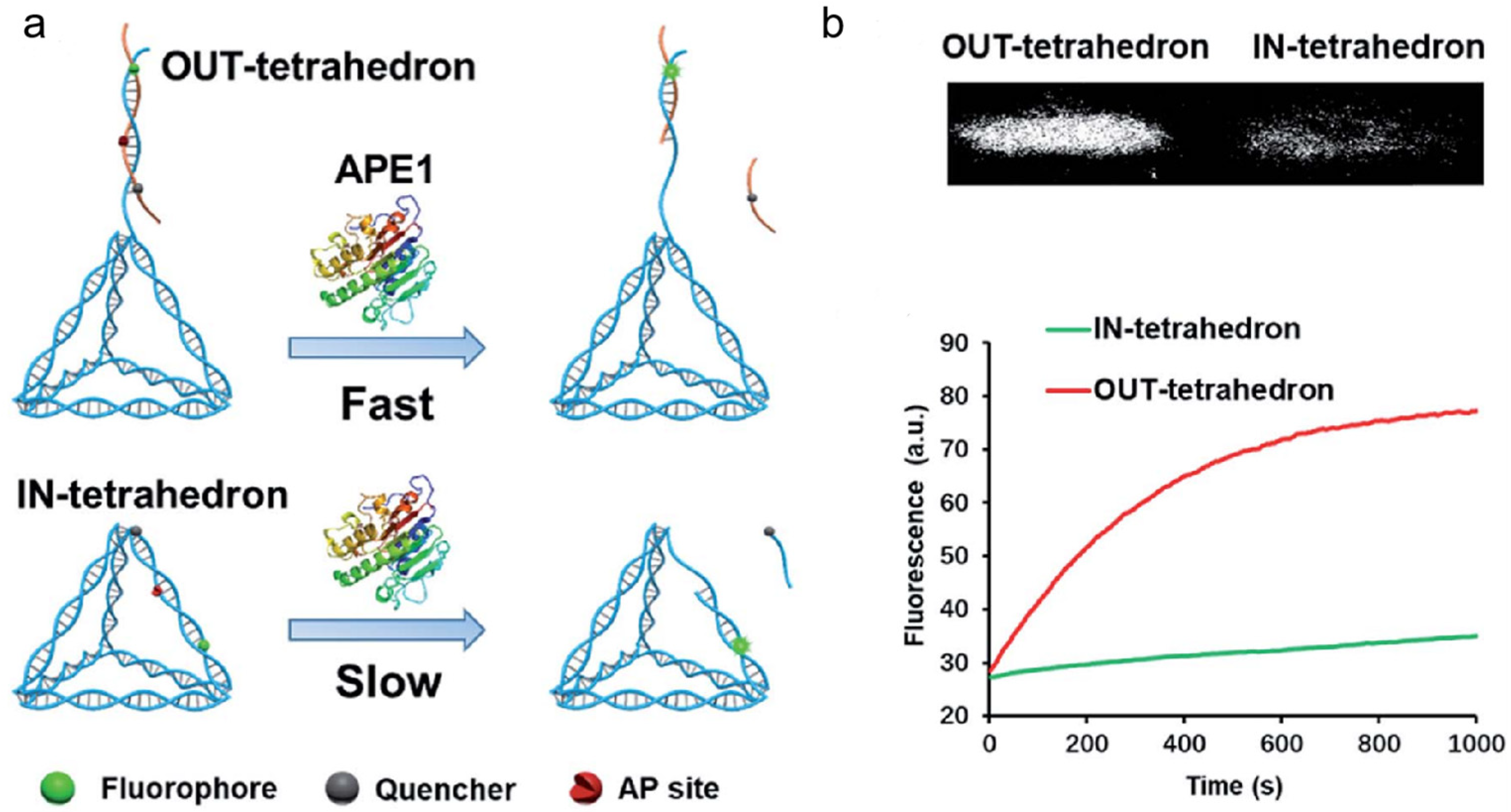
APE1 is also a desirable target for the development of anticancer drugs. Various small-molecule APE1 inhibitors have been studied in recent years to improve anticancer treatments. Small-molecule drugs, however, are typically linked to resistance and adverse effects [119]. DNA tetrahedrons have been widely used in the delivery of siRNA and small-molecule drugs, such as platinum drugs and Dox. Interestingly, DNA tetrahedrons can regulate APE1 activity. Using DNA tetrahedrons, Zhang et al. reported a unique method for examining and controlling cellular APE1 activity (shown in Figure 10) [120]. Contrary to typical molecular probes and inhibitors, DNA tetrahedrons function as both probes and inhibitors, and their functions may be changed by modifying the AP site. The tetrahedron with an antenna that contains an AP site demonstrated good APE1 sensitivity and specificity. On the other hand, the tetrahedron with an AP site on its scaffold displayed a substantial affinity for APE1 binding. This research offered a unique strategy for controlling enzyme activity as well as a fresh understanding of how enzymes and substrates interact. It had potential implications in the inhibition of APE1 activity and the treatment of cancer.
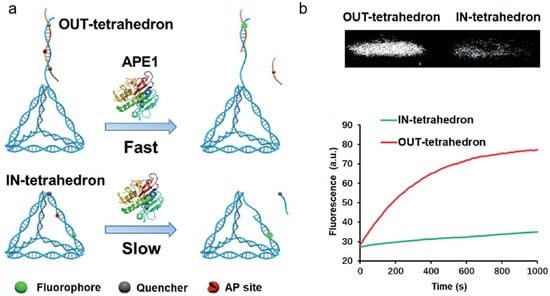
Figure 10.
(a) Schematic illustration of the OUT-tetrahedron and IN-tetrahedron DNA nanostructures in the APE1 enzymatic reaction; (b) PAGE analysis of the reaction products of the OUT-tetrahedron and IN-tetrahedron nanostructures with APE1; real-time monitoring of the reaction of the two DNA nanostructures. (Reprinted with permission from Ref. [120]. Copyright 2019 Royal Society of Chemistry).
All three of the above probes have been reviewed for the imaging of APE1 activity. In fact, linear DNA probes, composite DNA nanomaterials, and 3D DNA nanostructures can also be combined for APE1 imaging. Lv and co-workers described a stochastic bipedal DNA walker that traveled on GNPs by itself via the CHA reaction [121]. They designed a hairpin chain containing the AP site to serve as a substrate for APE1 and a stochastic bipedal DNA walker that autonomously traversed on GNPs. To improve the internalization efficiency, lipofectamine 3000 encapsulated probes were incubated with cells. This allowed for the very sensitive, activatable fluorescence imaging of intracellular BER activity. Additionally, to precisely monitor intracellular APE1 activity, Zhou et al. created the DNAzyme-modified tetrahedral DNA nanostructure (DZ-TDN) and a molecular beacon (MB)-integrated biosensing system [122]. Thanks to liposomes, the device can be successfully internalized into live cells to monitor changes in intracellular APE1 activity. In 2022, APE1 was successfully detected and imaged intracellularly by Zhou and co-workers. They used a tetrahedral DNA walker with four arms that moved autonomously on the GNPs via CHA. With the help of lipofectamine 3000, the DNA nanomaterials were transfected into tumor cells [123]. This design offered a unique DNA walker for the sensitive detection of APE1.
4. Discussion and Perspectives
In this review, we summarized recent advances in the DNA-nanotechnology-empowered fluorescence imaging of APE1 activity. APE1, which is important in the BER pathway and in the regulation of redox signaling in cells, is an essential multifunctional protein in mammals. The abnormal expression and disruptions in the biological functions of APE1 are linked to a variety of diseases. Since it can specifically recognize and cleave AP sites in DNA strands, DNA nanotechnology is commonly used for APE1 imaging. Linear-DNA-based nucleic acid probes are the most basic type of nucleic acid probes used for APE1 imaging. However, since naked linear DNA probes are small and have a negatively charged backbone, they exhibit ineffective cellular internalization. Also, they are quickly broken down once within cells, preventing them from arriving at the target areas. In order to transport DNA probes intracellularly and achieve a desired performance in bioanalysis and cancer therapies, nanocarriers have been widely employed. Despite the unique properties of these nanomaterials, the fabrication process of nanomaterials is complex and time-consuming, which severely restricts their practical applicability. With the rapid development of DNA nanotechnology, the advantages of the programmability and high biocompatibility of DNA have spurred extensive research on the design and assembly of a variety of 3D nanostructures with well-defined structures for the delivery of AP sites in recent years.
Despite current achievements, there is still plenty of room in the DNA-nanotechnology-empowered fluorescence imaging of APE1 activity. Firstly, complex biological matrices may seriously interfere with the selectivity of probes for imaging APE1 in living cells and in vivo, causing false-positive results. Thus, the specificity of APE1 detection can be improved using the following strategies: (1) introducing a small loop structure to reduce nonspecific cleavage by other nucleases in the cell or replacing traditional dsDNA with DNA/RNA probes; (2) using more complex DNA nanostructures. Secondly, the use of DNA nanotechnology for imaging APE1 is mainly based on its endonuclease activity. The measurement of APE1 expression in cells is also important for understanding disease progression. APE1 aptamer may be an excellent tool for APE1 assays [124]. For example, Qin et al. used APE1 aptamer to inhibit APE1, and they designed a partial complementary strand to enable APE1 release and activity restoration [125]. Thirdly, the cytoplasmic/nuclear distribution of APE1 is crucial for tumor initiation and metastasis. For example, mitochondrial APE1 may reflect the level of cellular response to oxidative stress [94], so it is important to visualize APE1 at high subcellular resolution.
Author Contributions
Conceptualization, investigation, and writing—original draft preparation, H.H.; conceptualization and investigation, X.L. and Y.W.; investigation, L.Q., J.H., Y.Z. and J.Z.; general guidance, project directing, and manuscript revisions, K.W. and X.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of China (22174044).
Data Availability Statement
Data available in a publicly accessible repository and cited in accordance with journal guidelines.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Izumi, T.; Mitra, S. Deletion analysis of human AP-endonuclease: Minimum sequence required for the endonuclease activity. Carcinogenesis 1998, 19, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.J.; Rodriguez, J.A.; Banuelos, S. Molecular mechanisms regulating the DNA repair protein APE1: A focus on its flexible N-terminal tail domain. Int. J. Mol. Sci. 2021, 22, 6308. [Google Scholar] [CrossRef] [PubMed]
- Demple, B.; Herman, T.; Chen, D.S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: Definition of a family of DNA repair enzymes. Proc. Natl. Acad. Sci. USA 1991, 88, 11450–11454. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.T.; Coutinho, L.G.; de Oliveira, L.O.A.; de Souza Timoteo, A.R.; Farias, G.C.; Agnez-Lima, L.F. APE1/Ref-1 role in inflammation and immune response. Front. Immunol. 2022, 13, 793096. [Google Scholar] [CrossRef] [PubMed]
- Barnes, T.; Kim, W.C.; Mantha, A.K.; Kim, S.E.; Izumi, T.; Mitra, S.; Lee, C.H. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009, 37, 3946–3958. [Google Scholar] [CrossRef]
- Tell, G.; Pellizzari, L.; Cimarosti, D.; Pucillo, C.; Damante, G. Ref-1 controls pax-8 DNA-binding activity. Biochem. Biophys. Res. Commun. 1998, 252, 178–183. [Google Scholar] [CrossRef]
- Berquist, B.R.; McNeill, D.R.; Wilson III, D.M. Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J. Mol. Biol. 2008, 379, 17–27. [Google Scholar] [CrossRef]
- Cohen, I. DNA damage talks to inflammation. Cytokine Growth Factor Rev. 2017, 33, 35–39. [Google Scholar] [CrossRef]
- Zou, G.M.; Luo, M.H.; Reed, A.; Kelley, M.R.; Yoder, M.C. Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood 2007, 109, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Fishel, M.L.; He, Y.; Reed, A.M.; Chin-Sinex, H.; Hutchins, G.D.; Mendonca, M.S.; Kelley, M.R. Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair 2008, 7, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Fishel, M.L.; Jiang, Y.; Rajeshkumar, N.V.; Scandura, G.; Sinn, A.L.; He, Y.; Shen, C.; Jones, D.R.; Pollok, K.E.; Ivan, M.; et al. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol. Cancer Ther. 2011, 10, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Dianov, G.; Lindahl, T. Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 1994, 4, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Schermerhorn, K.M.; Delaney, S. A chemical and kinetic perspective on base excision repair of DNA. Acc. Chem. Res. 2014, 47, 1238–1246. [Google Scholar] [CrossRef]
- Zou, G.M.; Karikari, C.; Kabe, Y.; Handa, H.; Anders, R.A.; Maitra, A. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: Therapeutic implications in tumor angiogenesis. J. Cell. Physiol. 2009, 219, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Xu, M.; Wang, Y.; Xu, J.; Yuan, Y. An APE1 inhibitor reveals critical roles of the redox function of APE1 in KSHV replication and pathogenic phenotypes. PLoS Pathog. 2017, 13, e1006289. [Google Scholar] [CrossRef] [PubMed]
- Pines, A.; Perrone, L.; Bivi, N.; Romanello, M.; Damante, G.; Gulisano, M.; Kelley, M.R.; Quadrifoglio, F.; Tell, G. Activation of APE1/Ref-1 is dependent on reactive oxygen species generated after purinergic receptor stimulation by ATP. Nucleic Acids Res. 2005, 33, 4379–4394. [Google Scholar] [CrossRef] [PubMed]
- Mohni, K.N.; Wessel, S.R.; Zhao, R.; Wojciechowski, A.C.; Luzwick, J.W.; Layden, H.; Eichman, B.F.; Thompson, P.S.; Mehta, K.P.M.; Cortez, D. HMCES maintains genome integrity by shielding abasic sites in single-strand DNA. Cell 2019, 176, 144–153. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Lindahl, T.; Nyberg, B. Rate of depurination of native deoxyribonucleic acid. Biochemistry 1972, 11, 3610–3618. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Wong, R.S.; McKnight, J.N.; Bowman, G.D.; Greenberg, M.M. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc. Natl. Acad. Sci. USA 2010, 107, 22475–22480. [Google Scholar] [CrossRef]
- Hoitsma, N.M.; Whitaker, A.M.; Beckwitt, E.C.; Jang, S.; Agarwal, P.K.; Houten, B.V.; Freudenthal, B.D. AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop. Nucleic Acids Res. 2020, 48, 7345–7355. [Google Scholar] [CrossRef] [PubMed]
- Drohat, A.C.; Coey, C.T. Role of base excision “repair” enzymes in erasing epigenetic marks from DNA. Chem. Rev. 2016, 116, 12711–12729. [Google Scholar] [CrossRef]
- Lindahl, T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 1974, 71, 3649–3653. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Nash, R.A.; Klungland, A.; Schar, P.; Barnes, D.E.; Lindahl, T. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 1996, 15, 6662–6670. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Demple, B. Knockout and inhibition of Ape1: Roles of Ape1 in base excision DNA repair and modulation of gene expression. Antioxidants 2022, 11, 1817. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, S.; Xing, D. New horizons for the roles and association of APE1/Ref-1 and ABCA1 in atherosclerosis. J. Inflamm. Res. 2021, 14, 5251–5271. [Google Scholar] [CrossRef]
- Weaver, T.M.; Hoitsma, N.M.; Spencer, J.J.; Gakhar, L.; Schnicker, N.J.; Freudenthal, B.D. Structural basis for APE1 processing DNA damage in the nucleosome. Nat. Commun. 2022, 13, 5390. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, A.M.; Freudenthal, B.D. APE1: A skilled nucleic acid surgeon. DNA Repair 2018, 71, 93–100. [Google Scholar] [CrossRef]
- Fan, Z.; Beresford, P.J.; Zhang, D.; Xu, Z.; Novina, C.D.; Yoshida, A.; Pommier, Y.; Lieberman, J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 2003, 4, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Hong, J.; Peng, D.; Bhat, A.A.; Chen, Z.; Zaika, A.; El-Rifai, W.M. A new function of APE1 in barrett’s esophagus and esophageal adenocarcinoma: APE1 upregulates MMP2 and MMP14 to promote invasion. Gastroenterology 2017, 152, S237. [Google Scholar] [CrossRef]
- Antoniali, G.; Serra, F.; Lirussi, L.; Tanaka, M.; D’Ambrosio, C.; Zhang, S.; Radovic, S.; Dalla, E.; Ciani, Y.; Scaloni, Y.; et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat. Commun. 2017, 8, 797. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, X.; Lu, X.; Dai, N.; Zhang, S.; Cheng, Y.; Zhang, L.; Yang, Y.; Liu, Y.; Yang, Z.; et al. APE1 deficiency promotes cellular senescence and premature aging features. Nucleic Acids Res. 2018, 46, 5664–5677. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shi, W.; Li, X.; Ma, H. Recognition moieties of small molecular fluorescent probes for bioimaging of enzymes. Acc. Chem. Res. 2019, 52, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Dischler, A.M.; Maslar, D.; Zhang, C.; Qin, Y. Development and characterization of a red fluorescent protein-based sensor RZnP1 for the detection of cytosolic Zn2+. ACS Sens. 2022, 7, 3838–3845. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Willner, B.; Willner, I. DNA nanotechnology: From sensing and DNA machines to drug-delivery systems. ACS Nano 2013, 7, 8320–8332. [Google Scholar] [CrossRef]
- Xie, N.; Huang, J.; Yang, X.; Yang, Y.; Quan, K.; Ou, M.; Fang, H.; Wang, K. Competition-mediated FRET-switching DNA tetrahedron molecular beacon for intracellular molecular detection. ACS Sens. 2016, 1, 1445–1452. [Google Scholar] [CrossRef]
- Lei, Y.; Qian, Z.; Tang, J.; He, X.; Shi, H.; Ye, X.; Yan, L.; He, D.; Wang, K. DNA nanotriangle-scaffolded activatable aptamer probe with ultralow background and robust stability for cancer theranostics. Theranostics 2018, 8, 4062–4071. [Google Scholar] [CrossRef]
- Zou, S.; Lei, Y.; Ma, W.; Chen, B.; Cheng, H.; Jia, R.; Li, Z.; He, X.; Wang, K. Extracellular pH-manipulated in situ reconfiguration of aptamer functionalized DNA monomer enables specifically improved affinity, detection and drug delivery. Analyst 2020, 145, 2562–2569. [Google Scholar] [CrossRef]
- Huang, J.; Wu, Y.; He, H.; Ma, W.; Liu, J.; Cheng, H.; Sun, H.; He, X.; Wang, K. Acidic microenvironment triggered in situ assembly of activatable three-arm aptamer nanoclaw for contrast-enhanced imaging and tumor growth inhibition in vivo. Theranostics 2022, 12, 3474–3487. [Google Scholar] [CrossRef]
- Wang, F.; Willner, B.; Willner, I. DNA nanotechnology with one-dimensional self-assembled nanostructures. Curr. Opin. Biotechnol. 2013, 24, 562–574. [Google Scholar] [CrossRef]
- Liu, B.; Yang, Z.; Huang, T.; Li, M.M.; Duan, W.; Xie, B.; Chen, J.X.; Dai, Z.; Chen, J. Label-free and highly sensitive APE1 detection based on rolling circle amplification combined with G-quadruplex. Talanta 2022, 244, 123404. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ma, Y.; Li, Y.; Xiong, M.; Li, X.; Zhang, L.; Zhao, S. Sensitive and label-free fluorescence detection of apurinic/apyrimidinic endonuclease 1 activity based on isothermal amplified-generation of G-quadruplex. New J. Chem. 2017, 41, 1893–1896. [Google Scholar] [CrossRef]
- Jans, H.; Huo, Q. Gold nanoparticle-enabled biological and chemical detection and analysis. Chem. Soc. Rev. 2012, 41, 2849–2866. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.; Wright, D.W. Size-dependent cellular uptake of DNA functionalized gold nanoparticles. Small 2016, 12, 5592–5600. [Google Scholar] [CrossRef]
- Liang, M.; Li, N.; Liu, F.; Zeng, N.; Yu, C.; Li, S. Apurinic/apyrimidinic endonuclease triggered doxorubicin-releasing DNA nanoprism for target therapy. Cell Cycle 2022, 21, 2627–2634. [Google Scholar] [CrossRef] [PubMed]
- Atamna, H.; Cheung, I.; Ames, B.N. A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. USA 2000, 97, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Khodyreva, S.; Lavrik, O. Non-canonical interaction of DNA repair proteins with intact and cleaved AP sites. DNA Repair 2020, 90, 102847. [Google Scholar] [CrossRef]
- Abe, Y.S.; Sasaki, S. DNA cleavage at the AP site via β-elimination mediated by the AP site-binding ligands. Bioorg. Med. Chem. 2016, 24, 910–914. [Google Scholar] [CrossRef]
- Boiteux, S.; Guillet, M. Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 2004, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.S.; Sasaki, S. The adduct formation between the thioguanine-polyamine ligands and DNA with the AP site under UVA irradiated and non-irradiated conditions. Bioorg. Med. Chem. 2019, 27, 115160. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Kinetic features of 3′-5′ exonuclease activity of human AP-endonuclease APE1. Molecules 2018, 23, 2101. [Google Scholar] [CrossRef] [PubMed]
- Long, K.; Gu, L.; Zhang, Z.; Li, E.; Zhang, Y.; He, L.; Pan, F.; Guo, Z.; Hu, Z. Small-molecule inhibition of APE1 induces apoptosis, pyroptosis, and necroptosis in non-small cell lung cancer. Cell Death Dis. 2021, 12, 503. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, S.B.; Samanta, D.; Mirkin, C.A. DNA-based nanostructures for live-cell analysis. J. Am. Chem. Soc. 2020, 142, 11343–11356. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, N.; Fu, S.; Deng, Y.; Yu, C.; Su, X. Base excision repair-inspired DNA motor powered by intracellular apurinic/apyrimidinic endonuclease. Nanoscale 2019, 11, 1343–1350. [Google Scholar] [CrossRef]
- Strohsahl, C.M.; Krauss, T.D.; Miller, B.L. Identification of high-stringency DNA hairpin probes by partial gene folding. Biosens. Bioelectron. 2007, 23, 233–240. [Google Scholar] [CrossRef]
- Bidar, N.; Amini, M.; Oroojalian, F.; Baradaran, B.; Hosseini, S.S.; Shahbazi, M.A.; Hashemzaei, M.; Mokhtarzadeh, A.; Hamblin, M.R.; de la Guardia, M. Molecular beacon strategies for sensing purpose. TrAC Trend. Anal. Chem. 2021, 134, 116143. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, W.; Luo, H.Q.; Li, N.B. Label-free cascade amplification strategy for sensitive visual detection of thrombin based on target-triggered hybridization chain reaction-mediated in situ generation of DNAzymes and Pt nanochains. Biosens. Bioelectron. 2016, 80, 463–470. [Google Scholar] [CrossRef]
- Yang, D.; Ning, L.; Gao, T.; Ye, Z.; Li, G. Enzyme-free dual amplification strategy for protein assay by coupling toehold-mediated DNA strand displacement reaction with hybridization chain reaction. Electrochem. Commun. 2015, 58, 33–36. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, X.; Yang, X.; Liu, P.; Wang, K.; Huang, J.; Liu, J.; Song, C.; Wang, J. Colorimetric detection of mercury ion based on unmodified gold nanoparticles and target-triggered hybridization chain reaction amplification. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 136, 283–287. [Google Scholar] [CrossRef]
- Zhou, Q.; Lin, Y.; Lin, Y.; Wei, Q.; Chen, G.; Tang, D. In situ amplified electrochemical aptasensing for sensitive detection of adenosine triphosphate by coupling target-induced hybridization chain reaction with the assembly of silver nanotags. Talanta 2016, 146, 23–28. [Google Scholar] [CrossRef]
- Gao, Z.; Qiu, Z.; Lu, M.; Shu, J.; Tang, D. Hybridization chain reaction-based colorimetric aptasensor of adenosine 5′-triphosphate on unmodified gold nanoparticles and two label-free hairpin probes. Biosens. Bioelectron. 2017, 89, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Miao, X.; Zhu, A.; Ling, L. Hybridization chain reaction and gold nanoparticles dual signal amplification for sensitive glucose detection. Biochem. Eng. J. 2015, 103, 205–210. [Google Scholar] [CrossRef]
- Dai, J.; Xing, C.; Lin, Y.; Huang, Y.; Yang, Y.; Chen, Z.; Lu, C.; Yang, H. Localized DNA catalytic hairpin assembly reaction on DNA origami for tumor-associated microRNA detection and imaging in live cells. Sens. Actuators B Chem. 2021, 344, 130195. [Google Scholar] [CrossRef]
- Miao, P. Magnetic multipedal DNA walking nanomachine driven by catalytic hairpin assembly. Anal. Chem. 2023, 95, 6760–6764. [Google Scholar] [CrossRef] [PubMed]
- Okabe, K.; Harada, Y.; Zhang, J.; Tadakuma, H.; Tani, T.; Funatsu, T. Real time monitoring of endogenous cytoplasmic mRNA using linear antisense 2′-O-methyl RNA probes in living cells. Nucleic Acids Res. 2011, 39, e20. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Guan, R.; Guo, M.; Ning, P.; Shao, R.; Meng, X. A FRET based two-photon fluorescent probe for ratiometric detection of Pd2+ in living cells and in vivo. Sens. Actuators B Chem. 2018, 254, 949–955. [Google Scholar] [CrossRef]
- He, J.H.; Cheng, Y.Y.; Zhang, Q.Q.; Liu, H.; Huang, C.Z. Carbon dots-based fluorescence resonance energy transfer for the prostate specific antigen (PSA) with high sensitivity. Talanta 2020, 219, 121276. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, D.; Luo, Z.; Duan, Y. A dual-functional fluorescent biosensor based on enzyme-involved catalytic hairpin assembly for the detection of APE1 and miRNA-21. Analyst 2022, 147, 2834–2842. [Google Scholar] [CrossRef]
- Fang, S.; Chen, L.; Zhao, M. Unimolecular chemically modified DNA fluorescent probe for one-step quantitative measurement of the activity of human apurinic/apyrimidinic endonuclease 1 in biological samples. Anal. Chem. 2015, 87, 11952–11956. [Google Scholar] [CrossRef]
- Chai, Q.; Chen, J.; Zeng, S.; Zhu, T.; Chen, J.; Qi, C.; Mao, G.; Liu, Y. Closed cyclic DNA machine for sensitive logic operation and APE1 detection. Small 2023, 19, e2207736. [Google Scholar] [CrossRef]
- Simeonov, A.; Kulkarni, A.; Dorjsuren, D.; Jadhav, A.; Shen, M.; McNeill, D.R.; Austin, C.P.; Wilson III, D.M. Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS ONE 2009, 4, e5740. [Google Scholar] [CrossRef] [PubMed]
- Seiple, L.A.; Cardellina II, J.H.; Akee, R.; Stivers, J.T. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol. Pharmacol. 2008, 73, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xie, Q.; Qin, Y.; Tong, C.; Liu, B.; Wang, W. Real-time monitoring and effector screening of APE1 based on rGO assisted DNA nanoprobe. Anal. Biochem. 2021, 633, 114394. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Cao, X.; Zheng, J.; Zhu, C.; Zhang, R.; Sun, Y.; Yang, Z.; Tang, Z.; Wang, J.; Zhao, M. A DNA/RNA hybrid fluorescent probe for high-throughput quantification of the activity of human apurinic/apyrimidinic endonuclease 1 in subcellular extracts. Biosens. Bioelectron. 2023, 14, 100329. [Google Scholar] [CrossRef]
- Li, X.; Xiong, M.; Huang, Y.; Zhang, L.; Zhao, S. Simple label-free fluorescence detection of apurinic/apyrimidinic endonuclease 1 activity and its inhibitor using the abasic site-binding fluorophore. Anal. Methods 2019, 11, 739–743. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Tang, H.; Yang, B.; Zhao, Y.; Wu, P. Evaluation of the sequence-dependent relative activity of APE1 for optimal biosensing design. Biosens. Bioelectron. 2022, 214, 114539. [Google Scholar] [CrossRef]
- Moon, W.J.; Liu, J. Interfacing catalytic DNA with nanomaterials. Adv. Mater. Interfaces 2020, 7, 2001017. [Google Scholar] [CrossRef]
- Lv, Z.; Zhu, Y.; Li, F. DNA Functional nanomaterials for controlled delivery of nucleic acid-based drugs. Front. Bioeng. Biotechnol. 2021, 9, 720291. [Google Scholar] [CrossRef]
- Baron, R.; Willner, B.; Willner, I. Biomolecule-nanoparticle hybrids as functional units for nanobiotechnology. Chem. Commun. 2007, 4, 323–332. [Google Scholar] [CrossRef]
- Samanta, A.; Medintz, I.L. Nanoparticles and DNA-a powerful and growing functional combination in bionanotechnology. Nanoscale 2016, 8, 9037–9095. [Google Scholar] [CrossRef]
- Heuer-Jungemann, A.; Linko, V. Engineering inorganic materials with DNA nanostructures. ACS Cent. Sci. 2021, 7, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Z.; Willner, I. Dynamic reconfigurable DNA nanostructures, networks and materials. Angew. Chem. Int. Ed. 2023, 62, e202215332. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Lin, L.; Chao, Z.; Shao, C.; Chen, Z.; Wei, Z.; Lu, J.; Huang, Y.; Li, L.; Liu, Q.; et al. Organic spherical nucleic acids for the transport of a NIR-II-emitting dye across the blood-brain barrier. Angew. Chem. Int. Ed. 2020, 59, 9702–9710. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, D.; Calandra, P.; Pasqua, L.; Magazu, S. Self-assembly of organic nanomaterials and biomaterials: The bottom-up approach for functional nanostructures formation and advanced applications. Materials 2020, 13, 1048. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, S.; Mallick, S.; Bahadur, D. Electrochemical method to prepare graphene quantum dots and graphene oxide quantum dots. ACS Omega 2017, 2, 8343–8353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ba, S.; Yang, Z.; Wang, T.; Lee, J.Y.; Li, T.; Shao, F. Graphene quantum dot-based nanocomposites for diagnosing cancer biomarker APE1 in living cells. ACS Appl. Mater. Interfaces 2020, 12, 13634–13643. [Google Scholar] [CrossRef]
- Cai, Z.; Wang, A.; Wang, Y.; Qiu, Z.; Li, Y.; Yang, H.; Fu, M.; Liu, M.; Yu, Y.; Gao, F. Smart programmable scalable dual-mode diagnostic logic nanoflare strategy for dual-tumor marker detection. Anal. Chem. 2022, 94, 9715–9723. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y.; Wang, W.; Min, Q.; Zhang, J.R.; Zhu, J.J. A telomerase-assisted strategy for regeneration of DNA nanomachines in living cells. Angew. Chem. Int. Ed. 2023, 62, e202213884. [Google Scholar] [CrossRef]
- Du, K.; Feng, J.; Gao, X.; Zhang, H. Nanocomposites based on lanthanide-doped upconversion nanoparticles: Diverse designs and applications. Light Sci. Appl. 2022, 11, 222. [Google Scholar] [CrossRef]
- Liang, G.; Wang, H.; Shi, H.; Wang, H.; Zhu, M.; Jing, A.; Li, J.; Li, G. Recent progress in the development of upconversion nanomaterials in bioimaging and disease treatment. J. Nanobiotechnol. 2020, 18, 154. [Google Scholar] [CrossRef]
- Shah, F.; Logsdon, D.; Messmann, R.A.; Fehrenbacher, J.C.; Fishel, M.L.; Kelley, M.R. Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: From bench to clinic. NPJ Precis. Oncol. 2017, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhao, J.; Yuan, J.; Zhao, Y.; Li, L. Organelle-specific photoactivation of DNA nanosensors for precise profiling of subcellular enzymatic activity. Angew. Chem. Int. Ed. 2021, 60, 8923–8931. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Shao, Y.; Chai, X.; Zhao, Y.; Li, L. Spatially selective monitoring of subcellular enzyme dynamics in response to mitochondria-targeted photodynamic therapy. Angew. Chem. Int. Ed. 2022, 61, e202203238. [Google Scholar] [CrossRef]
- Qian, R.; Ding, L.; Ju, H. Switchable fluorescent imaging of intracellular telomerase activity using telomerase-responsive mesoporous silica nanoparticle. J. Am. Chem. Soc. 2013, 135, 13282–13285. [Google Scholar] [CrossRef] [PubMed]
- Torney, F.; Trewyn, B.G.; Lin, V.S.; Wang, K. Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat. Nanotechnol. 2007, 2, 295–300. [Google Scholar] [CrossRef] [PubMed]
- He, X.X.; Wang, K.; Tan, W.; Liu, B.; Lin, X.; He, C.; Li, D.; Huang, S.; Li, J. Bioconjugated nanoparticles for DNA protection from cleavage. J. Am. Chem. Soc. 2003, 125, 7168–7169. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Liu, Y.; Huang, S.; Fang, S.; Zhao, M. A specific DNA-nanoprobe for tracking the activities of human apurinic/apyrimidinic endonuclease 1 in living cells. Nucleic Acids Res. 2017, 45, e45. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Feng, X.; Hu, W.; Li, L. An activatable DNA nanodevice for correlated imaging of apoptosis-related dual proteins. Nanoscale 2022, 14, 6465–6470. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Du, W.F.; Liu, Y.N.; Wang, F.; Tang, L.J.; Jiang, J.H. Protein-scaffolded DNA nanostructures for imaging of apurinic/apyrimidinic endonuclease 1 activity in live cells. Anal. Chem. 2023, 95, 3551–3555. [Google Scholar] [CrossRef]
- Zhang, T.; Tian, T.; Zhou, R.; Li, S.; Ma, W.; Zhang, Y.; Liu, N.; Shi, S.; Li, Q.; Xie, X.; et al. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat. Protoc. 2020, 15, 2728–2757. [Google Scholar] [CrossRef]
- Xu, W.; He, W.; Du, Z.; Zhu, L.; Huang, K.; Lu, Y.; Luo, Y. Functional nucleic acid nanomaterials: Development, properties, and applications. Angew. Chem. Int. Ed. 2021, 60, 6890–6918. [Google Scholar] [CrossRef] [PubMed]
- Mou, Q.; Ma, Y.; Pan, G.; Xue, B.; Yan, D.; Zhang, C.; Zhu, X. DNA trojan horses: Self-assembled floxuridine-containing DNA polyhedra for cancer therapy. Angew. Chem. Int. Ed. 2017, 56, 12528–12532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ptacin, J.L.; Fischer, E.C.; Aerni, H.R.; Caffaro, C.E.; Jose, K.S.; Feldman, A.W.; Turner, C.R.; Romesberg, F.E. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, T.; Shi, S.; Gao, Y.; Cai, X.; Lin, Y. A dynamic DNA tetrahedron framework for active targeting. Nat. Protoc. 2023, 18, 1028–1055. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Liu, S.; Fang, H.; Yang, Y.; Quan, K.; Li, J.; Yang, X.; Wang, K.; Huang, J. Three-dimensional molecular transfer from DNA nanocages to inner gold nanoparticle surfaces. ACS Nano 2019, 13, 4174–4182. [Google Scholar] [CrossRef]
- Du, R.R.; Cedrone, E.; Romanov, A.; Falkovich, R.; Dobrovolskaia, M.A.; Bathe, M. Innate immune stimulation using 3D wireframe DNA origami. ACS Nano 2022, 16, 20340–20352. [Google Scholar] [CrossRef]
- Cai, R.; Zhang, S.; Chen, L.; Li, M.; Zhang, Y.; Zhou, N. Self-assembled DNA nanoflowers triggered by a DNA walker for highly sensitive electrochemical detection of staphylococcus aureus. ACS Appl. Mater. Interfaces 2021, 13, 4905–4914. [Google Scholar] [CrossRef]
- Zhang, L.; Abdullah, R.; Hu, X.; Bai, H.; Fan, H.; He, L.; Liang, H.; Zou, J.; Liu, Y.; Sun, Y.; et al. Engineering of bioinspired, size-controllable, self-degradable cancer-targeting DNA nanoflowers via the incorporation of an artificial sandwich base. J. Am. Chem. Soc. 2019, 141, 4282–4290. [Google Scholar] [CrossRef]
- Wang, D.X.; Wang, J.; Wang, Y.X.; Du, Y.C.; Huang, Y.; Tang, A.N.; Cui, Y.X.; Kong, D.M. DNA nanostructure-based nucleic acid probes: Construction and biological applications. Chem. Sci. 2021, 12, 7602–7622. [Google Scholar] [CrossRef]
- Lu, N.; Pei, H.; Ge, Z.; Simmons, C.R.; Yan, H.; Fan, C. Charge transport within a three-dimensional DNA nanostructure framework. J. Am. Chem. Soc. 2012, 134, 13148–13151. [Google Scholar] [CrossRef]
- Goodman, R.P.; Schaap, I.A.T.; Tardin, C.F.; Erben, C.M.; Berry, R.M.; Schmidt, C.F.; Turberfield, A.J. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 2005, 310, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.S.; Yin, H.; Erben, C.M.; Wood, M.J.; Turberfield, A.J. DNA cage delivery to mammalian cells. ACS Nano 2011, 5, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Li, J.; Li, Q.; Huang, Q.; Shi, J.; Yan, H.; Fan, C. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed. 2014, 53, 7745–7750. [Google Scholar] [CrossRef] [PubMed]
- Keum, J.W.; Bermudez, H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. 2009, 7, 7036–7038. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, G.; Wang, T.; Fu, J.; Li, R.; Song, L.; Wang, Z.G.; Ding, B.; Chen, H. Enhanced stability of DNA nanostructures by incorporation of unnatural base pairs. ChemPhysChem 2017, 18, 2977–2980. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, L.; Qin, Z.; Karges, J.; Xiao, H.; Su, X. Unraveling and overcoming platinum drug-resistant cancer tumors with DNA nanostructures. Adv. Funct. Mater. 2022, 33, 2208797. [Google Scholar] [CrossRef]
- Kou, Q.; Wang, L.; Zhang, L.; Ma, L.; Fu, S.; Su, X. Simulation-assisted localized DNA logical circuits for cancer biomarkers detection and imaging. Small 2022, 18, e2205191. [Google Scholar] [CrossRef]
- Tang, Y.; Lu, B.; Zhu, Z.; Li, B. Establishment of a universal and rational gene detection strategy through three-way junction-based remote transduction. Chem. Sci. 2018, 9, 760–769. [Google Scholar] [CrossRef]
- Dorjsuren, D.; Kim, D.; Vyjayanti, V.N.; Maloney, D.J.; Jadhav, A.; Wilson III, D.M.; Simeonov, A. Diverse small molecule inhibitors of human apurinic/apyrimidinic endonuclease APE1 identified from a screen of a large public collection. PLoS ONE 2012, 7, e47974. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, Y.; Wang, C.; Li, L.; Xu, L.; Yu, Y.; Su, X. Probing and regulating the activity of cellular enzymes by using DNA tetrahedron nanostructures. Chem. Sci. 2019, 10, 5959–5966. [Google Scholar] [CrossRef]
- Lv, M.M.; Liu, J.W.; Yu, R.Q.; Jiang, J.H. A bipedal DNA nanowalker fueled by catalytic assembly for imaging of base-excision repairing in living cells. Chem. Sci. 2020, 11, 10361–10366. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Luo, R.; Li, Y.; Fan, J.; Hu, Y.; Tong, C.; Liu, B.; Li, D. Activity assay and intracellular imaging of APE1 assisted with tetrahedral DNA nanostructure modified-dnazyme and molecular beacon. Sens. Actuat. B-Chem. 2020, 317, 128203. [Google Scholar] [CrossRef]
- Zhou, X.M.; Zhuo, Y.; Tu, T.T.; Yuan, R.; Chai, Y.Q. Construction of fast-walking tetrahedral DNA walker with four arms for sensitive detection and intracellular imaging of apurinic/apyrimidinic endonuclease 1. Anal. Chem. 2022, 94, 8732–8739. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Sun, H.; Xu, S.; Li, K.; Su, X. Screening and application of inhibitory aptamers for DNA repair protein apurinic/apyrimidinic endonuclease 1. Int. J. Biol. Macromol. 2023, 242, 124918. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Liu, Y.; Zhang, L.; Liu, J.; Su, X. Programming dissipation systems by DNA timer for temporally regulating enzyme catalysis and nanostructure assembly. ACS Nano 2022, 16, 14274–14283. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).