Abstract
Self-assembled DNA nanospheres, as versatile and ideal vehicles, have offered new opportunities to create intelligent delivery systems for precise bioimaging and cancer therapy, due to their good biostability and cell permeability, large loading capacity, and programmable self–assembly behaviors. DNA nanospheres can be synthesized by the self–assembly of Y–shaped DNA monomers, ultra–long single-stranded DNA (ssDNA), and even metal–DNA coordination. Interestingly, they are size–controllable by varying some parameters including concentration, reaction time, and mixing ratio. This review summarizes the design of DNA nanospheres and their extensive biomedical applications. First, the characteristics of DNA are briefly introduced, and different DNA nanostructures are mentioned. Then, the design of DNA nanospheres is emphasized and classified into three main categories, including Y–shaped DNA unit self-assembly by Watson–Crick base pairing, liquid crystallization and the dense packaging of ultra–long DNA strands generated via rolling circle amplification (RCA), and metal–DNA coordination–driven hybrids. Meanwhile, the advantages and disadvantages of different self–assembled DNA nanospheres are discussed, respectively. Next, the biomedical applications of DNA nanospheres are mainly focused on. Especially, DNA nanospheres serve as promising nanocarriers to deliver functional nucleic acids and drugs for biosensing, bioimaging, and therapeutics. Finally, the current challenges and perspectives for self-assembled DNA nanospheres in the future are provided.
1. Introduction
DNA is traditionally considered the basic genetic material of all living organisms. In 1982, Seeman first put forward the establishment of DNA–branched junctions based on Watson–Crick base–pairing rules, which broke the traditional concept and ushered in the new era of DNA nanotechnology [1]. The outstanding advantages of DNA such as good biocompatibility and low cytotoxicity, remarkable molecular recognition properties, and programmable self-assembly abilities make DNA as versatile potential building blocks to fabricate multifunctional DNA nanostructures [2,3,4]. In recent decades, various DNA nanostructures have been fabricated including branched DNA, DNA dendrimers, DNA origamis, DNA polyhedrons, and DNA nanospheres [5,6,7,8,9,10]. These DNA nanostructures exhibit considerable advantages including good biocompatibility, great biostability, large loading capacity, and outstanding cell permeability, and have potential and extensive applications in biosensing, bioimaging, and therapeutics [11,12,13,14,15].
Undoubtedly, the development of DNA nanostructures has a significant impact on material science and biomedicine. Especially, DNA nanostructures have emerged as versatile and promising nanocarriers for cargo delivery, which greatly boosts the advances of precise medicine. However, myriad DNA strands with complicated sequences and even computer simulation software often are required to assemble multifunctional DNA nanostructures for available biomedical applications [16,17,18,19,20]. Recently, the advent of self-assembled DNA nanospheres has significantly simplified the synthesis process, even without complicated sequence design and operation procedures. They are formed by the self-assembly of the smallest Y-shaped DNA units, ultra-long single-stranded DNA (ssDNA) generated via rolling circle amplification (RCA) as basic building blocks, or even simple metal–DNA coordination [21,22,23]. Apart from the simple design and synthesis, more importantly, the size of DNA nanospheres is tunable, ranging from a few micrometers to tens of nanometers by varying corresponding experimental parameters such as concentration, reaction time, and mixing ratio. Interestingly, DNA nanospheres with suitable size (~200 nm) can be effectively internalized by most cells. Meanwhile, DNA nanospheres possess excellent resistance against enzyme degradation, high loading capacity, and biostability, which make them as ideal and promising nanocarriers to deliver aptamers, bioimaging agents, and drugs for targeting specific cancer cells, bioimaging, and cancer therapy [24,25,26].
Herein, the scope of this overview is the design and applications of programmatically self-assembled DNA nanospheres. The design and construction of self-assembled DNA nanospheres are discussed in categories, which mainly involve the self-assembly of Y(X)-DNA monomers, ultra-long RCA products, or metal–DNA coordination. Meanwhile, the biological applications of DNA nanospheres are mainly focused on. Especially, DNA nanospheres serve as potential and ideal nanocarriers to deliver signaling molecules, bioimaging agents, and drugs for bioassays, bioimaging, and tumor therapy. Furthermore, the challenges and perspectives for the advancement of self-assembled DNA nanospheres in the future are emphasized. It is hoped that this overview can provide useful guidance for the design, construction, and biomedical application of self-assembled DNA nanospheres.
2. Design and Construction of DNA Nanospheres
With the gradual advance of DNA nanostructures, various DNA nanospheres have been used as carriers. However, the basic building methods still follow the rules of DNA programmable self-assembly. According to the category of self-assembly strategies, they can be mainly divided into three types, including the self-assembly of Y(X)-shaped monomers/short DNA strands via Watson–Crick base pairing, liquid crystallization and the dense packaging of long ssDNA generated via RCA, and metal–DNA coordination.
2.1. Self-Assembly of Y (X)-Shaped Monomers
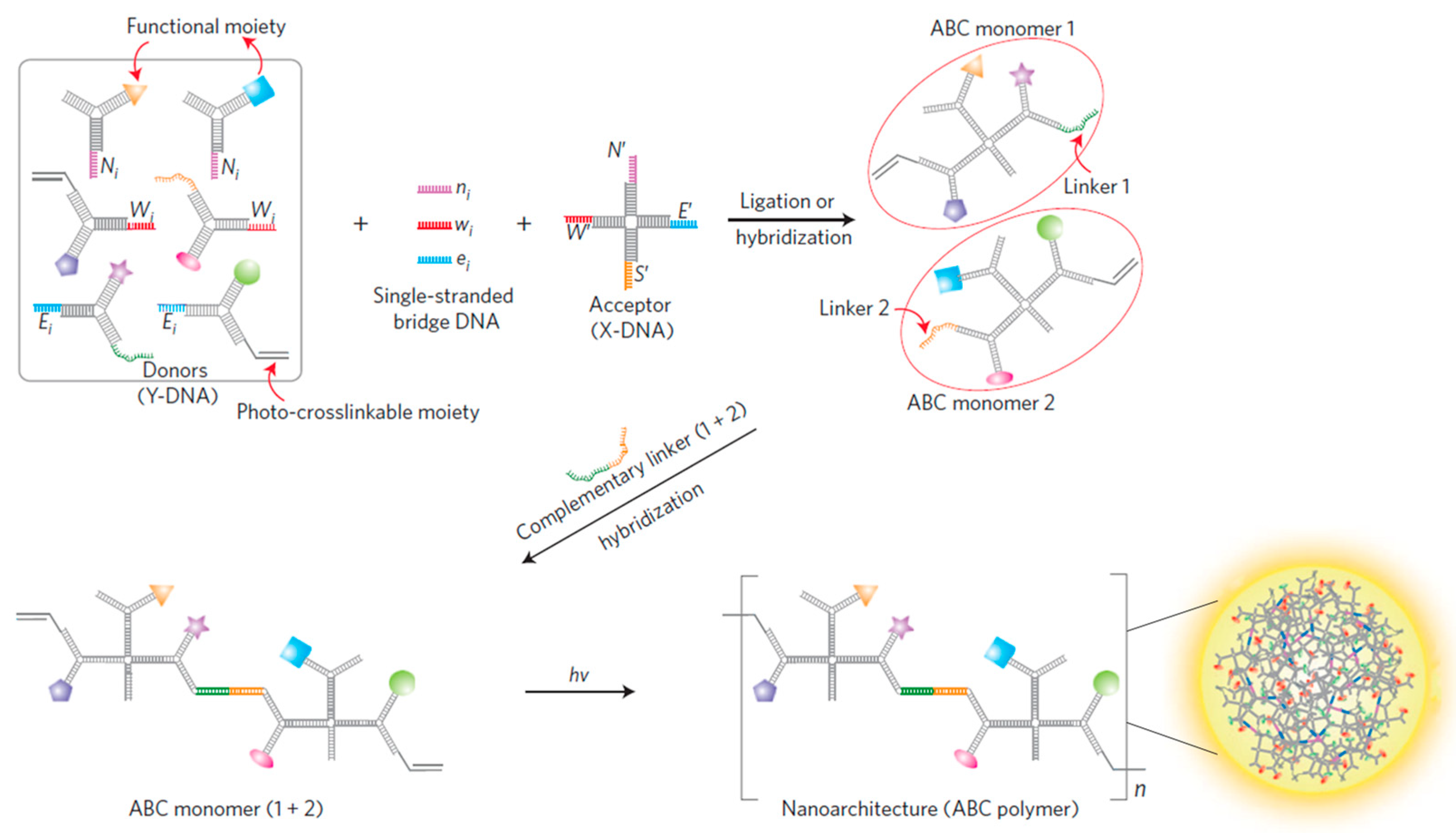
In 2004, Li et al. proposed the branched DNA, which was self-assembled by three ssDNA that were partially hybridized with each other [5]. The excellent properties of the Y-shaped scaffold including facile operation, cost-effective, and robust, endowed it as a basic building unit to establish various DNA nanospheres [6,27,28]. For example, the Luo’s group constructed a multifunctional nanosphere based on the self-assembly of an ABC monomer via photo-polymerization, in which the formation of the ABC monomer was mainly dependent on the complementary hybridization of Y-shaped DNA, X-shaped DNA, and single-stranded bridge DNA [29]. In this design, a single-stranded linker DNA that possessed a complementary sequence to Y-shaped DNA and X-shaped DNA was able to join Y-DNA and X-DNA. The resulting ABC monomers could be linked together with complementary DNA via base pairing and then photo-polymerized to form a DNA nanosphere (Figure 1). Interestingly, the diameter of DNA nanospheres could be tuned by varying the concentrations of the ABC monomer, and they could be internalized by most cells without an additional transfection reagent. Hence, the Tan’s group later reported a multifunctional DNA nanosphere through the photo-polymerization of basic building units that were formed by the complementary hybridization of different Y-shaped DNA units and X-shaped DNA connectors for target cancer therapy [30]. By adding diverse functional moieties, such as aptamers, antisense, and anticancer drugs into each building unit, the resulting DNA nanospheres had applicability to cell recognition, bioimaging, and cancer therapy. Afterward, to efficiently release cargo, their groups designed a stimuli-released DNA nanosphere for targeted gene regulation. The disulfide linkages were introduced to the Y-shaped DNA, which allowed intracellular glutathione (GSH) to cut them and induced the disassembly of DNA nanospheres to efficiently release therapeutic genes [31].
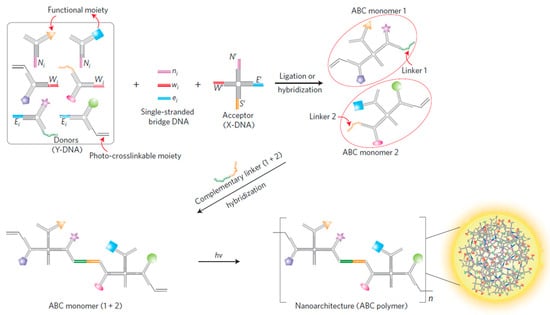
Figure 1.
A multifunctional DNA nanosphere formed through the photo-polymerization of ABC monomer building blocks from a complementary base pair of Y-shaped donor DNA and X-shaped acceptor DNA, reproduced with permission from Ref. [29]. Copyright 2009, Macmillan Publishers.
Although the DNA nanospheres mentioned above have made great progress, their formation still requires an X-shaped DNA connector/ssDNA linker and acrydite-modified DNA for photo-cross-linking, which may increase the cost and complexity of sequence design. To further reduce the assembly cost and sequence design complexity, a simple and straightforward strategy to construct DNA nanospheres that were self-assembled from Y-shaped DNA units or linear ssDNA was developed by many groups [21,32,33,34]. In this design, a self-complementary palindromic sequence was incorporated into a Y-shaped DNA or ssDNA end to catalyze the formation of DNA nanospheres through the hybridization-mediated cross-linkage of building units. For example, a 3D DNA nanosphere was reported by the Wu’s group, formed by palindrome-catalyzed in situ self-assembly, effectively avoiding the operation complexity [35]. Specifically, the same three palindromic sequences were separately applied to three hairpins for the formation of Y-shaped DNA units, and subsequent 3D DNA nanospheres were generated by the self-assembly of Y-shaped DNA units through intermolecular hybridization with each other under endogenous stimuli.
2.2. Self-Assembly of Long ssDNAs from RCA
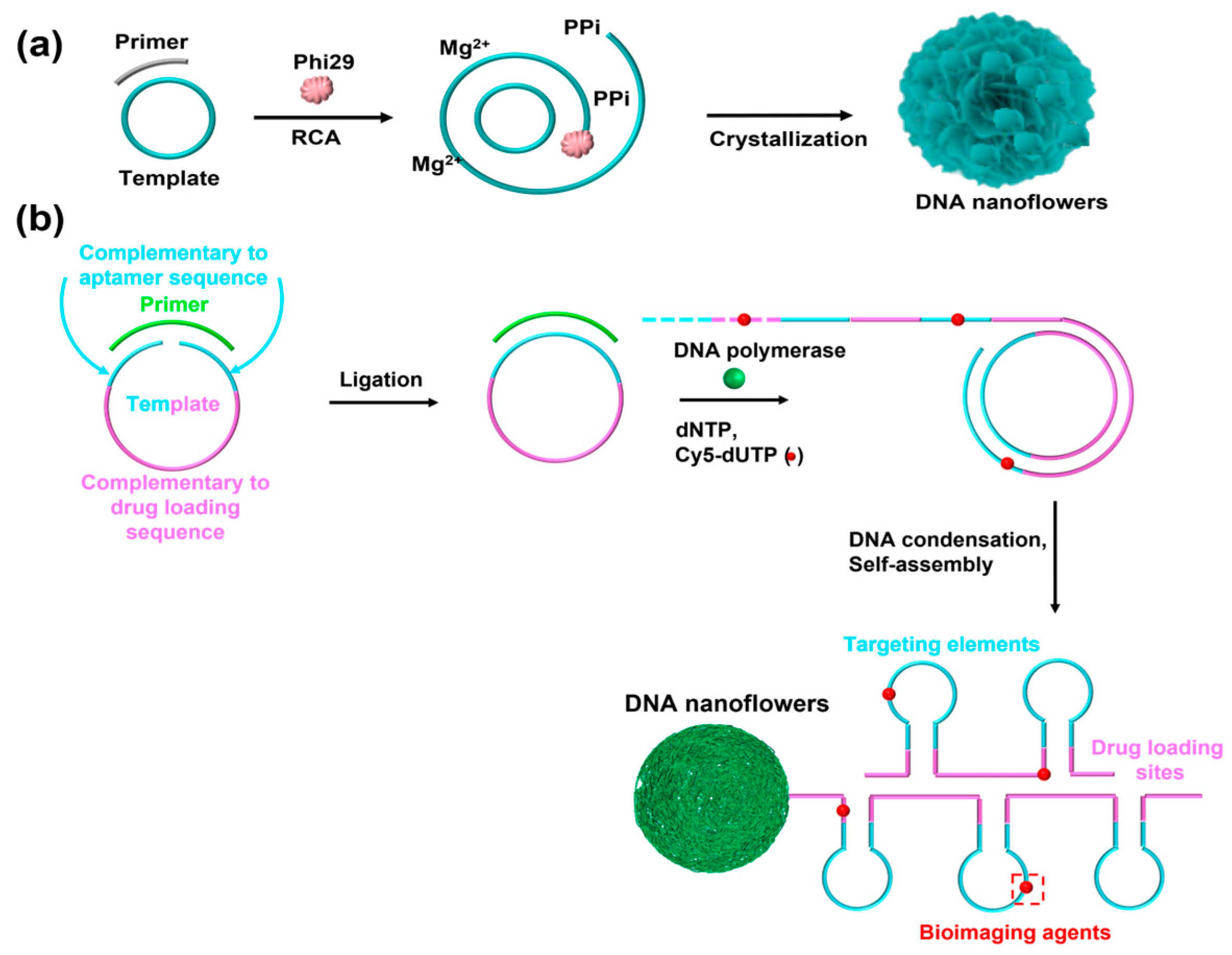
Recently, Zhu et al. reported a noncanonical and topological flower-like DNA nanosphere, also called a DNA nanoflower, which was self-assembled from long single-stranded DNA (lsDNA) generated via rolling circle replication (RCR)/rolling circle amplification (RCA) [25]. RCR is an efficient isothermal amplification process that only uses a template and a primer to generate a lsDNA with numerous repeated fragments [36,37]. In fact, the self-assembly of this DNA nanosphere mainly depended on liquid crystallization and compact packaging of lsDNA. In detail, Mg2+ is typically added to assist the RCR/RCA reaction, yielding a lsDNA and the byproduct of pyrophosphate anion (PPi4−). Notably, the resultant PPi4− reacts with Mg2+ to form magnesium pyrophosphate (Mg2PPi) in solution [38]. Then, the Mg2PPi begins to nucleate and crystallize with increasing concentrations. As a result of electrostatic adsorption, Mg2+ is initially located on the phosphate backbone of DNA strands. Thus, lsDNA spontaneously condenses on the surface of Mg2PPi to form a flower-like and uniform structure [39] (Figure 2a). During the assembly process, only the template and primer need to be designed, thus reducing the complexity of sequence design. In addition, these DNA nanospheres showed excellent biostability, size-controllability, and efficient resistance to nuclease degradation. More importantly, various functional elements such as aptamers, bioimaging agents, and even drug-loading sites were incorporated into template sequences, and the DNA nanospheres could be endowed with different functions including targeting cancer cells, bioimaging, and cancer therapy (Figure 2b). However, the molecular mechanism of the flower-like structure formation is still unclear and in-depth study is required.
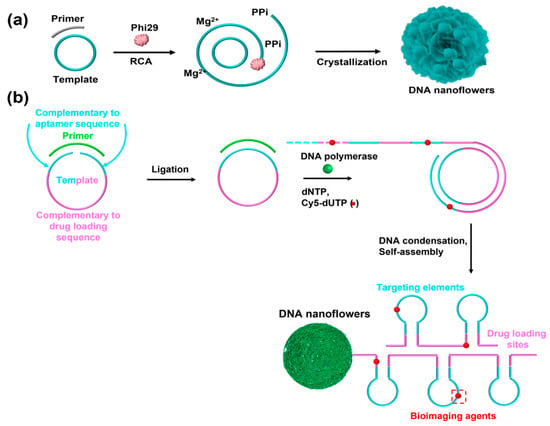
Figure 2.
(a) The self-assembly mechanism of DNA nanospheres based on ultra-long DNA strands as building blocks, reproduced with permission from Ref. [39]. Copyright 2019, American Chemical Society Publishers. (b) A multifunctional DNA nanosphere self-assembled by lsDNA building blocks incorporating different functional nucleic acids for multiple applications, reproduced with permission from Ref. [25]. Copyright 2013, American Chemical Society Publishers.
2.3. Self-Assembly of Metal—DNA Coordination
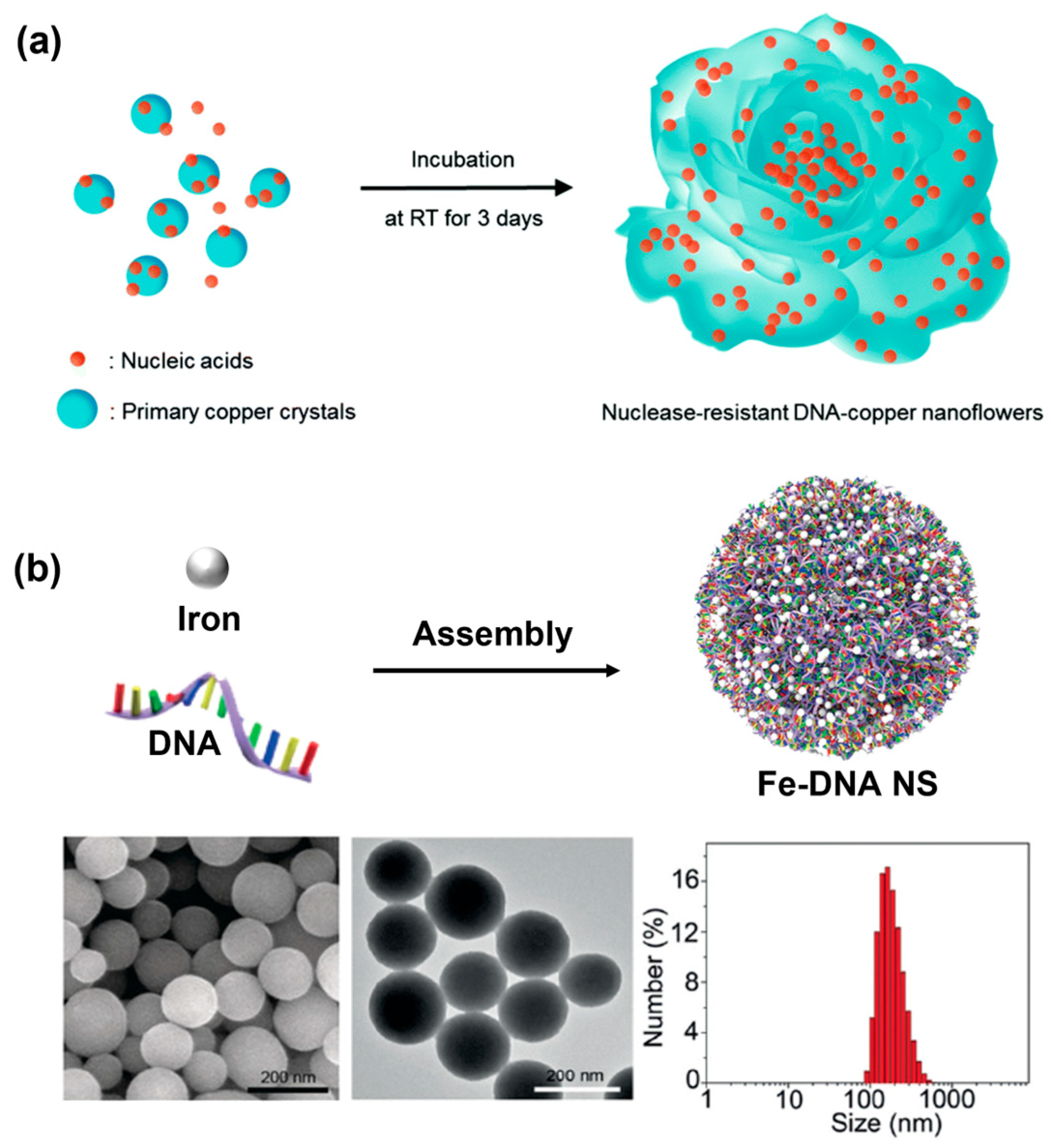
In 2017, the Park’s group discovered a DNA—copper hybrid nanosphere using DNA as an organic ligand [40]. These hybrid nanospheres were synthesized only by a simple one-pot process in which different DNA strands were incubated in phosphate-buffered saline containing copper (II) sulfate for 3 days at room temperature to generate DNA-Cu3(PO4)2 (Figure 3a). Notably, the differences in DNA sequence and length could affect the morphology of the nanospheres. The preparation could be conducted facilely under mild conditions based on the coordination interaction between metal ions (such as Cu, Zn, Fe, Co, Ca, and Mn) and amide and amine groups in DNA organic ligands [41], in which DNA–Mn hybrid nanospheres showed potential as a cell tracking agent and a contrast agent for magnetic resonance imaging (MRI) [22]. Compared with using DNA as a linker to assemble nanospheres, hybrid nanospheres provide many advantageous properties such as simplicity, resistance against nucleases, and large loading capacities [40]. More importantly, many metal ions have been reported to form DNA–inorganic hybrid nanospheres. For example, Lee et al. used Mn, Co, and Mg ions in the polymerization of DNA to generate DNA–inorganic hybrid nanospheres [42]. In addition, Baker et al., through the substitution of Mg2+ with Mn2+, Co2+, or Zn2+, prepared and characterized different DNA–metal hybrid nanospheres with tunable morphology and size by varying the conditions [43]. Interestingly, DNA–copper hybrid nanospheres exhibited excellent peroxidase mimicking activity, making them a promising tool for many fields including biosensing and biocatalysis. However, the average size was too large to be internalized by most cancer cells, and the synthesis time was too long, which hindered their further biomedical application.
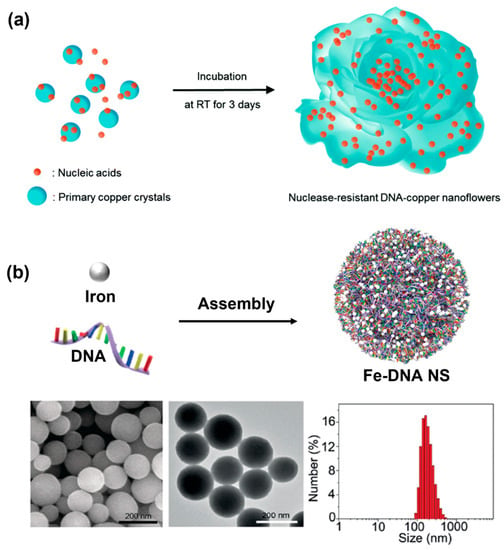
Figure 3.
(a) The formation of DNA–copper hybrid nanosphere through simple one-pot synthesis, reproduced with permission from Ref. [40]. Copyright 2017, Royal Society of Chemistry Publishers. (b) A coordination-driven Fe–DNA hybrid nanosphere and corresponding SEM imaging, TEM imaging, and dynamic light scattering analysis of Fe–DNA hybrid nanospheres, reproduced with permission from Ref. [23]. Copyright 2019, Wiley-VCH Publishers.
Fortunately, the Li’s group subsequently engineered a multifunctional Fe–DNA hybrid nanosphere based on the coordination interaction between FeII ions and random DNA sequences [23]. The synthesis procedure was very simple, only mixing FeII ion aqueous solution with random DNA solution and then incubating at 95 °C for 3 h. The assembly time was greatly reduced and any further purification was not required. The SEM image and TEM image of the Fe–DNA nanospheres also illustrated a uniform and defined spherical shape (Figure 3b). More importantly, the average hydrodynamic diameter of the Fe–DNA spheres was approximately ~200 nm, indicated by dynamic light scattering (DLS), which was essential to cancer cell internalization. The precise size of such DNA nanospheres could be manipulated by tuning the ratio of FeII ions and DNA or their total concentration during the assembly process. Furthermore, using functional nucleic acids such as CpG, DNAzyme, and antisense, as well as FeII or CuII ions to self-assemble via coordination interaction, a multifunctional DNA nanosphere could be obtained for bioimaging and cancer therapy.
3. Application of Various DNA Nanospheres
3.1. Biosensing
3.1.1. Electrochemical Biosensor
In recent years, more and more attention has been paid to electrochemical biosensors, owing to their high sensitivity, rapid response, and facile operation [44,45]. DNA nanospheres are often used as carriers for the loading of electroactive molecules. For example, Cai et al. built a dual-signal amplification electrochemical biosensor based on an exonuclease III-assisted DNA walker for the sensitive sensing of S. aureus, in which DNA nanospheres were employed as a carrier to load MB via electrostatic adsorption and intercalation binding for accelerating electron transfer between electrodes [46]. Similarly, by loading MB into DNA nanospheres, a novel ratiometric electrochemical biosensor was reported by Yuan’s group for the analysis of microRNA-21 (miRNA-21) based on target-induced recycling and Zn2+-mediated DNAzyme cleavage [47]. A small copy of miRNA-21 could yield abundant S1, and then the resultant S1 initiated RCA to produce DNA nanospheres containing DNAzyme and substrates. Here, abundant MB could be captured by in situ-generated DNA nanospheres on the electrode surface, generating a strong electrochemical signal (I1). When in the presence of Zn2+, DNA nanospheres are degraded through Zn2+-assisted DNAzyme cleavage, resulting in the liberation of MB and gaining a low electrochemical signal (I2). Thus, miRNA-21 could be sensitively detected by measuring the ratiometric response of I1/I2 with an attomole-level detection limit (Figure 4a).
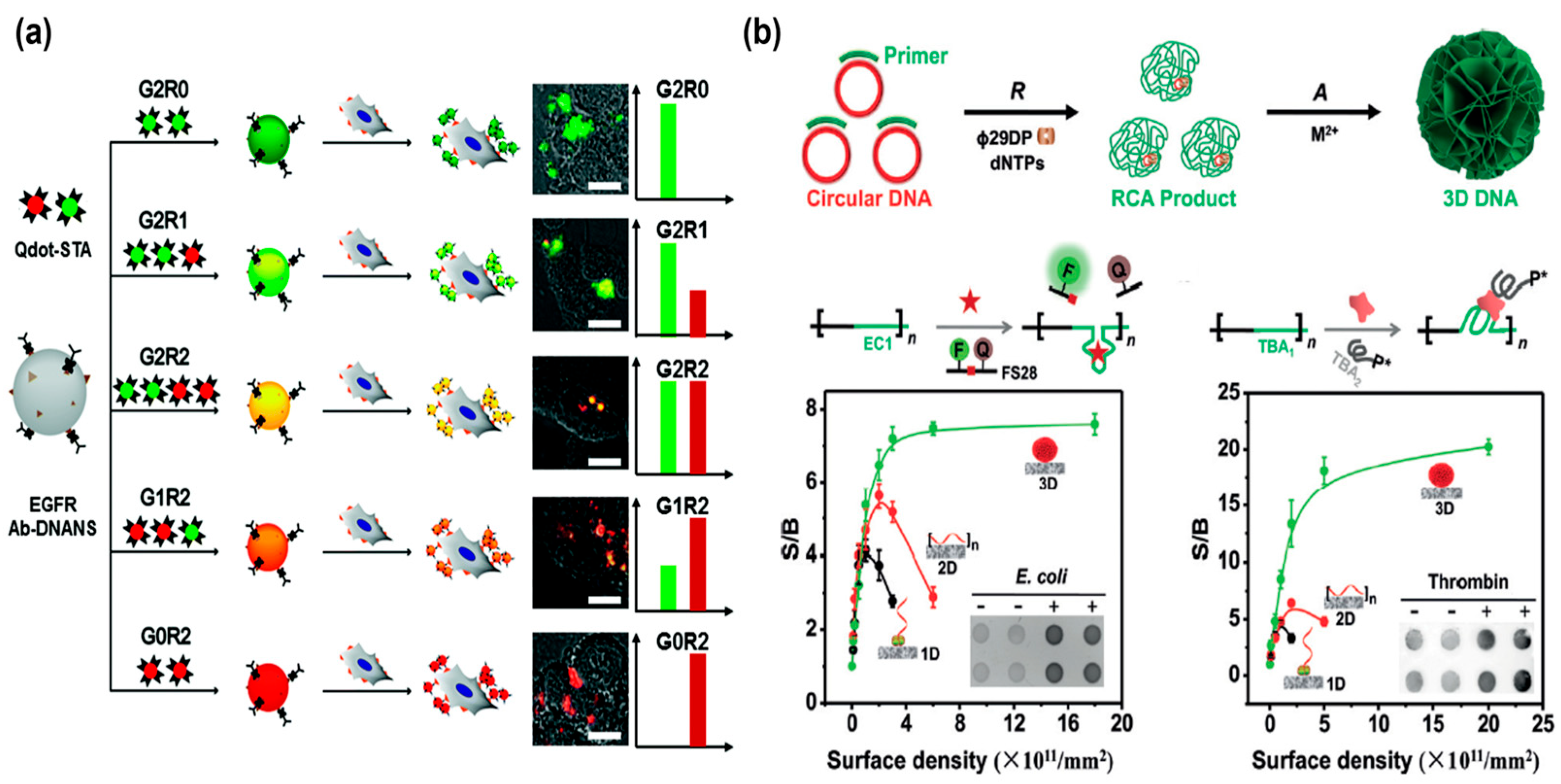
Electrochemiluminescence (ECL), as a powerful analytical method, has attracted extensive attention in biosensing [48]. Among them, ECL based on Ru(bpy)32+ possesses good stability and high efficiency [49]. Here, the Chen’s group designed an enhanced and stable ECL bioaptasensor based on restriction endonuclease-powered self-assembled 3D DNA nanospheres (EPDSs) from RCA products for the ultrasensitive detection of AFB 1 [50]. Similarly, numerous Ru (II) complexes (Ru(bpy)32+) were immobilized on DNA nanospheres to form EPDSs/Ru (II) complexes through the electrostatic adsorption interaction for signal enhancement and sensitivity improvement (Figure 4b). Furthermore, the ECL luminophore of the doxorubicin-N-(aminobutyl)-N-(ethylisoluminol) (Dox-ABEI) complex could be inserted into the dsDNA to output enhanced ECL signals. Li et al. created a sensitive ECL aptasensor based on the T7 exo-assisted cascade amplification strategy and the in situ assembly of DNA nanospheres on the electrode surface for sensing MUC1 [51]. Integrating DNA nanospheres with ECL luminophores could enhance the loading stability of ECL luminophores and greatly improve the detection sensitivity.
Photoelectrochemical (PEC) biosensors, as an emerging method, have gained considerable attention owing to their outstanding properties including good stability, low detection limit, and facile miniaturization [52,53,54]. Integrating 3D DNA nanospheres with a signal amplification strategy to construct a PEC biosensor has evoked tremendous research interest [55,56]. For example, the Tang’s group recently engineered a PEC biosensor for the sensitive assay of thrombin, employing graphene oxide-coated copper-doped zinc oxide quantum dots (Cu0.3Zn0.7O-GO QDs) as photoactive molecular and glucose oxidase-loaded DNA nanospheres (GOx-DSs) for signal amplification [57] (Figure 4c). In the presence of thrombin, magnetic microparticle (MMPs)/thrombin/GOx-DSs complexes were formed and glucose was oxidized to H2O2 by encapsulated GOx, leading to the enhanced photocurrent of the Cu0.3Zn0.7O-GO QDs-functionalized electrode. In addition, the dual-mode biosensor has emerged as a promising method, offering high accuracy and low false positive signals [58]. Deng et al. proposed a dual-mode biosensor based on perylene derivative PDA+-functionalized DNA nanospheres self-assembled by long RCA products for the sensitive and accurate sensing of miRNA-141 [59]. By integrating T7 endonuclease-assisted target recycle amplification with multifunctional DNA nanospheres, this dual-mode biosensor is endowed with a wide detection range and high accuracy.
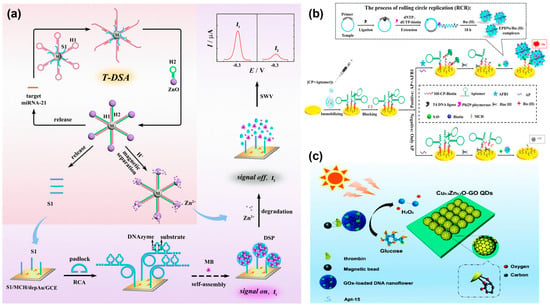
Figure 4.
(a) A novel ratiometric electrochemical biosensor with self-assembled DNA nanospheres loading MB for the sensitive analysis of miRNA-21, reproduced with permission from Ref. [47]. Copyright 2022, American Chemical Society Publishers. (b) An electrochemiluminescence bioaptasensor based on restriction endonuclease-powered self-assembled 3D DNA nanospheres (EPDSs) for the ultrasensitive quantification of AFB1, reproduced with permission from Ref. [50]. Copyright 2020, American Chemical Society Publishers. (c) A photoelectrochemical (PEC) biosensor based on glucose oxidase-loaded DNA nanospheres (GOx-DSs) for the sensitive assay of thrombin, reproduced with permission from Ref. [57]. Copyright 2021, Royal Society of Chemistry Publishers.
Figure 4.
(a) A novel ratiometric electrochemical biosensor with self-assembled DNA nanospheres loading MB for the sensitive analysis of miRNA-21, reproduced with permission from Ref. [47]. Copyright 2022, American Chemical Society Publishers. (b) An electrochemiluminescence bioaptasensor based on restriction endonuclease-powered self-assembled 3D DNA nanospheres (EPDSs) for the ultrasensitive quantification of AFB1, reproduced with permission from Ref. [50]. Copyright 2020, American Chemical Society Publishers. (c) A photoelectrochemical (PEC) biosensor based on glucose oxidase-loaded DNA nanospheres (GOx-DSs) for the sensitive assay of thrombin, reproduced with permission from Ref. [57]. Copyright 2021, Royal Society of Chemistry Publishers.
3.1.2. Colorimetric Assays
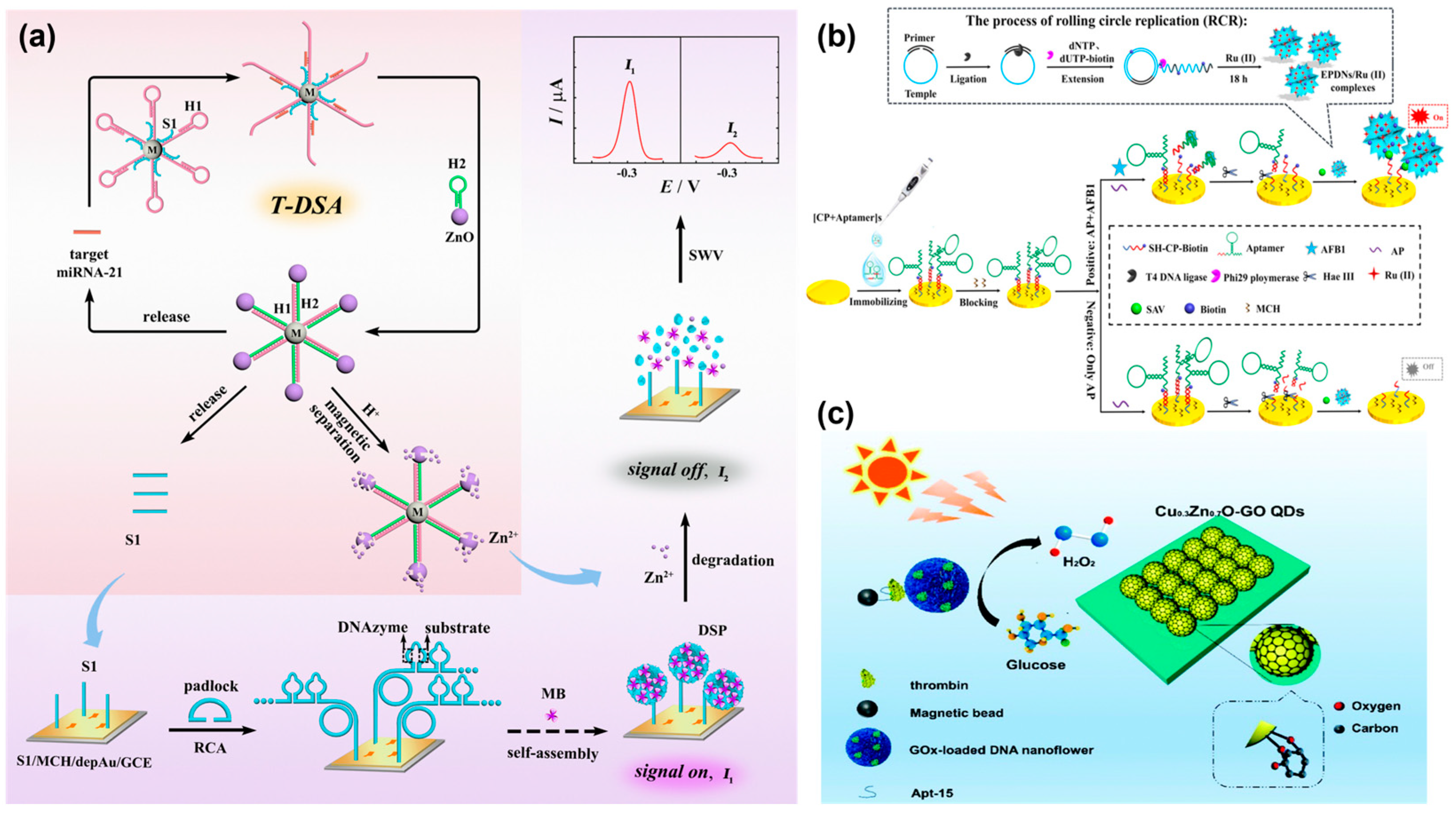
The colorimetric method is regarded as a simple, cost-effective, and convenient platform [60]. It is widely applied in point-of-care testing (POCT) because it does not require complicated and sophisticated instrumentation [61,62,63]. However, traditional colorimetric assay often requires natural enzymes for catalysis, and the poor stability of enzymes hinders its practical application [64]. Notably, 3D DNA nanospheres are highly stable and robust and can act as a carrier to encapsulate numerous protein enzymes and still retain their original catalytic activity. For example, Zeng et al. designed a versatile colorimetric biosensor based on horseradish peroxidase-encapsulated DNA nanospheres (HRP-DS) as recognition molecules and signal-reporting elements for the screening of cancer-derived exosomes [65] (Figure 5a). When target exosomes were applied to the detection system, a sandwiched structure was generated between the magnetic bead (MB), exosomes, and HRP-DSs, which enabled the oxidation of colorless ABTS into green-colored oxidized ABTS with the assistance of H2O2. Using similar assembly principles, Chang et al. reported a paper-based detection device with horseradish peroxidase (HRP)-encapsulated 3D DNA nanospheres for sensing alkaline phosphatase (ALP) in blood (Figure 5b) [66]. APL, as an enzyme, plays a vital role in dephosphorylation [67]. Thus, the HRP encapsulated DNA nanospheres could be controllably released via the catalysis reaction of ALP. As a result, in the presence of H2O2, the released HRP oxidized 3, 3′, 5, 5′-tetramethylbenzidine (TMB) into a colorless product. This device, with high stability, could rapidly detect ALP in blood samples within an hour. More importantly, complicated pre-treatment and sophisticated equipment were not required, making it as an ideal platform to fabricate POCT in areas with limited resources.
Without enzymes for catalysis, the Cao’s group designed a naked-eye colorimetric analysis platform for sensing telomerase activity, which integrated palindromic-catalyzed DNA nanospheres from Y-shaped DNA units with the color changes in gold nanoparticles (AuNPs) [68]. Here, a telomerase primer was introduced to a branched DNA, Y-shaped DNA units were formed via hybridization, and then the resulting Y-shaped DNA was linked to DNA nanospheres through the base pairing of their end palindromic sequence in the presence of telomerase. Subsequently, a large amount of SYBR Green I (SG) was inserted into the DNA nanospheres and the aggregation of AuNPs was inhibited, resulting in a red solution. However, the solution would change to blue, induced by free SG, in the absence of telomerase (Figure 5c). Therefore, a label-free and naked-eye colorimetric platform was constructed for cancer diagnosis.
In addition, DNA–copper hybrid nanospheres exhibited outstanding peroxidase mimicking activity, offering a promising application in colorimetric biosensors [40,69]. For example, the Kim’s group found that DNA–copper hybrid nanospheres showed an intrinsic laccase-mimicking activity [70]. Meanwhile, these hybrid nanospheres showed significantly increased stability toward pH, temperature, and ionic strength. Due to these excellent properties, DNA–copper hybrid nanospheres were employed on paper microfluidic devices for the colorimetric analysis of phenolic compounds. In the presence of targets, their oxidation could react with 4-aminoantipyrine to yield a colored product under the catalysis of DNA nanospheres and then be quantified using a portable device with image J software (Figure 5d).
Figure 5.
(a) A horseradish peroxidase-encapsulated DNA nanosphere (HRP-DS) for the colorimetric assays of cancer-derived exosomes, reproduced with permission from Ref. [65]. Copyright 2021, Elsevier Publishers. (b) A paper-based assay method based on horseradish peroxidase (HRP)-encapsulated DNA nanospheres for sensing alkaline phosphatase (ALP), reproduced with permission from Ref. [66]. Copyright 2023, Royal Society of Chemistry Publishers. (c) Label-free and naked-eye colorimetric assay based on palindromic DNA nanospheres and AuNPs, reproduced with permission from Ref. [68]. Copyright 2023, Elsevier Publishers. (d) DNA-copper hybrid nanospheres for the colorimetric assay of phenolic compounds in paper microfluidic devices, reproduced with permission from Ref. [70]. Copyright 2021, Elsevier Publishers.
Figure 5.
(a) A horseradish peroxidase-encapsulated DNA nanosphere (HRP-DS) for the colorimetric assays of cancer-derived exosomes, reproduced with permission from Ref. [65]. Copyright 2021, Elsevier Publishers. (b) A paper-based assay method based on horseradish peroxidase (HRP)-encapsulated DNA nanospheres for sensing alkaline phosphatase (ALP), reproduced with permission from Ref. [66]. Copyright 2023, Royal Society of Chemistry Publishers. (c) Label-free and naked-eye colorimetric assay based on palindromic DNA nanospheres and AuNPs, reproduced with permission from Ref. [68]. Copyright 2023, Elsevier Publishers. (d) DNA-copper hybrid nanospheres for the colorimetric assay of phenolic compounds in paper microfluidic devices, reproduced with permission from Ref. [70]. Copyright 2021, Elsevier Publishers.
3.1.3. Fluorescence Detection
The fluorescence method is a powerful tool for biosensing and bioimaging, due to its excellent characteristics including relative simplicity, high sensitivity, and selectivity [71,72,73,74]. Especially, bio-barcode technology has been utilized for nucleic acids and protein analysis due to its comparable sensitivity to polymerase chain reaction (PCR) [75,76,77]. The Lee’s group designed a novel bio-barcode strategy for sensing cells based on multi-functionalized DNA nanospheres (DNANSs) that contained various functional deoxynucleotide triphosphates (dNTPs) with dyes and biotin [78]. Different biomolecules, such as antibodies, peptides, or proteins could be incorporated into DNANSs by a biotin–streptavidin interaction for targeting cancer cells (Figure 6a). Meanwhile, various quantum dots with different fluorescence emissions could be immobilized on DNANS surfaces, thereby the DNANSs were stained with different colors and acted as barcodes. As a result, anti-epidermal growth factor receptor (anti-EGFR) antibodies (Ab) were functionalized on DNANSs for directly targeting tumor cells in the bloodstream. This DNANS barcode offered a new method for the simple and rapid detection of tumor cells, specifically.
In addition, both DNAzyme strand and substrate were simultaneously integrated into 3D DNA nanospheres, and a fluorescence sensing platform could be constructed in which DNAzyme could be activated by Escherichia coli (E. coli) and a substrate with a single ribonucleotide was labeled with fluorophores (FAMs) and dabcyl [79] (Figure 6b). In the initial state, the fluorescence intensity was very faint due to the close proximity of the fluorophores to the quencher. However, when E. coli appeared, it could activate DNAzyme and cleave the substrate, resulting in fluorescence enhancement. These 3D nanospheres as recognition elements could be utilized for printed paper sensors with low nonspecific protein adsorption, making such devices suitable for constructing POCT devices.
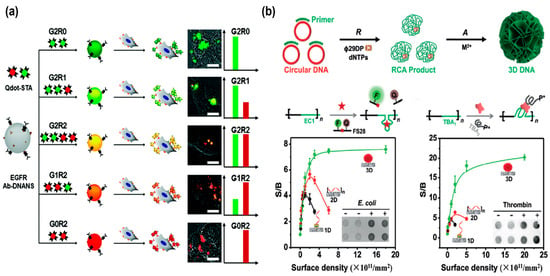
Figure 6.
(a) A bio-barcode system based on multi-functionalized DNA nanospheres for direct cell detection, reproduced with permission from Ref. [78]. Copyright 2017, Royal Society of Chemistry Publishers. (b) Three-dimensional nanospheres encoding DNAzyme with fluorophores (FAMs) and dabcyl for the fluorescence detection of Escherichia coli (E. coli), reproduced with permission from Ref. [79]. Copyright 2018, Wiley-VCH Publishers.
Figure 6.
(a) A bio-barcode system based on multi-functionalized DNA nanospheres for direct cell detection, reproduced with permission from Ref. [78]. Copyright 2017, Royal Society of Chemistry Publishers. (b) Three-dimensional nanospheres encoding DNAzyme with fluorophores (FAMs) and dabcyl for the fluorescence detection of Escherichia coli (E. coli), reproduced with permission from Ref. [79]. Copyright 2018, Wiley-VCH Publishers.
3.2. Bioimaging
3.2.1. Cancer Cell Recognition
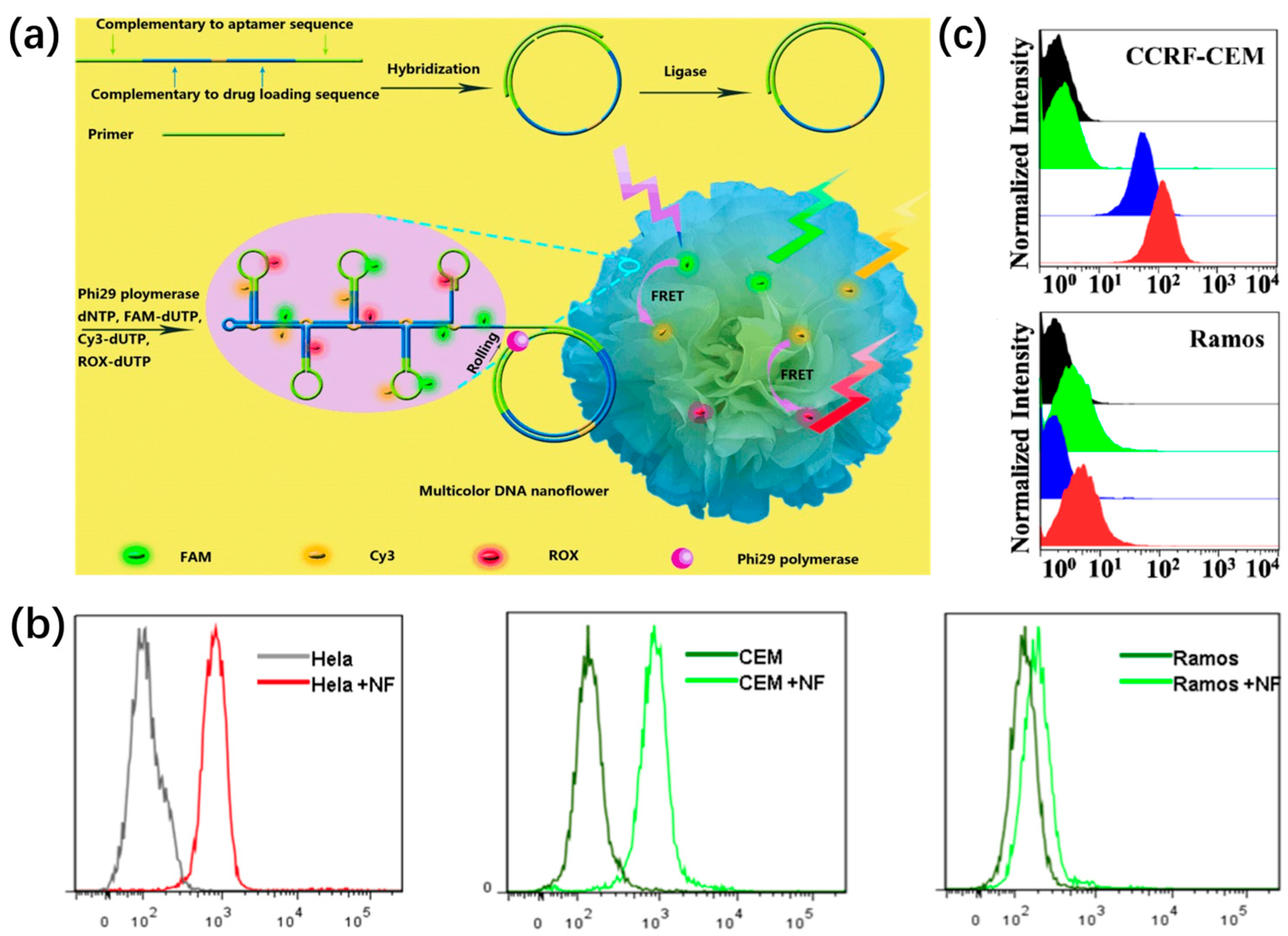
The specific targeting of certain membrane proteins allows for the selective recognition of different cancer cells [80]. As a proof of concept, Hu et al. selected aptamer sgc8 as a model to construct a multicolor DNA nanosphere for cancer cell recognition [81]. Sgc8 has been reported that specifically targets HeLa cells and CEM cells but not Ramos cells, which mainly depends on binding to protein tyrosine kinase 7 (PTK7) overexpressed on the surface of HeLa cells and CEM cells [82]. By introducing the sgc8 complementary sequence into the RCR template, multicolor DNA nanospheres specifically targeting HeLa cells and CEM cells were constructed, as shown in Figure 7a. The flow cytometry and confocal fluorescence images illustrate that the binding affinity and specificity of aptamer sgc8 were retained after being incorporated into the multicolor DNA nanospheres. A significant shift was displayed for CEM cells and HeLa cells, but no obvious shift was shown for Ramos cells, after treating these cells with multicolor DNA nanospheres (Figure 7b). It was fully evident that DNA nanospheres conjugating aptamers allowed for the specific recognition of cancer cells through binding to specific membrane proteins.
In addition, by conjugating sgc8 aptamer into one functional domain of Y-shaped building blocks, targeted DNA nanospheres self-assembled by Y-shaped DNA units could be established for the specific recognition of tumor cells [30]. The selective recognition capacity of sgc8-nanospheres was investigated by flow cytometry, which illustrated a 100-fold fluorescence signal shift for CEM cells compared to DNA nanospheres without sgc8 aptamer (Figure 7c). However, no obvious shift was seen for Ramos cells incubated with sgc8 nanospheres and pure nanospheres. Thus, sgc8-based nanospheres had the ability for selective targeting, giving DNA nanospheres potential applications in targeted cancer therapy.
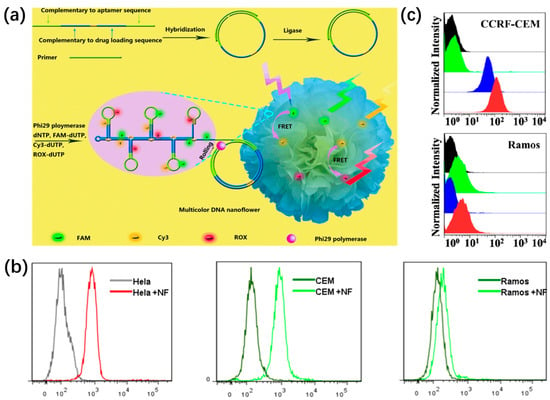
Figure 7.
(a) Sgc8 aptamer-encoded DNA nanospheres for cancer cell recognition and (b) corresponding flow cytometry analysis, reproduced with permission from Ref. [81]. Copyright 2014, Wiley Publishers. (c) The flow cytometry assays of sgc8 nanospheres assembled by Y-shaped DNA units, reproduced with permission from Ref. [30]. Copyright 2013, American Chemical Society Publishers.
Figure 7.
(a) Sgc8 aptamer-encoded DNA nanospheres for cancer cell recognition and (b) corresponding flow cytometry analysis, reproduced with permission from Ref. [81]. Copyright 2014, Wiley Publishers. (c) The flow cytometry assays of sgc8 nanospheres assembled by Y-shaped DNA units, reproduced with permission from Ref. [30]. Copyright 2013, American Chemical Society Publishers.
3.2.2. Intracellular Molecules Imaging
Endogenous biomolecules such as ATP, RNA, and telomerase play vital roles in maintaining intracellular balance and function, and their aberrant expression is closely associated with a variety of diseases [83,84,85]. Hence, the visualization and quantification of intracellular molecules are of great significance for an in-depth understanding of the pathogenesis of disease at the molecular level. For example, Kim et al. constructed a ratiometric aptasensing platform using DNA nanospheres (DNSs) as carriers to monitor intracellular ATP based on the Forster resonance energy transfer (FRET) in Figure 8a [86]. In this design, the DNA template was initially encoded with a prostate-specific membrane antigen for cell targeting, the binding site for the cholesterol-labeled DNA, and one aptamer for ATP sensing. When the DNS aptasensors entered cells, they specifically combined with ATP to induce the separation of acceptor dye from donor dye, yielding green fluorescence. Therefore, the quantitative analysis of intracellular ATP was achieved with different fluorescence ratios in colorful DNSs.
In addition, Li et al. developed sequence-encoded amplicons (SeqEA) based on DNSs for in situ multiplex RNA labeling, realizing multiplex RNA imaging in single cells (Figure 8b) [87]. SeqEA was designed with an encoded padlock probe containing recognition (R) and barcode (B1 and B2) modules. Target RNA could specifically recognize the R module, triggering RCA to generate a long DNA amplicon with numerous repeats of the padlock probes. Subsequently, fluorophore-labeled detection probes P1 and P2 were hybridized with the resultant RCA amplicon. Interestingly, SeqEA exhibited different fluorescence intensities by precisely modulating the hybridization efficiency of B1-P1, thereby determining the amount of detection probe P1 bound to the RCA amplicon. The cell experiment results indicated that SeqEA could simultaneously visualize nine mRNA species in single cells. Afterward, their groups further developed a splice-junction anchored padlock-probe-mediated rolling circle amplification assay (SpliceRCA) for single-cell imaging and distinguishing mRNA variants [88].
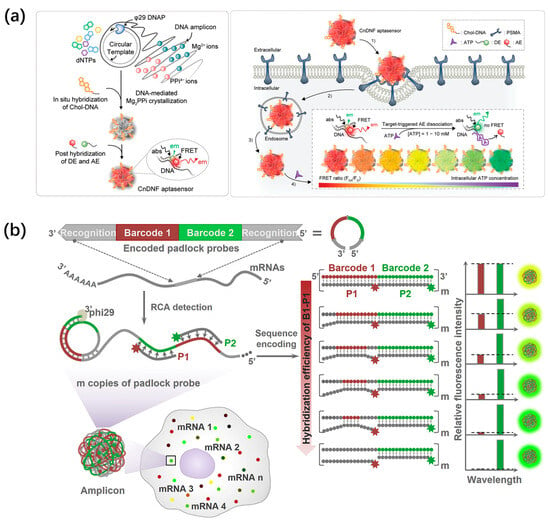
Figure 8.
(a) The fabrication and working principle of cholesterol-decorated DNS aptasensors for ratiometric ATP level monitoring, reproduced with permission from Ref. [86]. Copyright 2021, Wiley-VCH Publishers. (b) The schematic illustration of SeqEA for multiplexed RNA imaging in single cells, reproduced with permission from Ref. [87]. Copyright 2018, Elsevier Publishers.
Figure 8.
(a) The fabrication and working principle of cholesterol-decorated DNS aptasensors for ratiometric ATP level monitoring, reproduced with permission from Ref. [86]. Copyright 2021, Wiley-VCH Publishers. (b) The schematic illustration of SeqEA for multiplexed RNA imaging in single cells, reproduced with permission from Ref. [87]. Copyright 2018, Elsevier Publishers.
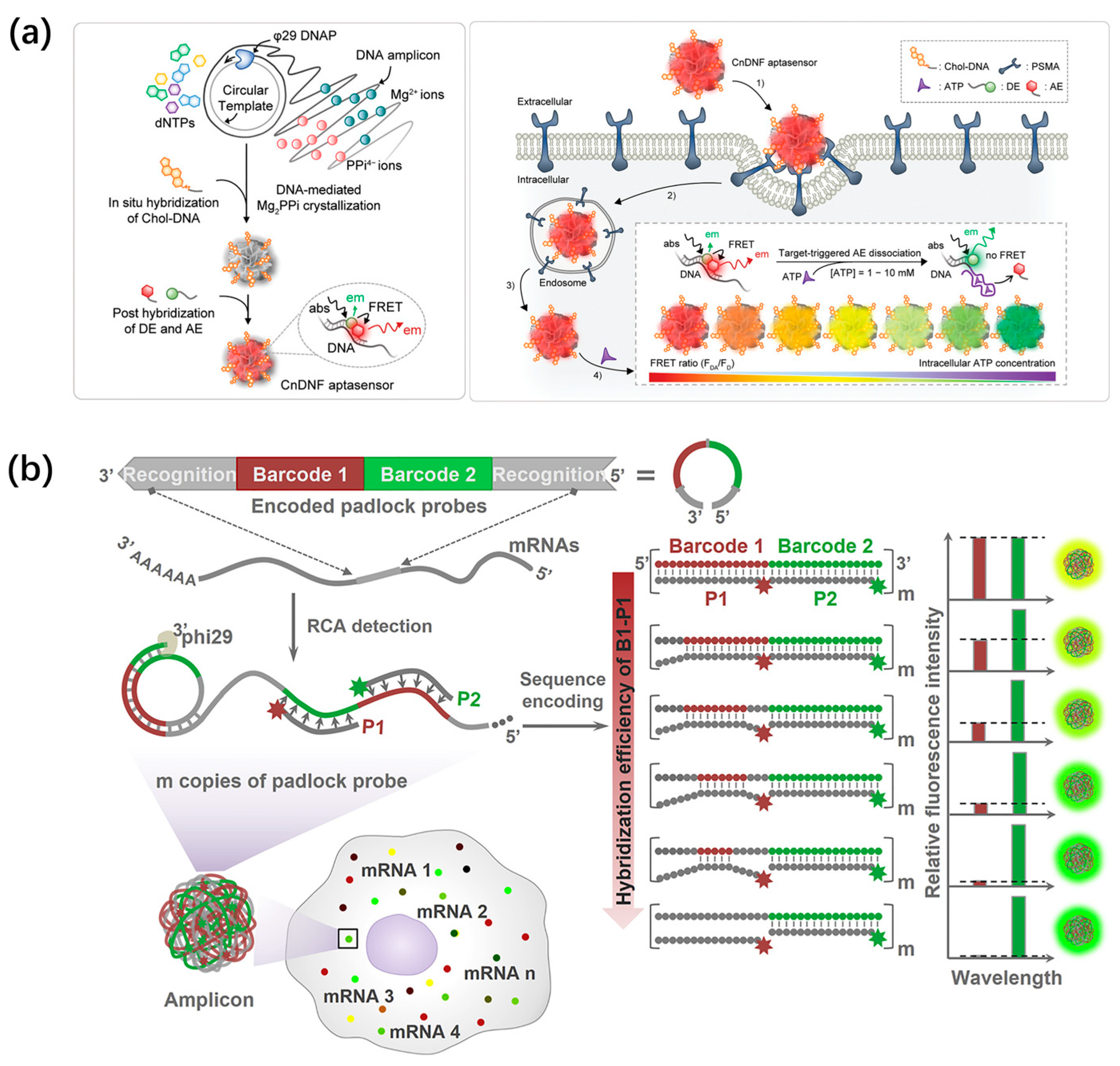
However, early-reported DNA nanospheres are often ineffective for activation, owing to the dense packing of bulky DNA strands. Recently, the Wang’s group designed stimulus-responsive DNA nanospheres based on the RCA long-strand self-assembly and the loading of ZnO nanoparticles via electrostatic adsorption for intracellular miRNA imaging (Figure 9a) [89]. I-R3 DNAzyme, as a self-hydrolyzing ribozyme, is easily introduced into RCA products for on-demand and controllable release cargo. Thus, I-R3 DNAzyme and 8–17 DNAzyme were incorporated into the RCA template for the formation of target-responsive DNA nanospheres, which could encapsulate ZnO nanoparticles through electrostatic adsorption. In an intracellular acidic environment, ZnO nanoparticles could be decomposed into Zn2+, which subsequently catalyzed the miRNA-21-activated self-catabolic degradation of I-R3 DNAzyme. As a result, the DNA nanospheres gradually collapsed, and 8–17 DNAzymes were released to cleave FAM-BHQ-labeled substrates, yielding amplified fluorescence signals. This target-responsive DNA nanosphere enables miRNA to be imaged in live cells, showing promising applications in clinical diagnosis.
Not only DNAzyme but also nonenzymatic and isothermal amplification techniques are widely used for intracellular RNA imaging, such as hybridization chain reactions (HCR), catalytical hairpin assembly (CHA), and entropy-driven catalytic (EDC) reactions [90,91,92]. A facile DNA/RNA hybrid nanosphere was proposed by Li et al. for the amplification imaging of telomerase RNA (TR) in living cells (Figure 9b) [93]. A long RNA strand produced via RCA served as a carrier to deliver multifunctional DNA, including H1, H2, and cholesterol–DNA, and the resulting RNA/DNA hybrid was then self-assembled into nanospheres. When the nanospheres entered the cells, intracellular RNase H specifically cleaved the RNA in the DNA/RNA hybrids and released H1 and H2 at high local concentrations, facilitating target-triggered HCR for the amplified imaging of TR in living cells. This DNA/RNA hybrid nanosphere exhibited a higher signal-to-noise ratio compared with the traditional HCR. According to a similar design, the Xu’s group designed a RNase H-responsive, self-assembled DNA/RNA nanosphere with dual-signal amplification for the analysis of miRNA, in which four different hairpins (THp, H1, H2, H3) were loaded on long-strand RNA through hybridization (Figure 9c) [94]. When intracellular RNase H appeared, the hairpin DNA was released and caused the miRNA-initiated cascade signal amplification of CHA and HCR for the sensitive sensing of the target.
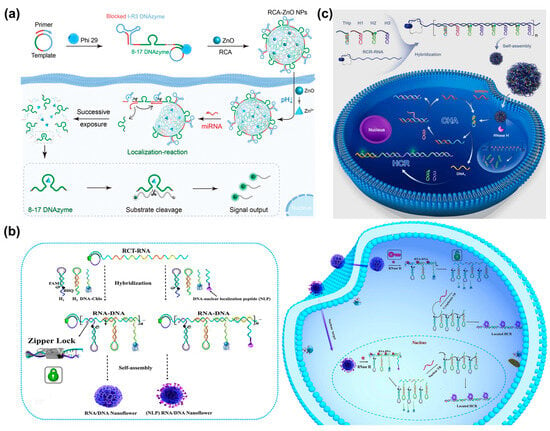
Figure 9.
(a) The preparation and working mechanism of RCA-ZnO nanospheres for miRNA-21 imaging, reproduced with permission from Ref. [89]. Copyright 2023, American Chemical Society Publishers. (b) The formation of DNA/RNA hybrid nanospheres and their applications in the amplified imaging of intracellular telomerase RNA (TR), reproduced with permission from Ref. [93]. Copyright 2020, Elsevier Publishers. (c) Self-assembled DNA/RNA nanospheres with cascade signal amplification for imaging intracellular miRNA, reproduced with permission from Ref. [94]. Copyright 2022, Elsevier Publishers.
Figure 9.
(a) The preparation and working mechanism of RCA-ZnO nanospheres for miRNA-21 imaging, reproduced with permission from Ref. [89]. Copyright 2023, American Chemical Society Publishers. (b) The formation of DNA/RNA hybrid nanospheres and their applications in the amplified imaging of intracellular telomerase RNA (TR), reproduced with permission from Ref. [93]. Copyright 2020, Elsevier Publishers. (c) Self-assembled DNA/RNA nanospheres with cascade signal amplification for imaging intracellular miRNA, reproduced with permission from Ref. [94]. Copyright 2022, Elsevier Publishers.
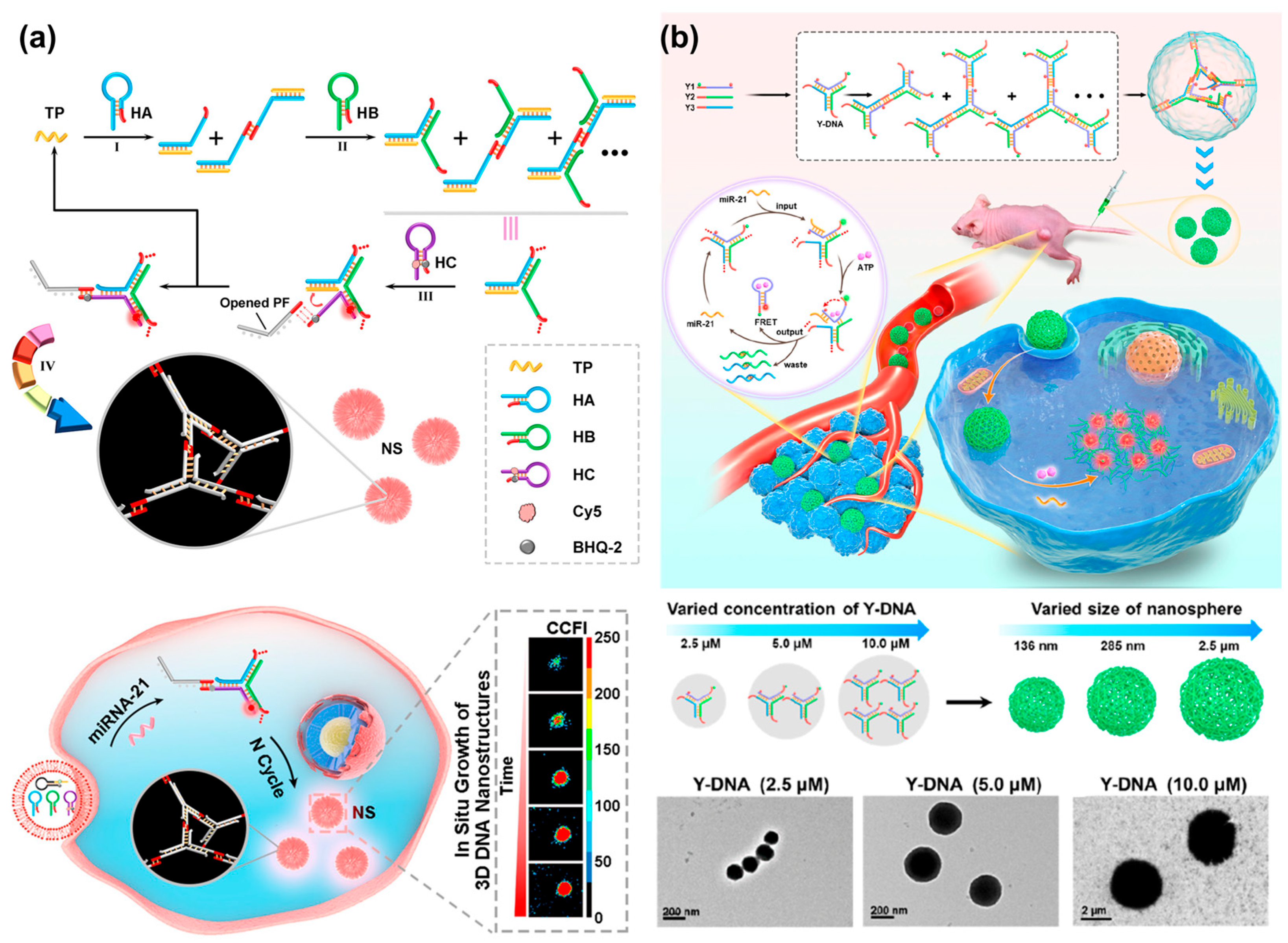
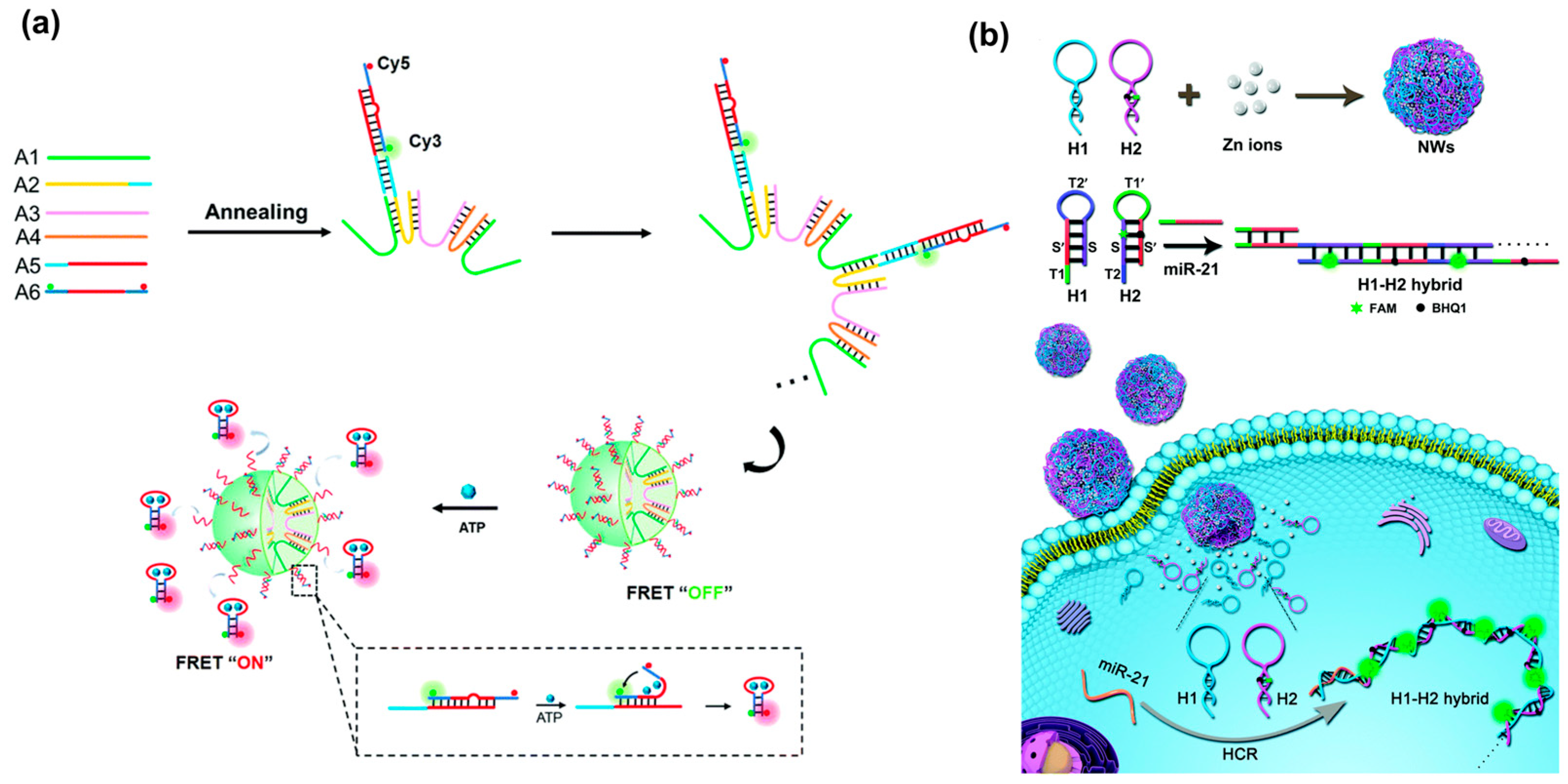
Without lsDNA as a building block, a palindrome-catalyzed self-assembly DNA nanosphere was constructed by the Wu’s group for in situ imaging of intracellular miRNA [35] (Figure 10a). When three hairpin DNAs with the same palindromic fragments were delivered into cells via lipofectamine 3000, a cascade strand displacement reaction occurred with the trigger of miRNA, accompanied by the release of miRNA and the formation of Y-shaped DNA units. The resultant Y-shaped DNA units were self-assembled into a 3D DNA nanosphere through the catalysis of palindromic fragments, yielding amplified fluorescence signals for in situ visualization of miRNA in real time. However, transfection was required, which might lead to complicated operation and damage to cells. Wang et al. recently reported a size-controllable DNA nanosphere based on Y-shaped DNA monomers with palindromic fragments for sensitive miRNA imaging in vivo, which could be effectively internalized into cells without additional transfection reagents [21] (Figure 10b). More importantly, only a type of Y-shaped DNA was designed and then self-assembled into DNA nanospheres through the base pairing of palindromic sequences. This design was simpler than previously reported DNA nanospheres assembled from Y-shaped units because DNA connectors and photo-cross-linking were not required. Meanwhile, the size of nanospheres was controllable by varying the concentration of Y-shaped DNA. Once the nanospheres entered the cells, miRNA would initiate the cyclic disassembly of DNA nanospheres with the fueling of ATP, leading to strong FRET signals.
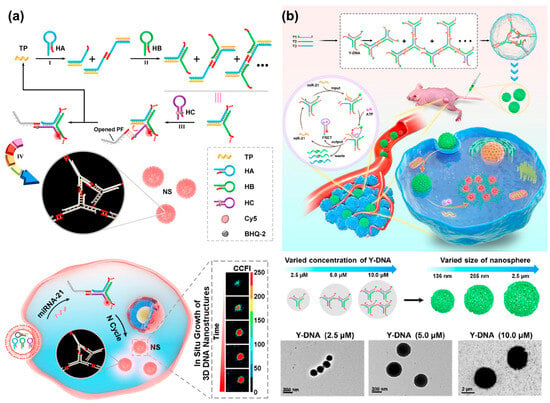
Figure 10.
(a) A palindrome-catalyzed self-assembly of 3D DNA nanospheres for in situ imaging of intracellular miRNA, reproduced with permission from Ref. [35]. Copyright 2020, American Chemical Society Publishers. (b) A size-controllable DNA nanosphere based on Y-shaped DNA monomers for amplified miRNA imaging in vivo and TEM images of DNA nanospheres with different sizes, reproduced with permission from Ref. [21]. Copyright 2022, American Chemical Society Publishers.
A simpler assembly method for DNA nanospheres was through short ssDNA self-assembly or metal–DNA coordination. For example, Cai et al. recently reported DNA nanospheres based on FRET nanoflare for intracellular ATP imaging, which were self-assembled through cyclic U-shaped hybridization between short ssDNA, as shown in Figure 11a [95]. Such DNA nanospheres were formed only using four ssDNA through simple annealing, and FRET nanoflare probes were loaded via hybridization. In the presence of ATP, it could specifically bind to its aptamer and release a hairpin probe, resulting in a high FRET signal. In addition to the self-assembly of ssDNA, Jia et al. synthesized nanowire balls (NWs) based on the metal–DNA coordination and HCR amplification technique through a one-pot annealing reaction [26]. By varying Zn ion/DNA molar ratios, highly monodispersed and uniformly sized DNA nanospheres were successfully synthesized. These bioinspired hybrid nanospheres could effectively deliver hairpin probes (H1 and H2) to living cells and in situ-triggered HCR for the amplified imaging of miRNA (Figure 11b). More interestingly, by integrating Y-shaped DNA with a metal–DNA coordination, a Y-shaped DNA@Cu3(PO4)2 hybrid nanosphere was reported by Yu et al. for TK1 mRNA imaging in living cells [96]. The introduction of Y-shaped DNA significantly decreased the size of Cu3(PO4)2, endowing it with a suitable size for cell uptake as a carrier. When the hybrid nanospheres were internalized into cells, they would be dissolved and liberated Y-shaped DNA probes to recognize TK1 mRNA via hybridization with the loop region of Y-DNA, resulting in red fluorescence. Interestingly, when biscyanine molecules were encapsulated in the DNA nanospheres via electrostatic adsorption, they could produce strong luminescent emission. Using this advantage, Wei et al. developed a label-free fluorescence probe for intracellular lysosome fluorescence imaging using the restriction of intramolecular rotation (RIR)-enhanced technique [97].
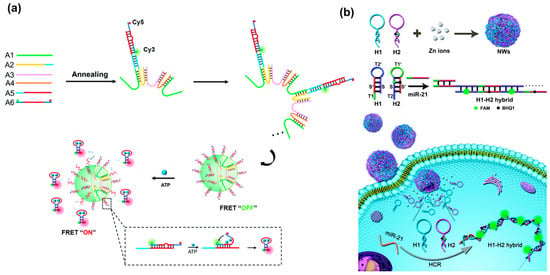
Figure 11.
(a) A DNA nanosphere self-assembled by cyclic U-shaped hybridization between short ssDNA for intracellular ATP imaging, reproduced with permission from Ref. [95]. Copyright 2021, Royal Society of Chemistry Publishers. (b) The preparation of nanowire balls (NWs) based on metal–DNA coordination for amplified miRNA imaging, reproduced with permission from Ref. [26]. Copyright 2020, Royal Society of Chemistry Publishers.
3.3. Cancer Therapy
3.3.1. Chemotherapy
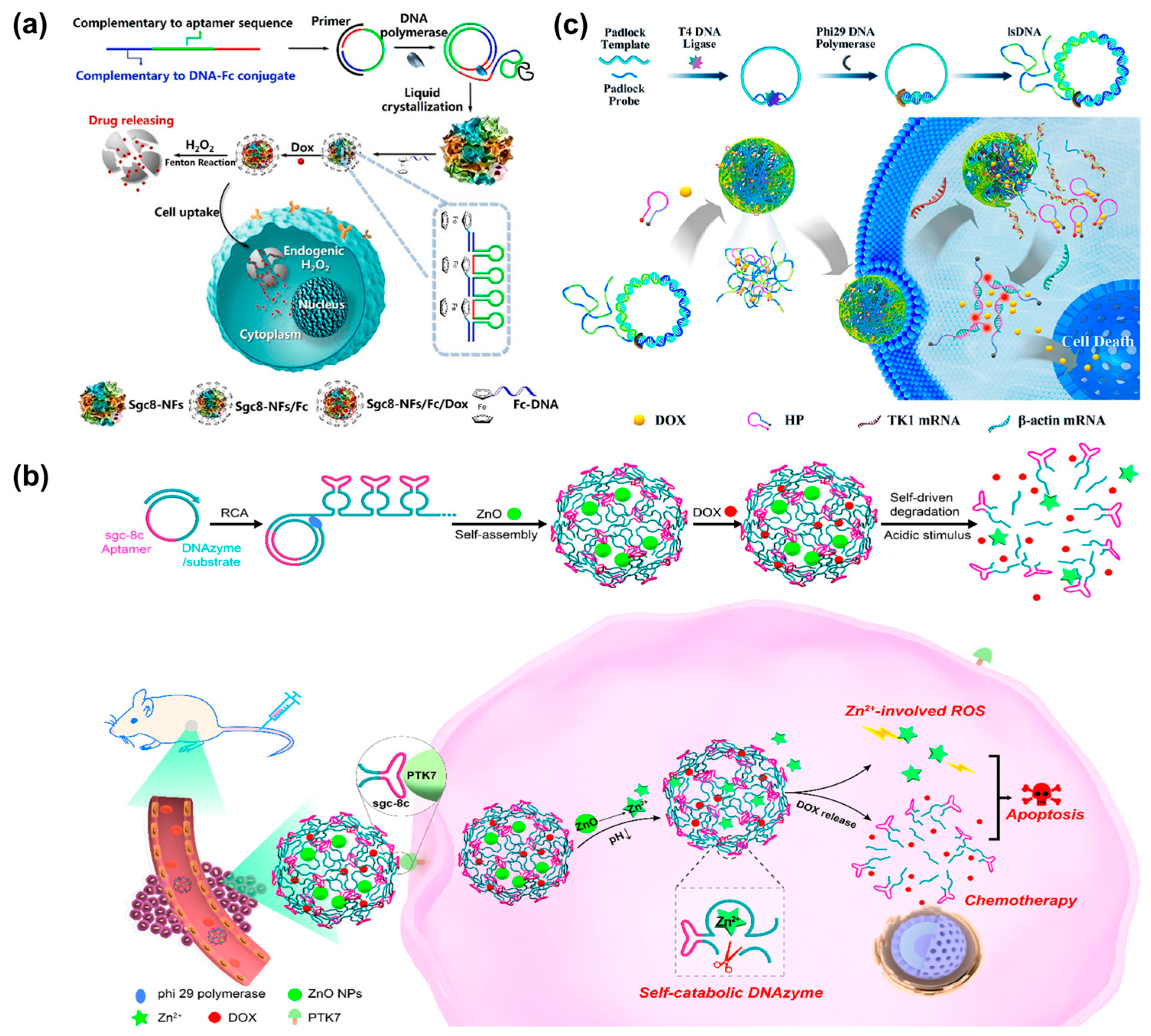
Doxorubicin (Dox), a class of broad-spectrum anticancer drugs, has been widely used in chemotherapy [98]. By mixing DNA nanospheres with Dox, NS-Dox composites would be generated. Benefiting from the specific recognition of cancer cells by sgc8-DNA nanospheres, the resultant NS-Dox composites could selectively deliver Dox to specific tumor sites for selective cancer therapy. Therefore, DNA nanospheres not only could specifically recognize cancer cells but could also be utilized as carriers for selective drug delivery in certain cancer cells [24]. To improve drug utilization and reduce undesirable side effects, it is necessary to develop a smart drug delivery system that can control the release of drugs in a stimuli-responsive manner. Herein, the Tan’s group designed a self-degradable DNA nanosphere (Sgc8-NSs-Fc/Dox) through the incorporation of a ferrocene (Fc) base, which could release drugs on-demand under the stimulation of H2O2 [99] (Figure 12a). Due to the introduction of the Fc base, Sgc8-NSs-Fc/Dox was size-controllable via hydrophobic interaction. Upon in the presence of H2O2, Sgc8-NSs-Fc/Dox would self-degrade through Fenton’s reaction induced by the Fc base, resulting in the controllable and efficient release of Dox and enhancing the therapeutic effect on cancer cells. In the same year, the Wang’s group reported a self-sufficient smart platform based on DNA nanospheres for precise drug delivery. DNA nanospheres were self-assembled by DNAzyme–substrate scaffolds generated via RCA, and then pH-responsive ZnO nanoparticles and anticancer drugs (Dox) were encapsulated into the nanospheres [100] (Figure 12b). Notably, the introduction of DNA-hydrolyzing DNAzyme allowed nanospheres to be degraded with the help of Zn2+ ions. Under the acidic microenvironment, ZnO was decomposed into Zn+ ions, which could be used as DNAzyme cofactors and induced the generation of reactive oxygen species (ROS). Thus, a DNAzyme-catalyzed self-cleavage reaction mediated by Zn2+ ion occurred, accompanied by the self-degradation of nanospheres and the precise release of Dox. As a result, the anticancer drug Dox, as well as Zn2+-involved ROS, synergistically enhanced therapeutic efficiency.
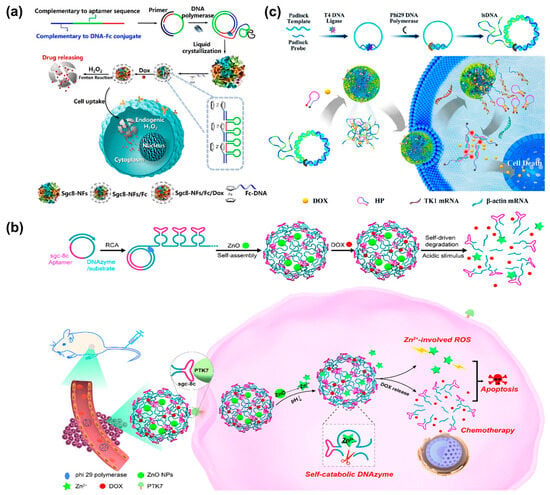
Figure 12.
(a) A self-degradable DNA nanosphere (Sgc8-NSs-Fc/Dox) for drug release under the stimulation of endogenous H2O2, reproduced with permission from Ref. [99]. Copyright 2019, American Chemical Society Publishers. (b) pH-responsive DNA nanospheres assembled by RCA products and ZnO nanoparticles for smart drug delivery, reproduced with permission from Ref. [100]. Copyright 2019, American Chemical Society Publishers. (c) mRNA-responsive DNA nanospheres self-assembled by RCA products for simultaneous bioimaging and cancer therapy, reproduced with permission from Ref. [101]. Copyright 2020, American Chemical Society Publishers.
Afterward, the Yang’s group proposed an intelligent drug delivery system (DDS) based on mRNA-responsive DNA nanospheres (DNA-NSs) self-assembled by RCA products, which could be loaded with hairpin probes and Dox for simultaneous bioimaging and cancer therapy [101] (Figure 12c). lsDNA with a specific recognition domain to TK1 mRNA was generated via RCA, and Dox was inserted into the G-C/C-G base pairs of functional hairpin probes labeled with Cy5 and BHQ3. Afterward, lsDNA, hairpins, and Dox were co-annealed to form DNA nanospheres (DNA-NSs). When TK1 mRNA appeared, it could specifically hybridize with lsDNA and induce the decomposition of DNA-NS, leading to the release of hairpin probes. As a result, the released hairpin probes were opened by β-actin mRNA, accompanied by the generation of fluorescence signals and the release of Dox. In addition, DNA nanospheres assembled by Y-shaped units or short ssDNA could also encapsulate Dox for antitumor chemotherapy [30]. For example, Liu et al. constructed a self-degradable DNA nanosphere via the hybridization of two cytosine-rich strands and two linker strands for the specific delivery of Dox to cancer cells [33]. In a slightly acidic environment, DNA nanospheres would be decomposed owing to the formation of i-motif structures, leading to the liberation of Dox. This pH-responsive targeted therapeutic effect has been validated at the cellular level and in vivo, providing a potential tool for tumor therapy.
3.3.2. Gene Therapy
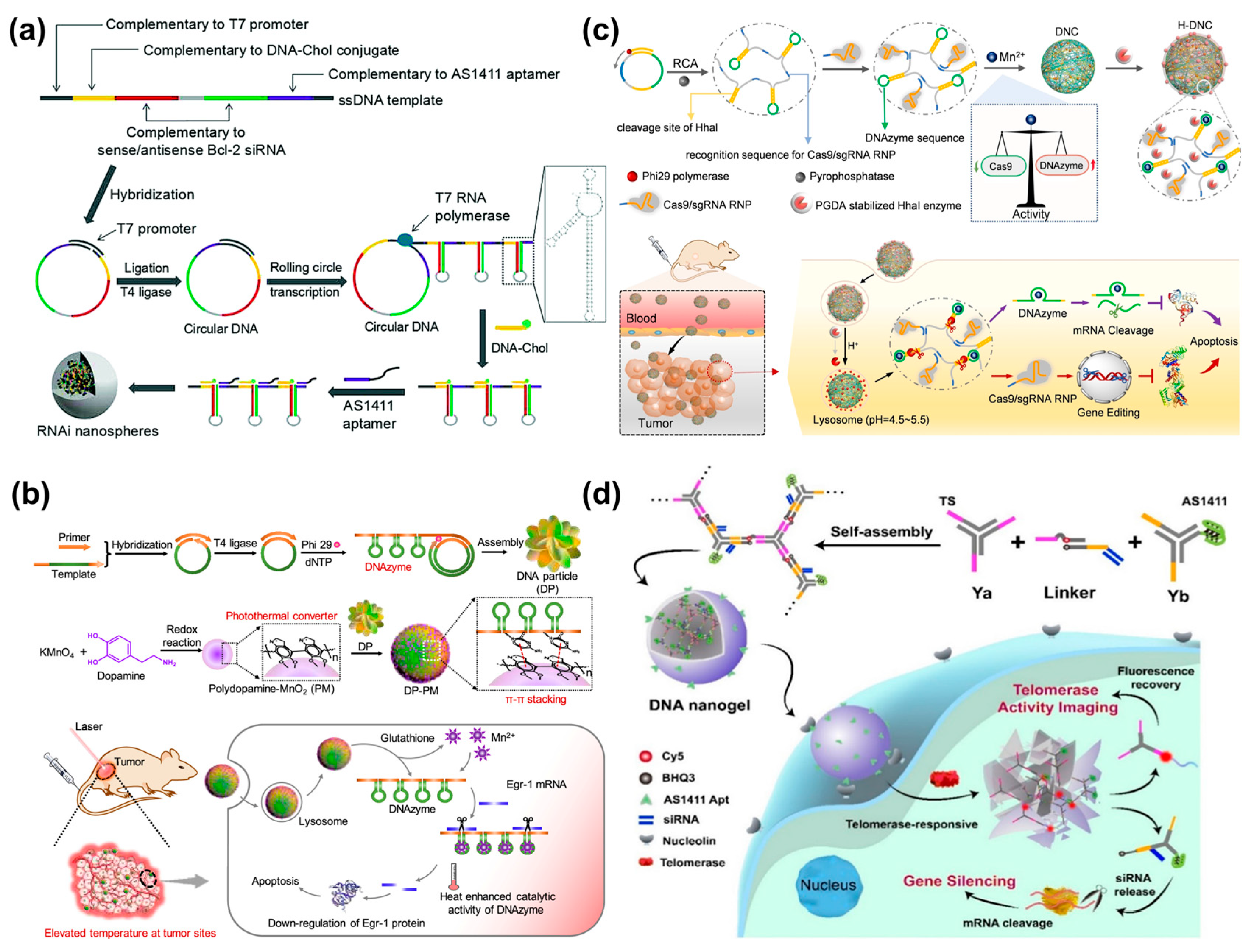
Small interfering RNA (siRNA) can specifically suppress the expression of target genes through RNA interference (RNAi) [102]. However, the efficient transport of exogenous siRNA into living systems is essential for the success of siRNA therapy. DNA nanospheres are considered as a class of ideal gene delivery carriers, owing to good biocompatibility, structural stability, high loading capacity, and excellent cell permeability [25,28,103]. Hammond’s group reported RNAi microsponges based on long RNA transcripts with numerous repeated siRNA precursors generated via rolling circle transcription (RCT) for efficient siRNA delivery [104]. However, the size of the microsponges was too large for cellular uptake. Interestingly, after coating cationic polyethylenimine (PEI) on the surface of particles through electrostatic interaction, the size of RNAi microsponges was dramatically reduced to 200 nm and still retained the same amount of siRNA precursors, achieving significant gene silencing efficiency in vivo. However, PEI could be cytotoxic to cells. To improve the safety of nanocarriers, Cheng et al. presented a safe and tumor-targetable RNAi nanosphere for siRNA delivery [105] (Figure 13a). AS1411 aptamer and cholesterol-modified DNA were introduced into long RNA transcripts via base paring, in which AS1411 aptamer was used for MCF-7 cell targeting and DNA-Chol could significantly decrease the size of RNAi microspheres by up to 120 nm that was suitable for cellular uptake. Once the RNAi nanospheres were internalized by target cells, large amounts of siRNA would be produced by Dicer enzyme cleavage, thereby inhibiting the gene expression of Bcl-2.
DNAzyme, as an ssDNA sequence with catalytic activity, has been developed as a promising therapeutic agent [106,107,108]. Recently, the Yang’s group developed a synergistic DNA-polydopamine-MnO2 nanoball (DP-PM) for DNAzyme-mediated gene therapy [109] (Figure 13b). The nanoballs were composed of a DNA particle (DP) and polydopamine-MnO2 (PM), in which DP was formed by the self-assembly of lsDNA containing repeated DNAzyme units generated via RCA, and PM was synthesized through a redox reaction of KMnO4 and dopamine. Under near-infrared-light irradiation, polydopamine caused a rise in temperature via photothermal conversion, and PM was reduced to Mn2+ by glutathione (GSH), resulting in DNAzyme cleaving Egr-1 mRNA for gene silencing. In addition, the enhancement in temperature induced heat stress to inhibit tumors.
In recent years, the emergence of clustered regularly interspaced short palindromic repeats (CRISPR) technology has attracted considerable attention in the field of gene editing [110,111]. Among them, Cas9/sgRNA ribonucleoproteins have been regarded as powerful therapeutic agents, which can downregulate target genes to inhibit the progression of tumors [112,113,114]. Thus, the Yang’s group constructed a proton-activatable DNA nanosphere to simultaneously transport Cas9/sgRNA and DNAzyme for multimodal gene therapy (Figure 13c) [115]. A lsDNA containing a repeated sgRNA complementary sequence, DNAzyme, and Hha I cleavage sites was produced by RCA as the building blocks of DNA nanospheres. Here, Mn2+ was introduced not only as a cofactor of DNAzyme but was also used to compress RCA products into nanospheres. Meanwhile, an acid-degradable polymer was employed to coat the Hha I enzyme, and then the resultants were assembled on the nanospheres’ surface to prepare proton-activatable DNA nanospheres. In an acidic lysosomal environment, the polymer was decomposed, and consequently, Hha I was exposed to cleave the specific sites, resulting in the liberation of Cas9/sgRNA, DNAzyme, and Mn2+. The released Cas9/sgRNA and DNAzyme could regulate gene expression and achieve combined gene therapy. Subsequently, Shi et al. developed miRNA-responsive DNA nanospheres to deliver Cas9/sgRNA to tumor sites for enhanced gene editing [116].
Similarly, self-assembled DNA nanospheres based on Y-shaped DNA units can also be utilized to deliver siRNA for gene therapy. For example, the Yang’s group developed a smart DNA nanosphere with targeting and telomerase-responsive capabilities for simultaneous telomerase activity imaging and gene therapy [117] (Figure 13d). Here, DNA nanospheres were assembled by a Y-shaped DNA with telomeric primer (Ya), a loading siRNA double-strands linker with fluorophores, and a Y-shaped DNA with AS1411 aptamer (Yb). When in the presence of telomerase, the DNA nanospheres collapsed due to the extension of the telomere primer and the separation of Ya from the linker. As a result, the fluorescence was recovered and siRNA was released, allowing for imaging of intracellular telomerase activity and effective gene regulation.
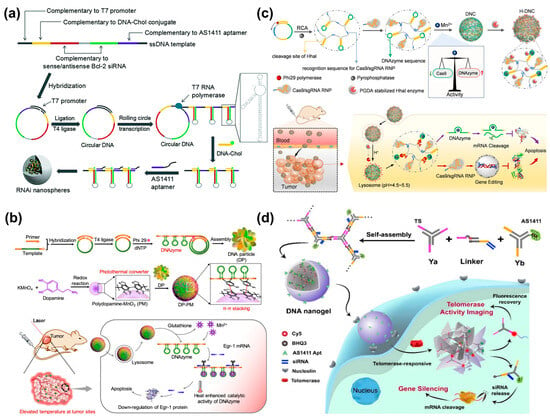
Figure 13.
(a) Safe and tumor-targetable RNAi nanospheres self-assembled by long RNA strands for siRNA delivery, reproduced with permission from Ref. [105]. Copyright 2018, Royal Society of Chemistry Publishers. (b) DNA-polydopamine-MnO2 nanospheres for DNAzyme-mediated gene therapy, reproduced with permission from Ref. [109]. Copyright 2021, American Chemical Society Publishers. (c) A proton-activatable DNA nanosphere to simultaneously transport Cas9/sgRNA and DNAzyme for multimodal gene therapy, reproduced with permission from Ref. [115]. Copyright 2022, Wiley Publishers. (d) Telomerase-responsive smart DNA nanospheres for simultaneous telomerase activity imaging and gene therapy, reproduced with permission from Ref. [117]. Copyright 2020, American Chemical Society Publishers.
Figure 13.
(a) Safe and tumor-targetable RNAi nanospheres self-assembled by long RNA strands for siRNA delivery, reproduced with permission from Ref. [105]. Copyright 2018, Royal Society of Chemistry Publishers. (b) DNA-polydopamine-MnO2 nanospheres for DNAzyme-mediated gene therapy, reproduced with permission from Ref. [109]. Copyright 2021, American Chemical Society Publishers. (c) A proton-activatable DNA nanosphere to simultaneously transport Cas9/sgRNA and DNAzyme for multimodal gene therapy, reproduced with permission from Ref. [115]. Copyright 2022, Wiley Publishers. (d) Telomerase-responsive smart DNA nanospheres for simultaneous telomerase activity imaging and gene therapy, reproduced with permission from Ref. [117]. Copyright 2020, American Chemical Society Publishers.
3.3.3. Photodynamic Therapy (PDT)
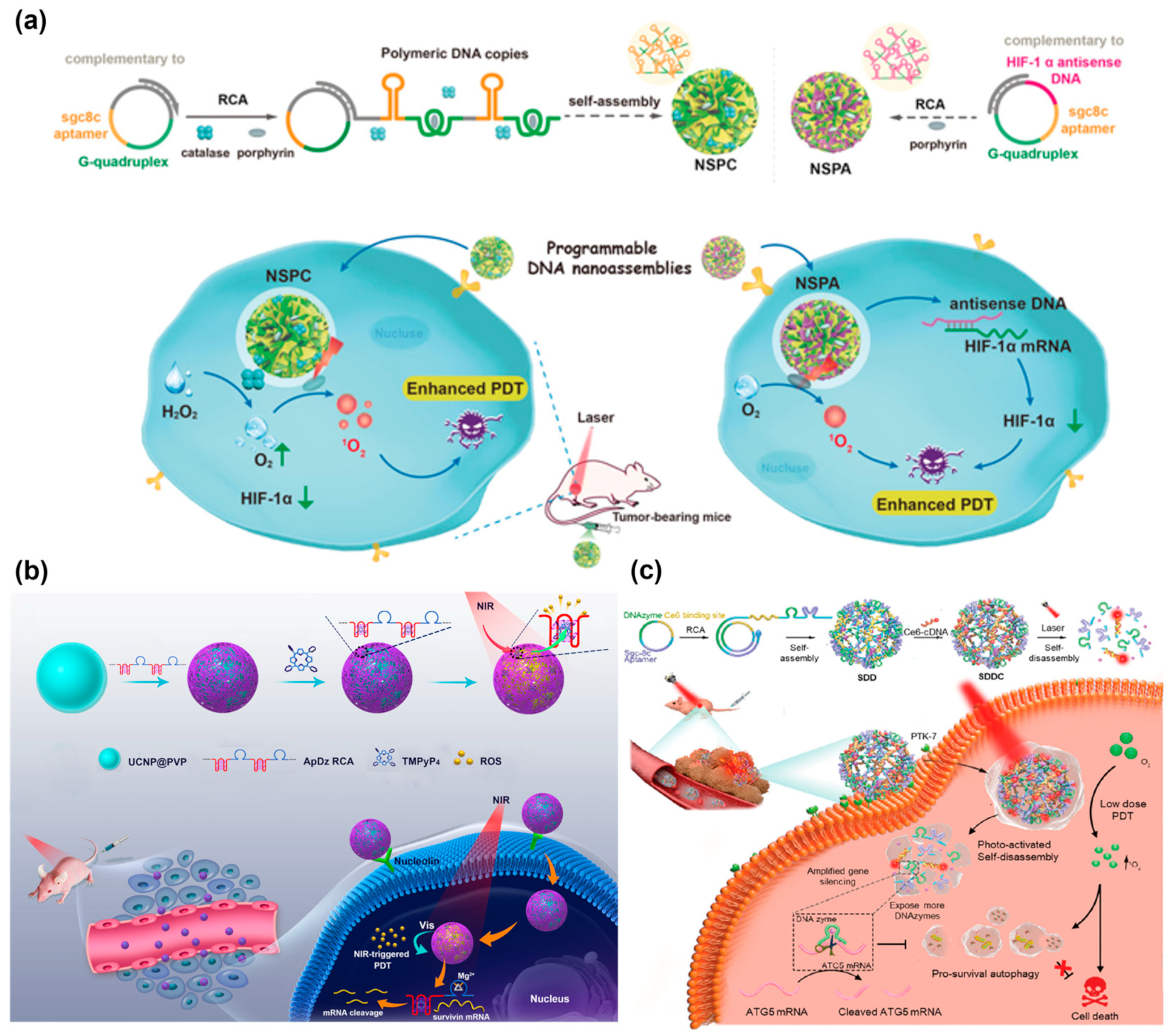
Photodynamic therapy (PDT) is considered as a safe and promising cancer therapy strategy due to its minimal or non-invasiveness, rapid therapeutic effect, and avoidance of drug resistance [118,119,120]. It mainly uses photosensitizers to generate singlet oxygen (SO) and reactive oxygen species (ROS) under light irradiation and then induce DNA damage and cell apoptosis, which involves the consumption of O2 [121]. However, the efficacy of PDT was unsatisfactory to a certain extent due to the hypoxic microenvironment of solid tumors that limited the supply of O2. The Liu’s group created an effective method to enhance the efficacy of PDT by two different DNA nanospheres (NSPC and NSPA) that were programmatically self-assembled by RCA products [122] (Figure 14a). For NSPC assembly, the complementary sgc8c aptamer and G-quadruplex were encoded in the RCA template for the specific targeting of cancer cells and intercalation with the porphyrin photosensitizer, TMPyP4, respectively. Subsequently, the catalase and photosensitizer were encapsulated into DNA nanospheres through the self-assembly of RCA products. For NSPA assembly, the complementary sgc8c aptamer, G-quadruplex, and hypoxia-inducible factor 1α (HIF-1α) antisense DNA were introduced into the RCA template, in which HIF-1α antisense DNA could regulate HIF-1α to enhance the sensitization of PDT. After being internalized by tumor cells, NSPC would catalyze H2O2 to O2, increasing the supply of O2 and thereby enhancing the efficacy of PDT. Meanwhile, HIF-1 α antisense DNA could bind HIF-1 α mRNA and downregulate its expression to improve the sensitization of PDT, further enhancing PDT.
Survivin, as an antiapoptosis protein, has been found to play a positive role in cancer progression [123]. Hence, Jin et al. presented a nanoplatform for the enhancement of PDT by downregulating the expression of survivin [124] (Figure 14b). A lsDNA containing AS1411 aptamer, G-quadruplex, and DNAzyme that specifically cleaved survivin mRNA was generated via RCA, and then it was loaded on the upconversion nanoparticles (UCNPs) through electrostatic interaction. Under NIR light irradiation, the photosensitizer TMPyP4 produced ROS to kill cancer cells. Meanwhile, the DNAzyme silenced the expression of survivin, further enhancing the efficiency of PDT.
However, a high dose of photosensitizer is required for efficient PDT, which causes certain side effects [125]. Thus, it is urgent to fabricate an intelligent delivery platform for low-dose photodynamic therapy. The Zhang’s group constructed a photoactivated DNA nanosphere with self-degradable properties, which could effectively impair cytoprotective autophagy via silencing autophagy-related genes [126] (Figure 14c). ATG5 mRNA is a vital autophagy-related protein, and inhibiting the expression of ATG5 mRNA has been reported to impair cytoprotective autophagy [127,128]. The DNA nanospheres were formed by the self-assembly of RCA lsDNA containing sgc8c aptamer, DNAzyme for silencing ATG5 mRNA, and photosensitizer (Ce6) binding sites. It was found that light irradiation could induce the self-disassembly of nanospheres, resulting in the release of Ce6 for PDT and the exposure of DNAzyme to cleave ATG5 mRNA for autophagy suppression. The in vivo experiments have demonstrated that DNA nanospheres could inhibit tumor growth with a very low injection dose of Ce6, which was 100 times lower than the generally applied dose.
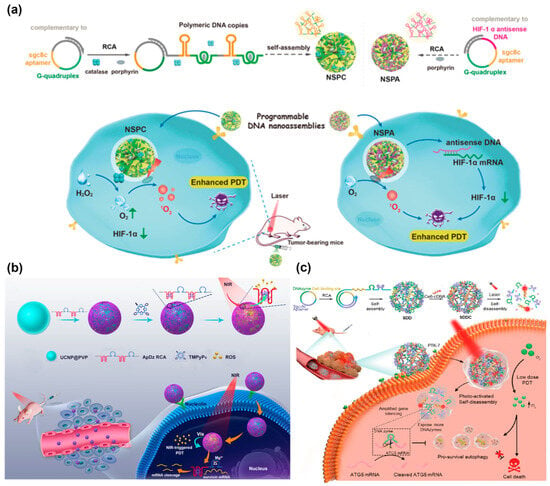
Figure 14.
(a) Two DNA nanospheres (NSPC and NSPA) programmatically self-assembled from long DNA strands generated via RCA to enhance the efficacy of PDT, reproduced with permission from Ref. [122]. Copyright 2020, Wiley-VCH Publishers. (b) NIR light-responsive DNA nanospheres for enhancing PDT by inhibiting the expression of survivin, reproduced with permission from Ref. [124]. Copyright 2020, American Chemical Society Publishers. (c) Photoactivated DNA nanospheres with self-degradable properties to silence autophagy-related genes for enhancing PDT with a low dose of photosensitizer, reproduced with permission from Ref. [126]. Copyright 2021, Wiley-VCH Publishers.
Figure 14.
(a) Two DNA nanospheres (NSPC and NSPA) programmatically self-assembled from long DNA strands generated via RCA to enhance the efficacy of PDT, reproduced with permission from Ref. [122]. Copyright 2020, Wiley-VCH Publishers. (b) NIR light-responsive DNA nanospheres for enhancing PDT by inhibiting the expression of survivin, reproduced with permission from Ref. [124]. Copyright 2020, American Chemical Society Publishers. (c) Photoactivated DNA nanospheres with self-degradable properties to silence autophagy-related genes for enhancing PDT with a low dose of photosensitizer, reproduced with permission from Ref. [126]. Copyright 2021, Wiley-VCH Publishers.
3.3.4. Immunotherapy
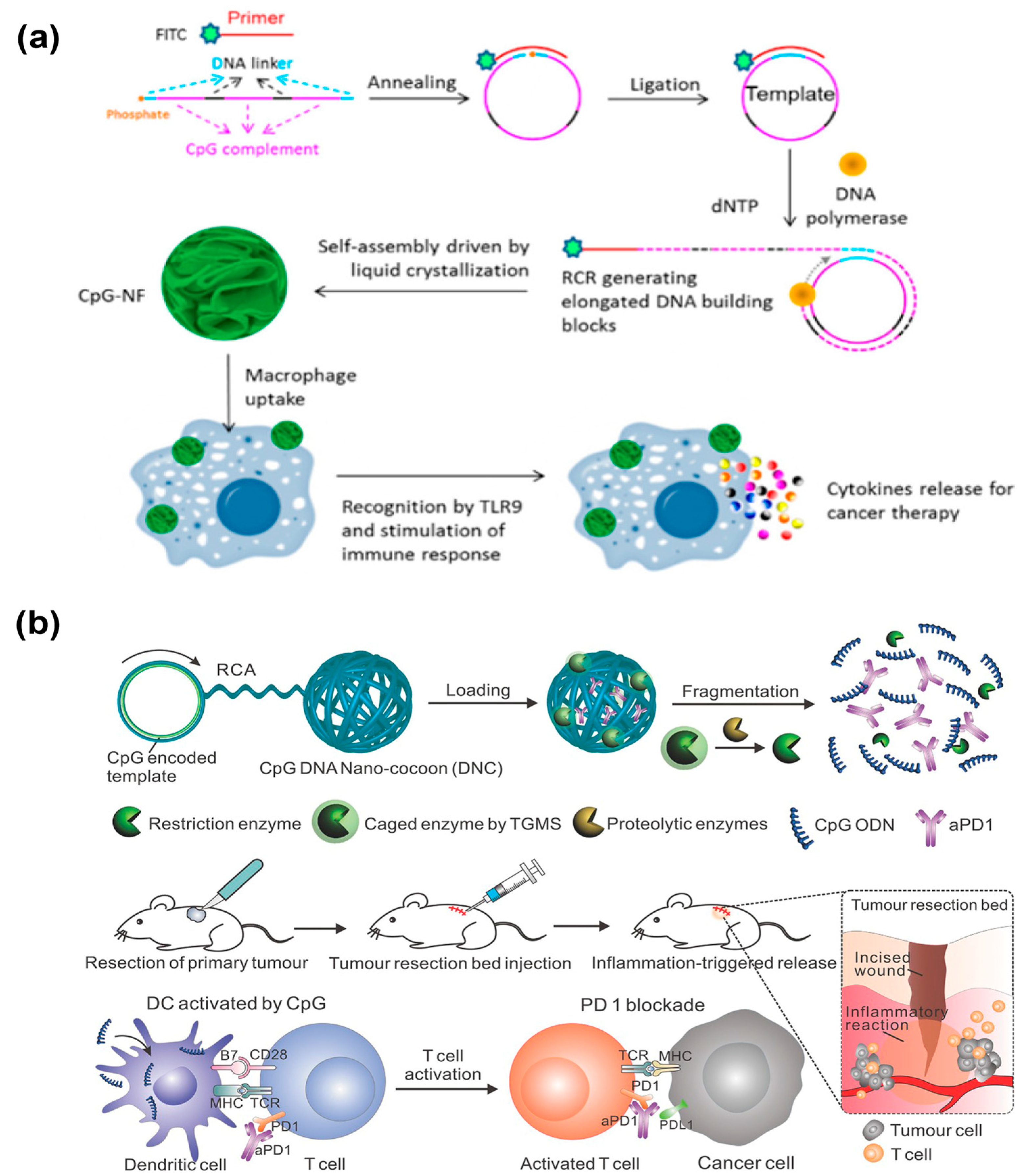
Different from the abovementioned therapy strategies, immunotherapy can modulate the function of immune cells and strengthen the immune system to specifically kill cancer cells [129,130]. The unmethylated cytosine–guanine oligodeoxynucleotide (CpG) has been regarded as a safe and effective immunomodulator to boost the immune response and combat cancers [131]. However, there are still several challenges to successfully delivering synthetic CpG to immune cells. In 2015, the Tan’s group employed DNA nanospheres that were self-assembled by lsDNA generated via RCR as vehicles to efficiently deliver CpG for potent immunostimulation (Figure 15a) [132]. Such CpG nanospheres have been demonstrated to stimulate the proliferation of immune cells through the activation of Toll-like receptor 9 (TLR 9), which could further secrete various immune regulators including TNF-α, IF-6, and IF-10 and induce cancer cells apoptosis and necrosis. Programmed cell death protein 1 (PD-1) has been found to be expressed in various immune cells. Using anti-PD antibodies to inhibit the interaction between PD-1 and PD-L1 (PD-1 and its ligand 1) can strengthen the immune response and kill cancer cells [133,134,135]. Therefore, Wang et al. developed an innovative DNA nanosphere to controllably release CpG and anti-PD1 antibody (aPD1) for enhanced cancer immunotherapy [136]. Similarly, DNA nanospheres were self-assembled by long-stranded RCA products encoding the CpG sequences and cleavage sites of restriction enzyme Hhal (Figure 15b). The caged restriction enzyme and aPD1 were incorporated into DNA nanospheres. Under the trigger of the inflammatory condition, the caged enzyme could be activated, and subsequently, the DNA nanospheres were degraded to release CpG as well as aPD1, facilitating anticancer immune response for cancer immunotherapy.
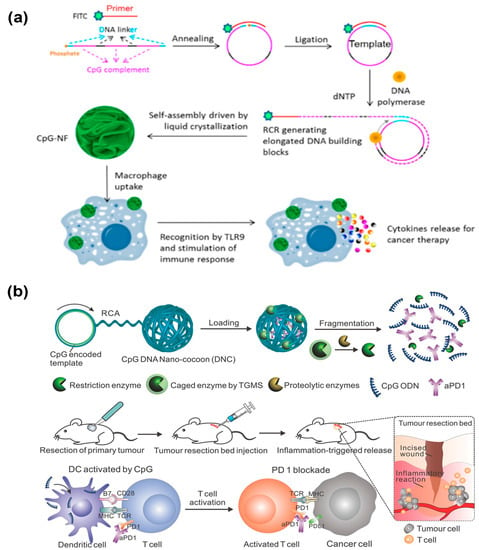
Figure 15.
(a) DNA nanospheres as vehicles to effectively deliver CpG for potent immunostimulation, reproduced with permission from Ref. [132]. Copyright 2015, American Chemical Society Publishers. (b) Stimulus-responsive DNA nanospheres to control the release of CpG and anti-PD1 antibody (aPD1) for enhanced immunotherapy, reproduced with permission from Ref. [136]. Copyright 2016, Wiley-VCH Publishers.
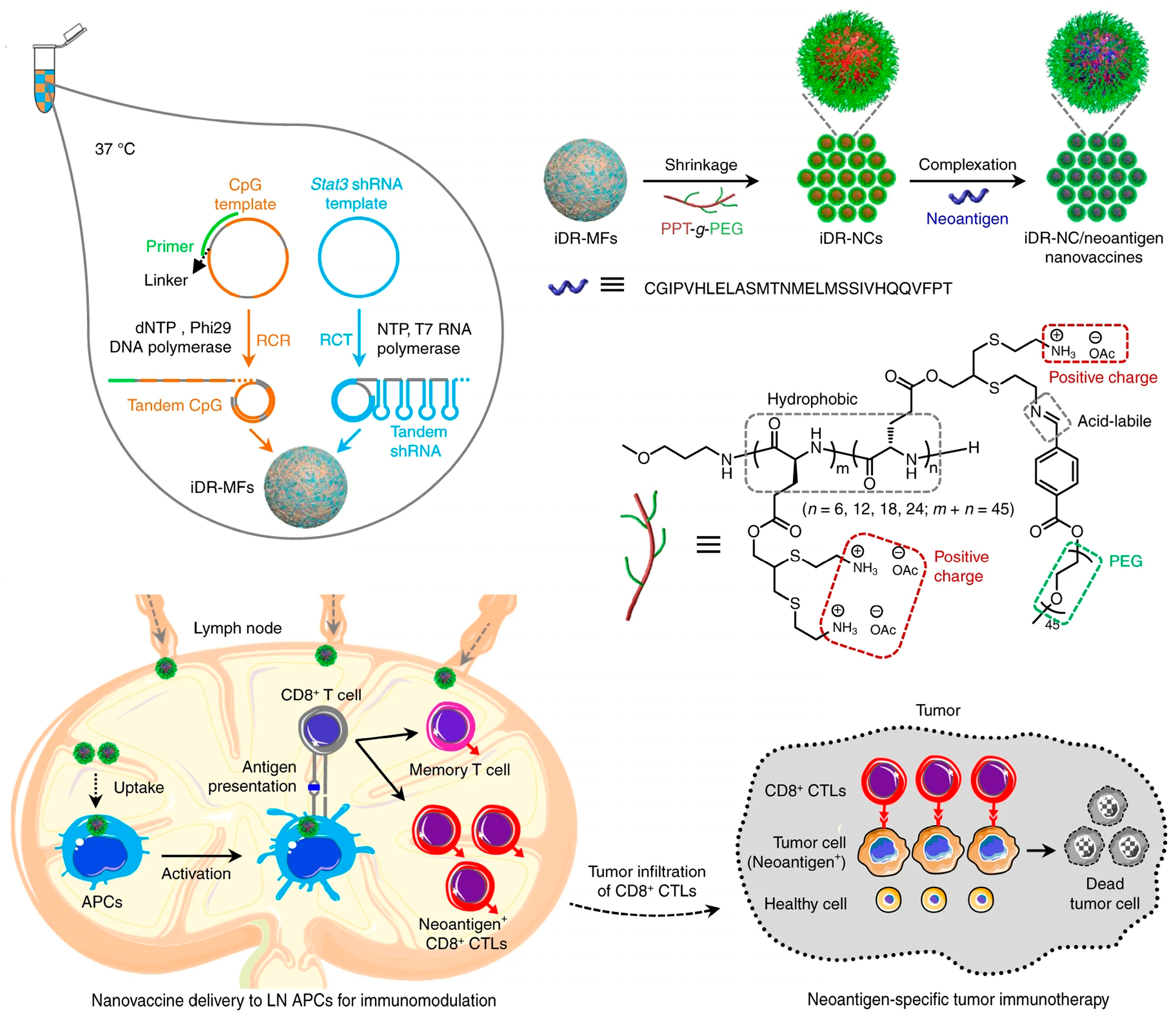
Tumor-specific neoantigens derived from mutations in tumors are considered as a class of safe immunotherapies and are expressed only in tumor cells but not in normal cells [137,138]. Therefore, using neoantigens as a vaccine can avoid autoimmunity against normal tissue. Zhu et al. developed intertwining DNA–RNA nanospheres (iDR-NSs) to efficiently deliver CpG, short hairpin (shRNA) adjuvants, and peptide neoantigens for cancer immunotherapy (Figure 16) [139]. Here, one lsDNA-encoding tandem CpG was generated via RCR, and the other RNA long-strand with shRNA sequences as building blocks was produced via RCT, respectively. The resultant lsDNA and RNA were self-assembled to intertwining DNA–RNA microspheres (iDR-MSs). In order to obtain a suitable size for cellular uptake, biocompatible PEG-grafted polypeptide (PPT-g-PEG) copolymers were employed to compress the microspheres (MSs) into iDR-NSs, and then peptide neoantigens were loaded via hydrophobic interactions. As a result, CpG and shRNA activated antigen-presenting cells (APCs) for antigen presentation, and neoantigen nanovaccines triggered frequent specific CD8+ T cells and induced T-cell memory, leading to the significant inhibition of tumor progression.
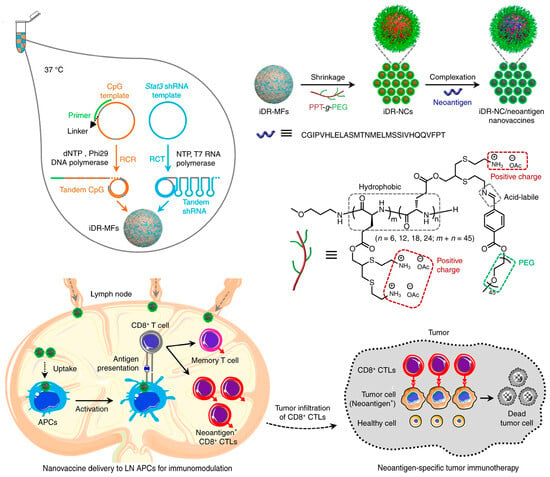
Figure 16.
Intertwining DNA–RNA nanospheres (iDR-NSs) via self-assembly to efficiently deliver CpG, short hairpin (shRNA) adjuvants, and peptide neoantigens for cancer immunotherapy, reproduced with permission from Ref. [139]. Copyright 2017, Nature Publishers.
To effectively deliver vaccine components, boost immune response, and improve therapeutic effect, the Liu’s group designed DNA nanospheres with applicable sizes to effectively deliver CpG for immune therapy [140]. Such DNA nanospheres were self-assembled by Y-shaped DNA encoding CpG sequences, L-DNA with CpG fragments, and E-DNA, which showed tunable sizes by varying the molar ratio of Y-DNA and E-DNA. More importantly, DNA nanospheres could efficiently accumulate in lymph nodes and enhance dendritic cell internalization, activating the immune response in mice.
3.3.5. Combined Therapy
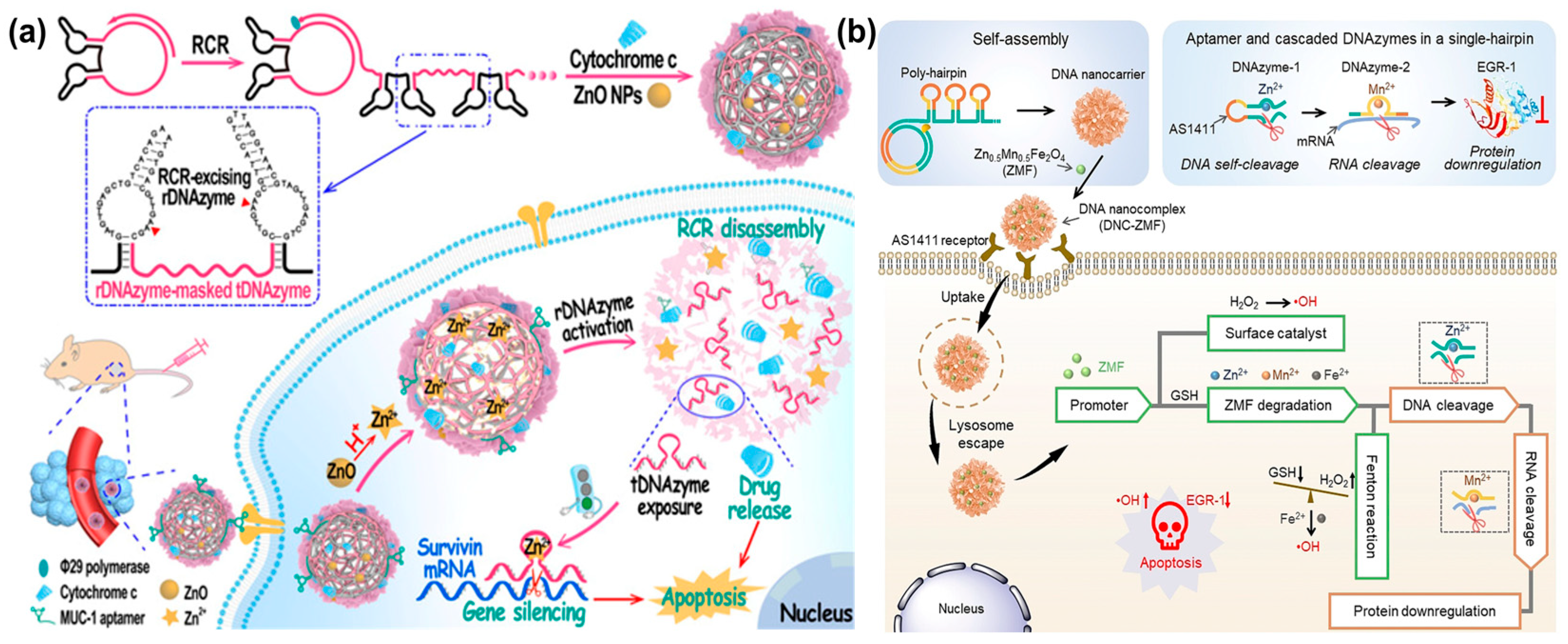
It is difficult to obtain a satisfactory therapeutic effect with a single-mode treatment strategy owing to the complexity of tumor pathogenesis. Thus, multimodal combination therapies have been positively pursued by many groups. For example, the Wang’s group designed a self-catabolic DNAzyme nanosphere for drug delivery and gene silencing, which was formed via the simple self-assembly of RCR products that encoded MUC-1 aptamer, self-hydrolyzing DNAzyme, and therapeutic DNAzyme [141] (Figure 17a). Meanwhile, ZnO nanoparticles (ZnO NPs) and pro-apoptotic cytochrome c protein were encapsulated into DNA nanospheres for supplying DNAzyme cofactors Zn2+ and activating cell apoptosis, respectively. When DNA nanospheres were internalized by specific cancer cells, ZnO NPs were dissolved to Zn2+ cofactors for DNAzyme cleavage under the stimulation of an acid microenvironment. As a result, self-hydrolyzing DNAzyme was activated to cleave RCR, releasing therapeutic DNAzyme and cytochrome c to cut survivin mRNA, thereby promoting cell apoptosis and achieving synergetic gene silencing and chemotherapy. In the same year, the Yang’s group reported a DNA nanosphere containing DNAzymes and promoter-like Zn-Mn-Ferrite (ZMF) for simultaneous gene/chemo-dynamic therapy [142] (Figure 17b). The DNA nanosphere was formed based on RCA lsDNA containing AS1411 aptamer, self-cleaving DNAzyme, and therapeutic DNAzyme. Notably, promoter-like ZMF was adsorbed on the DNA nanospheres via electrostatic interaction, which could be decomposed to Zn2+, Mn2+, and Fe2+ under the stimulation of H+ and glutathione (GSH). As a result, Zn2+ and Mn2+ ion-mediated DNAzyme cleaved Egr-1 mRNA for gene therapy, wherein Zn2+, Mn2+, and Fe2+ induced a Fenton reaction to produce free radicals for chemo-dynamic therapy.
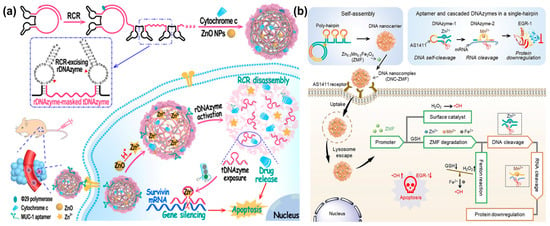
Figure 17.
(a) A self-catabolic DNAzyme nanosphere for drug delivery and gene silencing, reproduced with permission from Ref. [141]. Copyright 2021, Wiley-VCH Publishers. (b) A DNA nanosphere containing DNAzymes and promoter-like Zn-Mn-Ferrite (ZMF) for combined gene/chemo-dynamic therapy, reproduced with permission from Ref. [142]. Copyright 2021, Wiley-VCH Publishers.
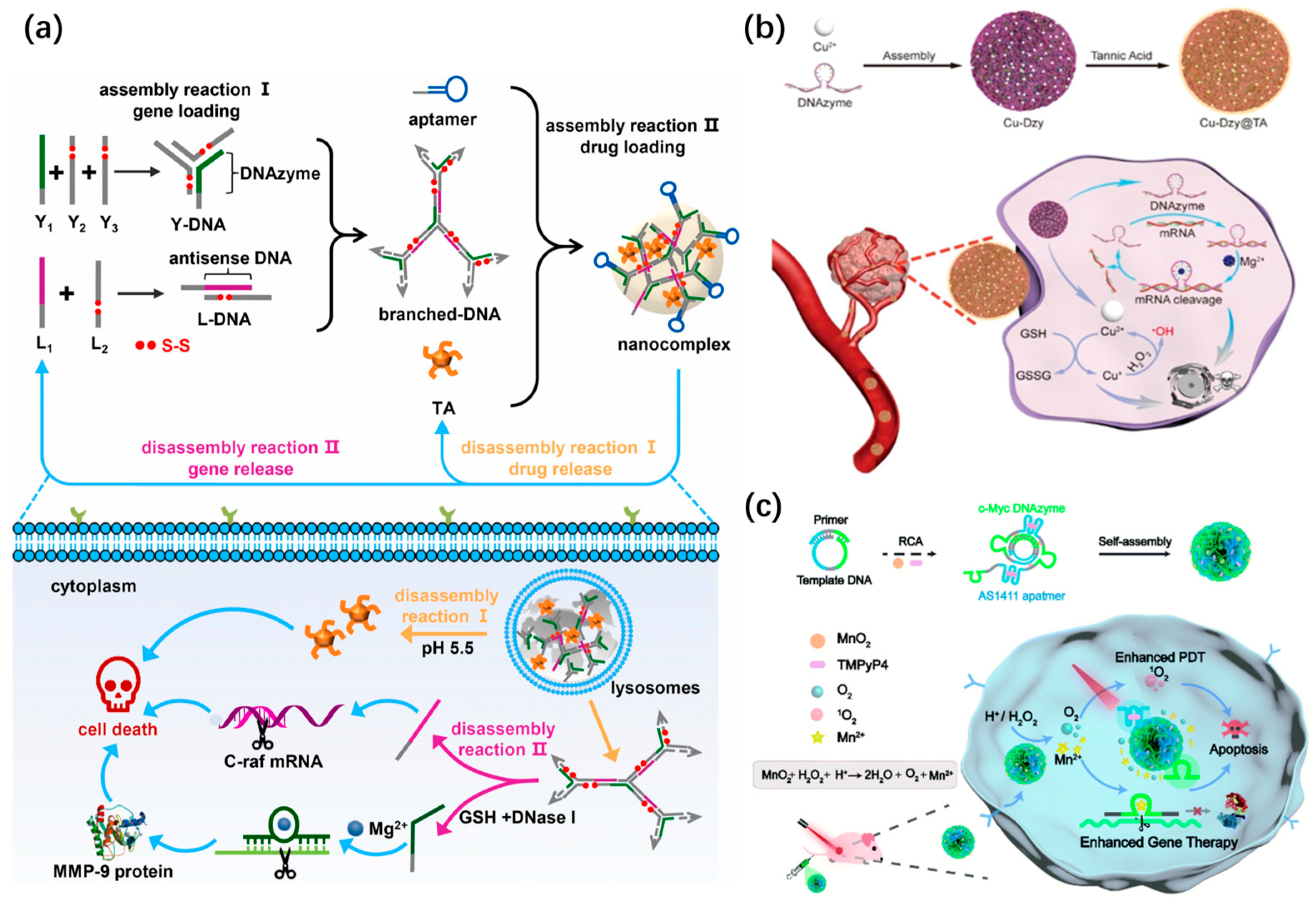
DNA nanospheres with self-assembly using Y-shaped DNA or X-shaped DNA units have also been constructed as vehicles for the delivery of drugs and gene agents [28]. For example, the Yang’s group developed a nanosphere with controllable disassembly properties for efficient gene/drug release [143] (Figure 18a). Specifically, Y-shaped DNA containing DNAzyme and disulfide bonds (-S-S-), as well as L-DNA duplex with antisense DNA, were hybridized into branched DNA. The resultant branched DNA, aptamer, and tannic acid (TA) were self-assembled into DNA nanospheres. The lysosomal acidic microenvironment triggered the disassembly of DNA nanospheres to liberate TA and branched DNA. Meanwhile, the glutathione (GSH) specifically cleaved disulfide bonds to precisely release two genes from branched DNA. The DNA nanospheres have been proven to achieve multiple-gene therapy and chemotherapy in vivo, in which TA promoted tumor cell apoptosis and two genes inhibited cell proliferation and migration.
Recently, metal–DNA coordination hybrid nanospheres have been widely applied for multimodal cancer therapy. In 2019, the Li’s group constructed a biomimetic coordination nanosphere via a simple and universal strategy for the codelivery of gene therapeutic agents and the drug doxorubicin [144]. G3139, as an antisense oligonucleotide, can regulate the expression of the antiapoptotic protein Bcl-2 [145]. Thus, G3139 was selected as a gene therapeutic agent. The hybrid nanospheres with uniform sizes were formed through simple mixing and annealing between G3139, FeII ions, and Dox. Such a coordination of nanospheres could be combined with gene therapy and chemotherapy for the synergistic inhibition of tumor growth. Later, the authors synthesized a copper–DNAzyme hybrid DNA nanosphere for combination gene and chemo-dynamic therapy [146] (Figure 18b). Notably, the released Cu2+ from DNA nanospheres could be reduced to Cu+ by glutathione, and then the resultant Cu+ allowed for the catalysis of endogenous H2O2 to produce highly toxic hydroxyl radicals (·OH) for chemo-dynamic therapy.
The Liu’s group recently designed a MnO2-encapsulated DNA nanosphere based on the self-assembly of RCA products to deliver nucleic acid drugs, a cofactor precursor (MnO2), and a photosensitizer (TMPyP4) for DNAzyme-mediated gene therapy and photodynamic therapy [147] (Figure 18c). In the tumor microenvironment, MnO2 could oxidize acidic H2O2 to produce Mn2+ and O2, in which Mn2+ catalyzed the cleavage of DNAzyme, and O2 promoted the generation of reactive oxygen species (ROS) by photosensitizers for photodynamic therapy. In the same year, Wang et al. integrated the immunomodulator CpG and a G-quadruplex structure into lsDNA generated via RCA to construct a photocontrolled DNA nanosphere for photodynamic immunotherapy, in which the G-quadruplex structure was used for inserting the photosensitizer (TMPyP4) [148].
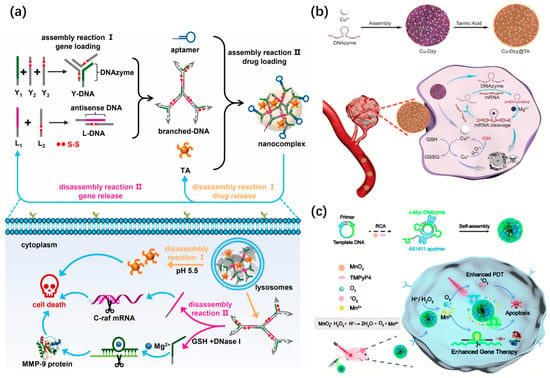
Figure 18.
(a) A controllably disassembled nanosphere for efficient gene/drug release, reproduced with permission from Ref. [143]. Copyright 2021, Elsevier Publishers. (b) A copper–DNAzyme hybrid DNA nanosphere for combination gene and chemo-dynamic therapy, reproduced with permission from Ref. [146]. Copyright 2021, Wiley-VCH Publishers. (c) A MnO2-encapsulated DNA nanosphere for DNAzyme-mediated gene therapy and photodynamic therapy, reproduced with permission from Ref. [147]. Copyright 2022, Royal Society of Chemistry Publishers.
Figure 18.
(a) A controllably disassembled nanosphere for efficient gene/drug release, reproduced with permission from Ref. [143]. Copyright 2021, Elsevier Publishers. (b) A copper–DNAzyme hybrid DNA nanosphere for combination gene and chemo-dynamic therapy, reproduced with permission from Ref. [146]. Copyright 2021, Wiley-VCH Publishers. (c) A MnO2-encapsulated DNA nanosphere for DNAzyme-mediated gene therapy and photodynamic therapy, reproduced with permission from Ref. [147]. Copyright 2022, Royal Society of Chemistry Publishers.
4. Conclusions and Perspectives
In this review, we divided DNA nanospheres into three main categories according to their different self-assembly types, including the Watson–Crick base pairing of Y-shaped DNA units, liquid crystallization and the dense packaging of long-stranded RCA products, and metal–DNA coordination. Due to its facile operation, low cost, and simple structure, Y-shaped DNA has become a building block for constructing DNA nanospheres via Watson–Crick base-pairing. To further reduce the complexity of sequence design and improve the biostability, DNA nanospheres, also called DNA nanoflowers, self-assembled by ultra-long ssDNA generated via RCA, have emerged as promising nanocarriers, owing to their excellent characteristics including being exceptionally resistant to nuclease degradation, attractive biostability, and high loading capacity. Without relying on base-pairing, such DNA nanospheres were assembled by magnesium pyrophosphate-mediated liquid crystallization and the dense packaging of lsDNA/RNA. Afterward, a simpler and more versatile method was developed based on metal–DNA coordination to create metal–DNA hybrid nanospheres. The synthesis of these metal–DNA hybrid nanospheres is very simple, carried out only through mixing and annealing between metal ions and DNA. Notably, by varying the ratio of metal ions and DNA, the size of hybrid DNA nanospheres can be precisely tuned.
Due to the high loading capacity and stability of the abovementioned DNA nanospheres, various biomolecules such as electroactive methylene blue (MB), Ru(bpy)32+, protein enzymes, and fluorescent dyes can be encapsulated into DNA nanospheres for electrochemical biosensors, colorimetric assays, and fluorescence detection. In addition, the diameter of DNA nanospheres can be tuned to a suitable size for cell uptake, which allows DNA nanospheres to be applied at the cellular level and in vivo. For example, by introducing fluorophores into building blocks, DNA nanospheres have the ability to image intracellular molecules (i.e., ATP, RNA, and telomerase). Furthermore, different functional nucleic acids such as aptamer, DNAzyme, and antisense DNA are incorporated into DNA nanospheres, and DNA nanospheres are capable of specific cell recognition, controllable drug release, and gene regulation. More importantly, diverse anticancer drugs including doxorubicin, siRNA, photosensitizer, and immunomodulators are encapsulated into DNA nanospheres, making DNA nanospheres ideal carriers to construct intelligent drug delivery systems for chemotherapy, gene therapy, photodynamic therapy, immunotherapy, and even multimodal combination therapies.
Although DNA nanospheres have many merits and great progress has been made, there are still some challenges that need to be addressed in the future. First, the formation mechanism of the flower-like structure is still unclear and requires in-depth study. Second, due to the dense packing of building blocks and the high stability of DNA nanospheres, the bioavailability and release efficiency of cargo are limited. Thus, stimuli-responsive DNA nanospheres should be constructed for intelligent cargo release. Third, DNA nanospheres cannot be generated on a large scale at a low cost, which impedes their clinical transformation. Therefore, a simple, low-cost, and versatile synthesis method should be developed to improve the production efficiency for large-scale applications. We believe that the advancement of DNA nanospheres will offer a new perspective for creating smart delivery nanoplatforms to boost the development of precision medicine.
Author Contributions
Conceptualization, J.H., Y.P. and J.L.; data collection, X.L., W.Z., M.C., J.W. and Y.C.; writing—original draft preparation, J.L., Q.J. and X.L.; writing—review and editing, J.L., X.L., Q.J. and J.H.; supervision, J.H. and Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
J.L. acknowledges the National Natural Science Foundation of China (No. 22204140); the Natural Science Foundation of Jiangsu Province (No. BK20220561); the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 22KJB150049); the China Postdoctoral Science Foundation (No. 2023M732982); the Scientific Research Foundation for High-Level Talents of Yangzhou University (No. 137012489) and the Green Yang Jinfeng Talent Program of Yangzhou (No. YZLYJFJH2022YXBS088). J.H. acknowledges the National Natural Science Foundation of China (No. 22174042); the Key Research and Development Program of Hunan Province of China (No. 2022GK2017) and the Natural Science Foundation for Distinguished Young Scholars of Hunan Province (2021JJ10011).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Seeman, N.C. Nucleic Acid Junctions and Lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Zhang, F.; Nangreave, J.; Liu, Y.; Yan, H. Structural DNA Nanotechnology: State of the Art and Future Perspective. J. Am. Chem. Soc. 2014, 136, 11198–11211. [Google Scholar] [CrossRef]
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef]
- Wang, D.X.; Wang, J.; Wang, Y.X.; Du, Y.C.; Huang, Y.; Tang, A.N.; Cui, Y.X.; Kong, D.M. DNA Nanostructure-Based Nucleic Acid Probes: Construction and Biological Applications. Chem. Sci. 2021, 12, 7602–7622. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.G.; Tseng, Y.D.; Kwon, S.Y.; D’Espaux, L.; Bunch, J.S.; McEuen, P.L.; Luo, D. Controlled Assembly of Dendrimer-Like DNA. Nat. Mater. 2004, 3, 38–42. [Google Scholar] [CrossRef]
- Zhou, T.; Chen, P.; Niu, L.; Jin, J.; Liang, D.; Li, Z.; Yang, Z.; Liu, D. pH-Responsive Size-Tunable Self-Assembled DNA Dendrimers. Angew. Chem. Int. Ed. 2012, 51, 11271–11274. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Jiang, S.; Wang, T.; Liu, Y.; Yan, H. 3D Framework DNA Origami with Layered Crossovers. Angew. Chem. Int. Ed. 2016, 55, 12832–12835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ko, S.H.; Su, M.; Leng, Y.; Ribbe, A.E.; Jiang, W.; Mao, C. Symmetry Controls the Face Geometry of DNA Polyhedra. J. Am. Chem. Soc. 2009, 131, 1413–1415. [Google Scholar] [CrossRef]
- Roh, Y.H.; Lee, J.B.; Tan, S.J.; Kim, B.; Park, H.; Rice, E.J.; Luo, D. Photocrosslinked DNA Nanospheres for Drug Delivery. Macromol. Rapid Comm. 2010, 31, 1207–1211. [Google Scholar] [CrossRef]
- Dong, Y.; Yao, C.; Zhu, Y.; Yang, L.; Luo, D.; Yang, D. DNA Functional Materials Assembled from Branched DNA: Design, Synthesis, and Applications. Chem. Rev. 2020, 120, 9420–9481. [Google Scholar] [CrossRef]
- Ren, K.; Xu, Y.; Liu, Y.; Yang, M.; Ju, H. A Responsive “Nano String Light” for Highly Efficient mRNA Imaging in Living Cells via Accelerated DNA Cascade Reaction. ACS Nano 2018, 12, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Zhang, S.; Yu, X.; Hu, S.; Lu, Y.; Wu, Z.-S. Periodically Ordered, Nuclease-Resistant DNA Nanowires Decorated with Cell-Specific Aptamers as Selective Theranostic Agents. Angew. Chem. Int. Ed. 2020, 59, 17540–17547. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, C.; Zhang, S.; Xue, C.; Yu, X.; Xu, H.; Wang, Z.; Lu, Y.; Wu, Z.S. Precision-Guided Missile-Like DNA Nanostructure Containing Warhead and Guidance Control for Aptamer-Based Targeted Drug Delivery into Cancer Cells in Vitro and in Vivo. J. Am. Chem. Soc. 2020, 142, 1265–1277. [Google Scholar] [CrossRef]
- Zhu, J.J. Special Topic: Biomedical Application of DNA-Assembled Nanostructure. J. Anal. Test. 2021, 5, 93–94. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Z.; Ma, Y.; Han, Z.; Zhang, M.; Chen, H.; Gu, Y. A Telomerase-Responsive DNA Icosahedron for Precise Delivery of Platinum Nanodrugs to Cisplatin-Resistant Cancer. Angew. Chem. Int. Ed. 2018, 57, 5389–5393. [Google Scholar] [CrossRef]
- Green, L.N.; Subramanian, H.K.K.; Mardanlou, V.; Kim, J.; Hariadi, R.F.; Franco, E. Autonomous Dynamic Control of DNA Nanostructure Self-Assembly. Nat. Chem. 2019, 11, 510–520. [Google Scholar] [CrossRef]
- Zhao, S.; Tian, R.; Wu, J.; Liu, S.; Wang, Y.; Wen, M.; Shang, Y.; Liu, Q.; Li, Y.; Guo, Y.; et al. A DNA Origami-Based Aptamer Nanoarray for Potent and Reversible Anticoagulation in Hemodialysis. Nat. Commun. 2021, 12, 358. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Demirer, G.S.; Gonzalez-Grandio, E.; Fan, C.; Landry, M.P. Engineering DNA Nanostructures for SiRNA Delivery in Plants. Nat. Protoc. 2020, 15, 3064–3087. [Google Scholar] [CrossRef]
- Lee, D.S.; Qian, H.; Tay, C.Y.; Leong, D.T. Cellular Processing and Destinies of Artificial DNA Nanostructures. Chem. Soc. Rev. 2016, 45, 4199–4225. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Chen, Y.; Liu, R.; Wu, Y.; Liu, J.; Yang, X.; Wang, K.; Huang, J. Size-Controllable and Self-Assembled DNA Nanosphere for Amplified MicroRNA Imaging through ATP-Fueled Cyclic Dissociation. Nano Lett. 2022, 22, 8216–8223. [Google Scholar] [CrossRef]
- Zhao, H.; Lv, J.; Li, F.; Zhang, Z.; Zhang, C.; Gu, Z.; Yang, D. Enzymatical Biomineralization of DNA Nanoflowers Mediated by Manganese Ions for Tumor Site Activated Magnetic Resonance Imaging. Biomaterials 2021, 268, 120591. [Google Scholar] [CrossRef]
- Li, M.; Wang, C.; Di, Z.; Li, H.; Zhang, J.; Xue, W.; Zhao, M.; Zhang, K.; Zhao, Y.; Li, L. Engineering Multifunctional DNA Hybrid Nanospheres through Coordination-Driven Self-Assembly. Angew. Chem. Int. Ed. 2019, 58, 1350–1354. [Google Scholar] [CrossRef]
- Bi, S.; Dong, Y.; Jia, X.; Chen, M.; Zhong, H.; Ji, B. Self-assembled Multifunctional DNA Nanospheres for Biosensing and Drug Delivery into Specific Target Cells. Nanoscale 2015, 7, 7361–7367. [Google Scholar] [CrossRef]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical Self-Assembly of Multifunctional DNA Nanoflowers for Biomedical Applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef]
- Jia, Y.; Shen, X.; Sun, F.; Na, N.; Ouyang, J. Metal-DNA Coordination Based Bioinspired Hybrid Nanospheres for in Situ Amplification and Sensing of MicroRNA. J. Mater. Chem. B 2020, 8, 11074–11081. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.; Ree, B.J.; Lee, Y.M.; Park, H.; Lee, T.G.; Kim, J.H.; Kim, W.J. Self-Assembled Aptamer Nanoconstruct: A Highly Effective Molecule-Capturing Platform Having Therapeutic Applications. Adv. Ther. 2019, 2, 1800111. [Google Scholar] [CrossRef]
- Yue, S.; Qiao, Z.; Yu, K.; Hai, X.; Li, Y.; Li, Y.; Song, W.; Bi, S. DNA Addition Polymerization with Logic Operation for Controllable Self-Assembly of Three-Dimensional Nanovehicles and Combinatorial Cancer Therapy. Chem. Eng. J. 2021, 408, 127258. [Google Scholar] [CrossRef]
- Lee, J.B.; Roh, Y.H.; Um, S.H.; Funabashi, H.; Cheng, W.; Cha, J.J.; Kiatwuthinon, P.; Muller, D.A.; Luo, D. Multifunctional Nanoarchitectures from DNA-Based ABC Monomers. Nat. Nanotechnol. 2009, 4, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Han, D.; Chen, T.; Peng, L.; Zhu, G.; You, M.; Qiu, L.; Sefah, K.; Zhang, X.; Tan, W. Building a Multifunctional Aptamer-Based DNA Nanoassembly for Targeted Cancer Therapy. J. Am. Chem. Soc. 2013, 135, 18644–18650. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, C.; Cansiz, S.; Wu, C.; Xu, J.; Cui, C.; Liu, Y.; Hou, W.; Wang, Y.; Zhang, L.; et al. Self-assembly of DNA Nanohydrogels with Controllable Size and Stimuli-Responsive Property for Targeted Gene Regulation Therapy. J. Am. Chem. Soc. 2015, 137, 1412–1415. [Google Scholar] [CrossRef]
- Zikich, D.; Liu, K.; Sagiv, L.; Porath, D.; Kotlyar, A. I-Motif Nanospheres: Unusual Self-Assembly of Long Cytosine Strands. Small 2011, 7, 1029–1034. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Zhang, Z.; Deng, Z.; Bu, J.; Li, T.; Nie, J.; Qin, X.; Yang, Y.; Zhong, S. Self-Assembly of DNA Nanospheres with Controllable Size and Self-Degradable Property for Enhanced Antitumor Chemotherapy. Colloid. Surf. B 2023, 222, 113122. [Google Scholar] [CrossRef]
- Wang, D.X.; Wang, J.; Cui, Y.X.; Wang, Y.X.; Tang, A.N.; Kong, D.M. Nanolantern-Based DNA Probe and Signal Amplifier for Tumor-Related Biomarker Detection in Living Cells. Anal. Chem. 2019, 91, 13165–13173. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, S.; Zheng, T.T.; Chen, Y.R.; Wu, J.T.; Wu, Z.S. Intracellular Nonenzymatic in Situ Growth of Three-Dimensional DNA Nanostructures for Imaging Specific Biomolecules in Living Cells. ACS Nano 2020, 14, 9572–9584. [Google Scholar] [CrossRef]
- Zhao, W.; Ali, M.M.; Brook, M.A.; Li, Y. Rolling circle amplification: Applications in Nanotechnology and Biodetection with Functional Nucleic Acids. Angew. Chem. Int. Ed. 2008, 47, 6330–6337. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling Circle Amplification: A Versatile Tool for Chemical Biology, Materials Science and Medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Shopsowitz, K.E.; Roh, Y.H.; Deng, Z.J.; Morton, S.W.; Hammond, P.T. RNAi-Microsponges Form through Self-Assembly of the Organic and Inorganic Products of Transcription. Small 2014, 10, 1623–1633. [Google Scholar] [CrossRef]
- Kim, E.; Agarwal, S.; Kim, N.; Hage, F.S.; Leonardo, V.; Gelmi, A.; Stevens, M.M. Bioinspired Fabrication of DNA-Inorganic Hybrid Composites Using Synthetic DNA. ACS Nano 2019, 13, 2888–2900. [Google Scholar] [CrossRef]
- Park, K.S.; Batule, B.S.; Chung, M.; Kang, K.S.; Park, T.J.; Kim, M.I.; Park, H.G. A Simple and Eco-Friendly One-Pot Synthesis of Nuclease-Resistant DNA-Inorganic Hybrid Nanoflowers. J. Mater. Chem. B 2017, 5, 2231–2234. [Google Scholar] [CrossRef]
- Dang, T.V.; Kim, M.I. Diversified Component Incorporated Hybrid Nanoflowers: A Versatile Material for Biosensing and Biomedical Application. Korean J. Chem. Eng. 2023, 40, 302–310. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, H.; Jo, C.; Jeong, J.; Ko, J.; Han, S.; Lee, M.S.; Lee, H.Y.; Han, J.W.; Lee, J.; et al. Enzyme-Driven Hasselback-Like DNA-Based Inorganic Superstructures. Adv. Funct. Mater. 2017, 27, 1704213. [Google Scholar] [CrossRef]
- Barker, Y.R.; Chen, J.F.; Brown, J.; EI-Sagheer, A.H.; Wiseman, P.; Johnson, E.; Goddard, P.; Brown, T. Preparation and Characterization of Manganese, Cobalt and Zinc DNA Nanoflowers with Tunable Morphology, DNA Content and Size. Nucleic Acids Res. 2018, 46, 7495–7505. [Google Scholar] [CrossRef]
- Zhao, J.G.; Cao, J.; Wang, W.Z. Peptide-Based Electrochemical Biosensors and Their Applications in Disease Detection. J. Anal. Test. 2022, 6, 193–203. [Google Scholar] [CrossRef]
- Rubab, M.; Shahbaz, H.M.; Olaimat, A.N.; Oh, D.-H. Biosensors for Rapid and Sensitive Detection of Staphylococcus Aureus in Food. Biosens. Bioelectron. 2018, 105, 49–57. [Google Scholar] [CrossRef]
- Cai, R.; Zhang, S.; Chen, L.; Li, M.; Zhang, Y.; Zhou, N. Self-Assembled DNA Nanoflowers Triggered by a DNA Walker for Highly Sensitive Electrochemical Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2021, 13, 4905–4914. [Google Scholar] [CrossRef]
- Kong, L.; Li, H.; Zhang, X.; Zhuo, Y.; Chai, Y.; Yuan, R. A Novel Ratiometric Electrochemical Biosensor Using Only One Signal Tag for Highly Reliable and Ultrasensitive Detection of miRNA-21. Anal. Chem. 2022, 94, 5167–5172. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Ma, C.; Zhu, J.-J. DNA Technology-assisted Signal Amplification Strategies in Electrochemiluminescence Bioanalysis. J. Anal. Test. 2021, 5, 95–111. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, G.; Li, X.; Liu, L.; Cao, W.; Wei, Q. Electrochemiluminescent Competitive Immunosensor Based on Polyethyleneimine Capped SiO2 Nanomaterials as Labels to Release Ru (bpy)32+ Fixed in 3D Cu/Ni Oxalate for the Detection of Aflatoxin B1. Biosens. Bioelectron. 2018, 101, 290–296. [Google Scholar] [CrossRef]
- Yan, C.; Yang, L.; Yao, L.; Xu, J.; Yao, B.; Liu, G.; Cheng, L.; Chen, W. Ingenious Electrochemiluminescence Bioaptasensor Based on Synergistic Effects and Enzyme-Driven Programmable 3D DNA Nanoflowers for Ultrasensitive Detection of Aflatoxin B1. Anal. Chem. 2020, 92, 14122–14129. [Google Scholar] [CrossRef] [PubMed]
- Li, S.K.; Chen, A.Y.; Niu, X.X.; Liu, Z.T.; Du, M.; Chai, Y.Q.; Yuan, R.; Zhuo, Y. In Situ Generation of Electrochemiluminescent DNA Nanoflowers as a Signal Tag for Mucin 1 Detection Based on a Strategy of Target and Mimic Target Synchronous Cycling Amplification. Chem. Commun. 2017, 53, 9624–9627. [Google Scholar] [CrossRef]
- Zeng, R.; Luo, Z.; Su, L.; Zhang, L.; Tang, D.; Niessner, R.; Knopp, D. Palindromic Molecular Beacon Based Z-Scheme BiOCI-Au-CdS Photoelectrochemical Biodetection. Anal. Chem. 2019, 91, 2447–2454. [Google Scholar] [CrossRef]
- Zhou, Q.; Tang, D. Recent advances in photoelectrochemical biosensors for analysis of mycotoxins in food. TrAC Trends Anal. Chem. 2020, 124, 115814. [Google Scholar] [CrossRef]
- Guo, L.; Yin, H.; Xu, M.; Zheng, Z.; Fang, X.; Chong, R.; Zhou, Y.; Xu, L.; Xu, Q.; Li, J.; et al. In Situ Generated Plasmonic Silver Nanoparticle-Sensitized Amorphous Titanium Dioxide for Ultrasensitive Photoelectrochemical Sensing of Formaldehyde. ACS Sens. 2019, 4, 2724–2729. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Niu, S.; Ge, J.; Luan, Q.; Jie, G. 3D DNA Nanosphere-Based Photoelectrochemical Biosensor Combined with Multiple Enzyme-Free Amplification for Ultrasensitive Detection of Cancer Biomarkers. Biosens. Bioelectron. 2020, 147, 111778. [Google Scholar] [CrossRef]
- Xiao, M.J.; Ding, Q.; Deng, H.M.; Yuan, R.; Yuan, Y.L. Enzyme-Free Triple Cycles Triggered in-Situ Generation of Nanospheres on DNA Planar Tripod for Sensitive Photoelectrochemical Biosensor. Sens. Actuators B Chem. 2022, 358, 131491. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.; Gong, H.; Xu, J.; Yu, Z.; Wei, Q.; Tang, D. Graphene-Coated Copper-Doped ZnO Quantum Dots for Sensitive Photoelectrochemical Bioanalysis of Thrombin Triggered by DNA Nanoflowers. J. Mater. Chem. B 2021, 9, 6818–6824. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, H.; Fan, G.C.; Luo, X. Coupling Photoelectrochemical and Electrochemical Strategies in One Probe Electrode: Toward Sensitive and Reliable Dual-Signal Bioassay for Uracil-DNA Glycosylase Activity. Biosens. Bioelectron. 2019, 142, 111569. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Chai, Y.; Yuan, R.; Yuan, Y. In Situ Formation of Multifunctional DNA Nanospheres for a Sensitive and Accurate Dual-Mode Biosensor for Photoelectrochemical and Electrochemical Assay. Anal. Chem. 2020, 92, 8364–8370. [Google Scholar] [CrossRef]
- Huang, G.B.; Guo, Z.Y.; Ye, T.X.; Zhang, C.; Zhou, Y.M.; Yao, Q.H.; Chen, X. Colorimetric Determination of Chloridion in Domestic Water Based on the Wavelength Shift of CsPbBr3 Perovskite Nanocrystals via Halide Exchange. J. Anal. Test. 2021, 5, 3–10. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Butte, M.J.; Whitesides, G.M. Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays. Angew. Chem. Int. Ed. 2007, 46, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Yang, M.; Chang, D.; Zhao, W.; Zhang, S.; Du, Y.; Li, Y. A DNA Nanoflower-Assisted Separation-Free Nucleic Acid Detection Platform with a Commercial Pregnancy Test Strip. Angew. Chem. Int. Ed. 2021, 60, 24823–24827. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, L.; Zhu, J.; Li, B.; Jin, Y. Visual and Sensitive Detection of Telomerase Activity via Hydrogen Peroxide Test Strip. Biosens. Bioelectron. 2020, 156, 112132. [Google Scholar] [CrossRef]
- Ye, H.; Yang, K.; Tao, J.; Liu, Y.; Zhang, Q.; Habibi, S.; Nie, Z.; Xia, X. An Enzyme-Free Signal Amplification Technique for Ultrasensitive Colorimetric Assay of Disease Biomarkers. ACS Nano 2017, 11, 2052–2059. [Google Scholar] [CrossRef]
- Zeng, R.; Wang, J.; Wang, Q.; Tang, D.; Lin, Y. Horseradish Peroxidase-Encapsulated DNA Nanoflowers: An Innovative Signal-Generation Tag for Colorimetric Biosensor. Talanta 2021, 221, 121600. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Zhang, Q.; Xue, W.; Wu, Y.; Liu, Y.; Liu, M. Self-Assembly of Protein-DNA Superstructures for Alkaline Phosphatase Detection in Blood. Chem. Commun. 2023, 59, 3399–3402. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, X.; Walt, D.R. Single-Molecule Analysis Determines Isozymes of Human Alkaline Phosphatase in Serum. Angew. Chem. Int. Ed. 2020, 59, 18010–18015. [Google Scholar] [CrossRef]
- He, J.L.; Tang, L.; Liao, S.Q.; Guo, M.T.; Wu, L.; Song, Y.; Liu, S.; Cao, Z. Label-Free Palindromic DNA Nanospheres as Naked-Eye Colorimetric Assay Platform for Detection of Telomerase Activity. Talanta 2023, 253, 123990. [Google Scholar] [CrossRef]
- Thinh Viet, D.; Heo, N.S.; Cho, H.-J.; Lee, S.M.; Song, M.Y.; Kim, H.J.; Kim, M.I. Colorimetric Determination of Phenolic Compounds Using Peroxidase Mimics Based on Biomolecule-Free Hybrid Nanoflowers Consisting of Graphitic Carbon Nitride and Copper. Microchim. Acta 2021, 188, 293. [Google Scholar]
- Tran, T.D.; Nguyen, P.T.; Le, T.N.; Kim, M.I. DNA-Copper Hybrid Nanoflowers as Efficient Laccase Mimics for Colorimetric Detection of Phenolic Compounds in Paper Microfluidic Devices. Biosens. Bioelectron. 2021, 182, 113187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kang, W.K.; Wang, Y.; Wang, X.Z.; Lu, L.H. An Enzymatic Reaction Modulated Fluorescence-on Omethoate Biosensor Based on Fe3O4@GO and Copper Nanoparticles. J. Anal. Test. 2022, 6, 3–11. [Google Scholar] [CrossRef]
- Liu, W.; Zhu, X. A Novel Fluorescence “Turn Off-On” Sensor Based on N-Doped Graphene Quantum Dots in Amino Acid Ionic Liquid Medium and Its Application. Talanta 2019, 197, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Liu, S.; Xie, N.; Quan, K.; Yang, Y.; Yang, X.; Huang, J.; Wang, K. Amplified FRET Nanoflares: An Endogenous mRNA-Powered Nanomachine for Intracellular MicroRNA Imaging. Angew. Chem. Int. Ed. 2020, 59, 20104–20111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, S.; Wang, J.; Liu, R.; Yang, X.; Wang, K.; Huang, J. Photocaged Amplified FRET Nanoflares: Spatiotemporal Controllable of mRNA-Powered Nanomachines for Precise and Sensitive MicroRNA Imaging in Live Cells. Nucleic Acids Res. 2022, 50, e40. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.M.; Thaxton, C.S.; Mirkin, C.A. Nanoparticle-Based Bio-Bar Codes for the Ultrasensitive Detection of Proteins. Science 2003, 301, 1884–1886. [Google Scholar] [CrossRef]
- Nam, J.M.; Stoeva, S.I.; Mirkin, C.A. Bio-Bar-Code-Based DNA Detection with PCR-Like Sensitivity. J. Am. Chem. Soc. 2004, 126, 5932–5933. [Google Scholar] [CrossRef]
- Zhao, Q.; Pan, B.; Long, W.; Pan, Y.; Zhou, D.; Luan, X.; He, B.; Wang, Y.; Song, Y. Metal Organic Framework-Based Bio-Barcode CRISPR/Cas12a Assay for Ultrasensitive Detection of MicroRNAs. Nano Lett. 2022, 22, 9714–9722. [Google Scholar] [CrossRef]
- Han, S.; Lee, J.S.; Lee, J.B. Synthesis of a Multi-Functional DNA Nanosphere Barcode System for Direct Cell Detection. Nanoscale 2017, 9, 14094–14102. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, Q.; Kannan, B.; Botton, G.A.; Yang, J.; Soleymani, L.; Brennan, J.D.; Li, Y. Self-Assembled Functional DNA Superstructures as High-Density and Versatile Recognition Elements for Printed Paper Sensors. Angew. Chem. Int. Ed. 2018, 57, 12440–12443. [Google Scholar] [CrossRef]
- Chang, X.; Zhang, C.; Lv, C.; Sun, Y.; Zhang, M.; Zhao, Y.; Yang, L.; Han, D.; Tan, W. Construction of a Multiple-Aptamer-Based DNA Logic Device on Live Cell Membranes via Associative Toehold Activation for Accurate Cancer Cell Identification. J. Am. Chem. Soc. 2019, 141, 12738–12743. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, X.; Zhao, Z.; Zhu, G.; Chen, T.; Fu, T.; Tan, W. DNA Nanoflowers for Multiplexed Cellular Imaging and Traceable Targeted Drug Delivery. Angew. Chem. Int. Ed. 2014, 53, 5821–5826. [Google Scholar] [CrossRef]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Zhao, J.; Di, Z.; Huang, Y.; Zhao, Y.; Li, L. Spatially Resolved in Vivo Imaging of Inflammation-Associated mRNA via Enzymatic Fluorescence Amplification in a Molecular Beacon. Nat. Biomed. Eng. 2022, 6, 1074–1084. [Google Scholar] [CrossRef]
- Zhao, J.; Gao, J.; Xue, W.; Di, Z.; Xing, H.; Lu, Y.; Li, L. Upconversion Luminescence-Activated DNA Nanodevice for ATP Sensing in Living Cells. J. Am. Chem. Soc. 2018, 140, 578–581. [Google Scholar] [CrossRef]
- Ran, X.; Wang, Z.; Pu, F.; Ju, E.; Ren, J.; Qu, X. Nucleic Acid-Driven Aggregation-Induced Emission of Au Nanoclusters for Visualizing Telomerase Activity in Living Cells and in Vivo. Mater. Horiz. 2021, 8, 1769–1775. [Google Scholar] [CrossRef]
- Kim, N.; Kim, E.; Kim, H.; Thomas, M.R.; Najer, A.; Stevens, M.M. Tumor-Targeting Cholesterol-Decorated DNA Nanoflowers for Intracellular Ratiometric Aptasensing. Adv. Mater. 2021, 33, 2007738. [Google Scholar] [CrossRef]
- Deng, R.; Zhang, K.; Wang, L.; Ren, X.; Sun, Y.; Li, J. DNA-Sequence-Encoded Rolling Circle Amplicon for Single-Cell RNA Imaging. Chem 2018, 4, 1373–1386. [Google Scholar] [CrossRef]
- Ren, X.; Deng, R.; Zhang, K.; Sun, Y.; Teng, X.; Li, J. SpliceRCA: In Situ Single-Cell Analysis of mRNA Splicing Variants. ACS Cent. Sci. 2018, 4, 680–687. [Google Scholar] [CrossRef]
- Shang, J.; Yu, S.; Li, R.; He, Y.; Wang, Y.; Wang, F. Bioorthogonal Disassembly of Hierarchical DNAzyme Nanogel for High-Performance Intracellular microRNA Imaging. Nano Lett. 2023, 23, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Quan, K.; Li, J.; Wang, J.; Xie, N.; Wei, Q.; Tang, J.; Yang, X.; Wang, K.; Huang, J. Dual-MicroRNA-Controlled Double-Amplified Cascaded Logic DNA Circuits for Accurate Discrimination of Cell Subtypes. Chem. Sci. 2019, 10, 1442–1449. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Huang, J.; Li, J.; Wang, J.; Yang, X.; Liu, J.; Wang, K. A DNA Nanowire Based Localized Catalytic Hairpin Assembly Reaction for MicroRNA Imaging in Live Cells. Chem. Sci. 2018, 9, 7802–7808. [Google Scholar] [CrossRef]
- He, L.; Lu, D.; Liang, H.; Xie, S.; Zhang, X.; Liu, O.; Yuan, Q.; Tan, W. mRNA-Initiated, Three-Dimensional DNA Amplifier Able to Function inside Living Cells. J. Am. Chem. Soc. 2018, 140, 258–263. [Google Scholar] [CrossRef]
- Li, X.; Yin, F.; Xu, X.; Liu, L.; Xue, Q.; Tong, L.; Jiang, W.; Li, C. A Facile DNA/RNA Nanoflower for Sensitive Imaging of Telomerase RNA in Living Cells Based on “Zipper Lock-and-Key” Strategy. Biosens. Bioelectron. 2020, 147, 111788. [Google Scholar] [CrossRef]
- Song, J.; Mou, H.Z.; Li, X.Q.; Liu, Y.; Yang, X.J.; Chen, H.Y.; Xu, J.J. Self-Assembled DNA/RNA Nanospheres with Cascade Signal Amplification for Intracellular MicroRNA Imaging. Sens. Actuators B Chem. 2022, 360, 131644. [Google Scholar] [CrossRef]
- Cai, S.; Wang, J.; Li, J.; Zhou, B.; He, C.; Meng, X.; Huang, J.; Wang, K. A Self-Assembled DNA Nanostructure as a FRET Nanoflare for Intracellular ATP Imaging. Chem. Commun. 2021, 57, 6257–6260. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hu, L.; He, H.; Zhang, F.; Wang, M.; Wei, W.; Xia, Z. Y-Shaped DNA-Mediated Hybrid Nanoflowers as Efficient Gene Carriers for Fluorescence Imaging of Tumor-Related mRNA in Living Cells. Anal. Chim. Acta 2019, 1057, 114–122. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, X.; Shang, Y.; Jiang, Q.; Li, C.; Ding, B. Visualization of the Intracellular Location and Stability of DNA Flowers with a Label-Free Fluorescent Probe. RSC Adv. 2019, 9, 15205–15209. [Google Scholar] [CrossRef]
- Li, W.; Yang, X.; He, L.; Wang, K.; Wang, Q.; Huang, J.; Liu, J.; Wu, B.; Xu, C. Self-Assembled DNA Nanocentipede as Multivalent Drug Carrier for Targeted Delivery. ACS Appl. Mater. Interfaces 2016, 8, 25733–25740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Abdullah, R.; Hu, X.; Bai, H.; Fan, H.; He, L.; Liang, H.; Zou, J.; Liu, Y.; Sun, Y.; et al. Engineering of Bioinspired, Size-Controllable, Self-Degradable Cancer-Targeting DNA Nanoflowers via the Incorporation of an Artificial Sandwich Base. J. Am. Chem. Soc. 2019, 141, 4282–4290. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Wang, H.; He, S.; Li, R.; Deng, Z.; Liu, X.; Wang, F. Nonviolent Self-Catabolic DNAzyme Nanosponges for Smart Anticancer Drug Delivery. ACS Nano 2019, 13, 5852–5863. [Google Scholar] [CrossRef]
- Jiang, Y.; Xu, X.; Fang, X.; Cai, S.; Wang, M.; Xing, C.; Lu, C.; Yang, H. Self-Assembled mRNA-Responsive DNA Nanosphere for Bioimaging and Cancer Therapy in Drug-Resistant Cells. Anal. Chem. 2020, 92, 11779–11785. [Google Scholar] [CrossRef] [PubMed]
- Kanasty, R.; Dorkin, J.R.; Vegas, A.; Anderson, D. Delivery Materials for SiRNA Therapeutics. Nat. Mater. 2013, 12, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lv, Z.; Zhang, X.; Dong, Y.; Ding, X.; Li, Z.; Li, S.; Yao, C.; Yang, D. Supramolecular Self-Assembled DNA Nanosystem for Synergistic Chemical and Gene Regulations on Cancer Cells. Angew. Chem. Int. Ed. 2021, 60, 25557–25566. [Google Scholar] [CrossRef]
- Lee, J.B.; Hong, J.; Bonner, D.K.; Poon, Z.; Hammond, P.T. Self-Assembled RNA Interference Microsponges for Efficient SiRNA Delivery. Nat. Mater. 2012, 11, 316–322. [Google Scholar] [CrossRef]
- Cheng, H.; Hong, S.; Wang, Z.; Sun, N.; Wang, T.; Zhang, Y.; Chen, H.; Pei, R. Self-Assembled RNAi Nanoflowers via Rolling Circle Transcription for Aptamer-Targeted SiRNA Delivery. J. Mater. Chem. B 2018, 6, 4638–4644. [Google Scholar] [CrossRef] [PubMed]
- Fokina, A.A.; Stetsenko, D.A.; Francois, J.C. DNA Enzymes as Potential Therapeutics: Towards Clinical Application of 10–23 DNAzymes. Expert Opin. Biol. Ther. 2015, 15, 689–711. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Catalyic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef]
- Su, J.; Sun, C.; Du, J.; Xing, X.; Wang, F.; Dong, H. RNA-Cleaving DNAzyme-Based Amplification Strategies for Biosensing and Therapy. Adv. Healthc. Mater. 2023, 2300367. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, Z.; Zuo, D.; Li, L.; Li, F.; Yang, D. A Synergistic DNA-Polydopamine-MnO2 Nanocomplex for Near-Infrared-Light-Powered DNAzyme-Mediated Gene Therapy. Nano Lett. 2021, 21, 5377–5385. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Platt, R.J.; Zhang, F. Therapeutic Genome Editing: Prospects and Challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, C.; Gao, S.; Li, P.; Kong, Y.; Li, T.; Li, Y.; Xu, F.-J.; Du, J. CRISPR/Cas9 Delivery Mediated with Hydroxyl-Rich Nanosystems for Gene Editing in Aorta. Adv. Sci. 2019, 6, 1900386. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yang, J.; Luan, X.; Liu, X.; Li, X.; Yang, J.; Huang, T.; Sun, L.; Wang, Y.; Lin, Y.; et al. Near-Infrared Upconversion-Activated CRISPR-Cas9 System: A Remote-Controlled Gene Editing Platform. Sci. Adv. 2019, 5, eaav7199. [Google Scholar] [CrossRef]
- Zhuo, C.; Zhang, J.; Lee, J.H.; Jiao, J.; Cheng, D.; Liu, L.; Kim, H.-W.; Tao, Y.; Li, M. Spatiotemporal Control of CRISPR/Cas9 Gene Editing. Signal Transduct. Target. Ther. 2021, 6, 238. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Song, N.; Dong, Y.; Li, S.; Li, L.; Liu, Y.; Li, Z.; Yang, D. A Proton-Activatable DNA-Based Nanosystem Enables Co-Delivery of CRISPR/Cas9 and DNAzyme for Combined Gene Therapy. Angew. Chem. Int. Ed. 2022, 61, e202116569. [Google Scholar] [CrossRef]
- Shi, J.; Yang, X.; Li, Y.; Wang, D.; Liu, W.; Zhang, Z.; Liu, J.; Zhang, K. MicroRNA-Responsive Release of Cas9/sgRNA from DNA Nanoflower for Cytosolic Protein Delivery and Enhanced Genome Editing. Biomaterials 2020, 256, 120221. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, Y.; Yang, Y.; Jiang, L.; Xing, C.; Li, J.; Lu, C.; Yang, H. Functional Self-Assembled DNA Nanohydrogels for Specific Telomerase Activity Imaging and Telomerase-Activated Antitumor Gene Therapy. Anal. Chem. 2020, 92, 15179–15186. [Google Scholar] [CrossRef]
- Lucky, S.S.; Soo, K.C.; Zhang, Y. Nanoparticles in Photodynamic Therapy. Chem. Rev. 2015, 115, 1990–2042. [Google Scholar] [CrossRef]
- Luby, B.M.; Walsh, C.D.; Zheng, G. Advanced Photosensitizer Activation Strategies for Smarter Photodynamic Therapy Beacons. Angew. Chem. Int. Ed. 2019, 58, 2558–2569. [Google Scholar] [CrossRef]
- Dolmans, D.; Fukumura, D.; Jain, R.K. Photodynamic Therapy for Cancer. Nat. Rev. Cancer 2003, 3, 380–387. [Google Scholar] [CrossRef]
- Li, X.; Lee, S.; Yoon, J. Supramolecular Photosensitizers Rejuvenate Photodynamic Therapy. Chem. Soc. Rev. 2018, 47, 1174–1188. [Google Scholar] [CrossRef]
- Pan, M.; Jiang, Q.; Sun, J.; Xu, Z.; Zhou, Y.; Zhang, L.; Liu, X. Programming DNA Nanoassembly for Enhanced Photodynamic Therapy. Angew. Chem. Int. Ed. 2020, 59, 1897–1905. [Google Scholar] [CrossRef]
- Zaffaroni, N.; Daidone, M.G. Survivin Expression and Resistance to Anticancer Treatments: Perspectives for New Therapeutic Interventions. Drug Resist. Updates 2002, 5, 65–72. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, H.; Li, X.; Zhu, H.; Sun, D.; Sun, X.; Liu, H.; Zhang, Z.; Cao, L.; Gao, C.; et al. Multifunctional DNA Polymer-Assisted Upconversion Therapeutic Nanoplatform for Enhanced Photodynamic Therapy. ACS Appl. Mater. Interfaces 2020, 12, 26832–26841. [Google Scholar] [CrossRef]
- van Straten, D.; Mashayekhi, V.; de Bruijn, H.S.; Oliveira, S.; Robinson, D.J. Oncologic Photodynamic Therapy: Basic Principles, Current Clinical Status and Future Directions. Cancers 2017, 9, 19. [Google Scholar] [CrossRef]
- Shi, J.; Wang, D.; Ma, Y.; Liu, J.; Li, Y.; Reza, R.; Zhang, Z.; Liu, J.; Zhang, K. Photoactivated Self-Disassembly of Multifunctional DNA Nanoflower Enables Amplified Autophagy Suppression for Low-Dose Photodynamic Therapy. Small 2021, 17, 2104722. [Google Scholar] [CrossRef]
- Ouyang, D.Y.; Xu, L.H.; He, X.H.; Zhang, Y.T.; Zeng, L.H.; Cai, J.Y.; Ren, S. Autophagy is Differentially Induced in Prostate Cancer LNCaP, DU145 and PC-3 Cells via Distinct Splicing Profiles of ATG5. Autophagy 2013, 9, 20–32. [Google Scholar] [CrossRef]
- Andrzejak, M.; Price, M.; Kessel, D.H. Apoptotic and Autophagic Responses to Photodynamic Therapy in 1c1c7 Murine Hepatoma Cells. Autophagy 2011, 7, 979–984. [Google Scholar] [CrossRef]
- Dougan, M.; Dranoff, G. Immune Therapy for Cancer. Annu. Rev. Immunol. 2009, 27, 83–117. [Google Scholar] [CrossRef]
- Tian, R.; Shang, Y.; Wang, Y.; Jiang, Q.; Ding, B. DNA Nanomaterials-Based Platforms for Cancer Immunotherapy. Small Methods 2023, 7, 2201518. [Google Scholar] [CrossRef]
- Sun, Q.; Barz, M.; De Geest, B.G.; Diken, M.; Hennink, W.E.; Kiessling, F.; Lammers, T.; Shi, Y. Nanomedicine and Macroscale Materials in Immuno-Oncology. Chem. Soc. Rev. 2019, 48, 351–381. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, G.; Mei, L.; Wu, C.; Qiu, L.; Cui, C.; Liu, Y.; Teng, I.T.; Tan, W. Self-Assembled DNA Immunonanoflowers as Multivalent CpG Nanoagents. ACS Appl. Mater. Interfaces 2015, 7, 24069–24074. [Google Scholar] [CrossRef] [PubMed]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.-J.; et al. Checkpoint Blockade Cancer Immunotherapy Targets Tumour-Specific Mutant Antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Wang, C.; Ye, Y.; Hochu, G.M.; Sadeghifar, H.; Gu, Z. Enhanced Cancer Immunotherapy by Microneedle Patch-Assisted Delivery of Anti-PD1 Antibody. Nano Lett. 2016, 16, 2334–2340. [Google Scholar] [CrossRef]
- Wang, C.; Sun, W.; Wright, G.; Wang, A.Z.; Gu, Z. Inflammation-Triggered Cancer Immunotherapy by Programmed Delivery of CpG and Anti-PD1 Antibody. Adv. Mater. 2016, 28, 8912–8920. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in Cancer Immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Kuai, R.; Ochyl, L.J.; Bahjat, K.S.; Schwendeman, A.; Moon, J.J. Designer Vaccine Nanodiscs for Personalized Cancer Immunotherapy. Nat. Mater. 2017, 16, 489–496. [Google Scholar] [CrossRef]
- Zhu, G.; Mei, L.; Vishwasrao, H.D.; Jacobson, O.; Wang, Z.; Liu, Y.; Yung, B.C.; Fu, X.; Jin, A.; Niu, G.; et al. Intertwining DNA-RNA Nanocapsules Loaded with Tumor Neoantigens as Synergistic Nanovaccines for Cancer Immunotherapy. Nat. Commun. 2017, 8, 1482. [Google Scholar] [CrossRef]
- Sun, J.; Pan, M.; Liu, F.; Yu, W.; Wang, W.; Mo, F.; Yu, S.; Zhou, Y.; Liu, X. Immunostimulatory DNA Nanogel Enables Effective Lymphatic Drainage and High Vaccine Efficacy. ACS Mater. Lett. 2020, 2, 1606–1614. [Google Scholar] [CrossRef]
- Wang, J.; Yu, S.; Wu, Q.; Gong, X.; He, S.; Shang, J.; Liu, X.; Wang, F. A Self-Catabolic Multifunctional DNAzyme Nanosponge for Programmable Drug Delivery and Efficient Gene Silencing. Angew. Chem. Int. Ed. 2021, 60, 10766–10774. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Qi, H.; Jia, X.; Xu, Y.; Tong, Z.; Gu, Z.; Yang, D. A DNA Nanocomplex Containing Cascade DNAzymes and Promoter-Like Zn-Mn-Ferrite for Combined Gene/Chemo-dynamic Therapy. Angew. Chem. Int. Ed. 2022, 61, e202113619. [Google Scholar] [CrossRef]
- Han, J.; Cui, Y.; Gu, Z.; Yang, D. Controllable Assembly/Disassembly of Polyphenol-DNA Nanocomplex for Cascade-Responsive Drug Release in Cancer Cells. Biomaterials 2021, 273, 120846. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Hu, F.; Zhang, J.; Wang, C.; Li, L. A Biomimetic Coordination Nanoplatform for Controlled Encapsulation and Delivery of Drug-Gene Combinations. Angew. Chem. Int. Ed. 2019, 58, 8804–8808. [Google Scholar] [CrossRef]
- Klasa, R.J.; Gillum, A.M.; Klem, R.E.; Frankel, S.R. Oblimersen Bcl-2 Antisense: Facilitating Apoptosis in Anticancer Treatment. Antisense Nucleic Acid Drug Dev. 2002, 12, 193–213. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Y.; Zhao, J.; Wang, Y.; Shao, Y.; Gu, Z.; Li, L.; Zhao, Y. Self-Assembly of Copper-DNAzyme Nanohybrids for Dual-Catalytic Tumor Therapy. Angew. Chem. Int. Ed. 2021, 60, 14324–14328. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Lin, X.; Zhao, Y.; Hu, J.; Mo, F.; Song, G.; Zou, Z.; Wang, F.; Liu, X. A Dynamic DNA Nanosponge for Triggered Amplification of Gene-Photodynamic Modulation. Chem. Sci. 2022, 13, 5155–5163. [Google Scholar] [CrossRef]
- Wang, D.; Liu, J.; Duan, J.; Ma, Y.; Gao, H.; Zhang, Z.; Liu, J.; Shi, J.; Zhang, K. Photocontrolled Spatiotemporal Delivery of DNA Immunomodulators for Enhancing Membrane-Targeted Tumor Photodynamic Immunotherapy. ACS Appl. Mater. Interfaces 2022, 14, 44183–44198. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).