Condensed DNA Nanosphere for DNA Origami Cryptography
Abstract
:1. Introduction
2. Materials and Instruments
2.1. Materials and Reagents
| M1 | AGTGTACTTGAAAGTATTAAGAGGCCGCCACCTTT-bio |
| M2 | CCGCCAGCCATTGCAACAGGAAAAATATTTTTTTT-bio |
| M3 | TTTTTATAAGTATAGCCCGGCCGTCGAGTTT-bio |
| M4 | GCGCATTATTTTGCTTATCCGGTATTCTAAATCAGATTT-bio |
| M5 | CCCCGATTTAGAGCTTGACGGGGAAATCAAAATTT-bio |
| M6 | CAATGACACTCCAAAAGGAGCCTTACAACGCCTTT-bio |
| M7 | GCTAAATCTTTTCTGTAGCTCAACATGTATTGCTGATTT-bio |
| M8 | CAGCGAAAATTTTACTTTCAACAGTTTCTGGGATTTTTT-bio |
| I1 | GACCTGACGACATAGACTTGAGAGAGCGACTTT-bio |
| I2 | ACTTGAGAGAGCGACTCGACGACTACTGACTTT-bio |
| I3 | GCGGACATTCGCTGACCTCTCACCCACCATTTT-bio |
2.2. Experimental Instruments
3. Methods
3.1. Preparation of DNA Nanospheres
3.2. Compression Ratio and Release Rate of the DNA Nanospheres
3.3. Synthesis of Protected DNA Nanosphere
3.4. DNA Release
3.5. Denaturing Polyacrylamide Gel Electrophoresis (dPAGE)
3.6. Reading of DNA Information
3.7. Characterization of Patterns Using AFM
4. Result and Discussion
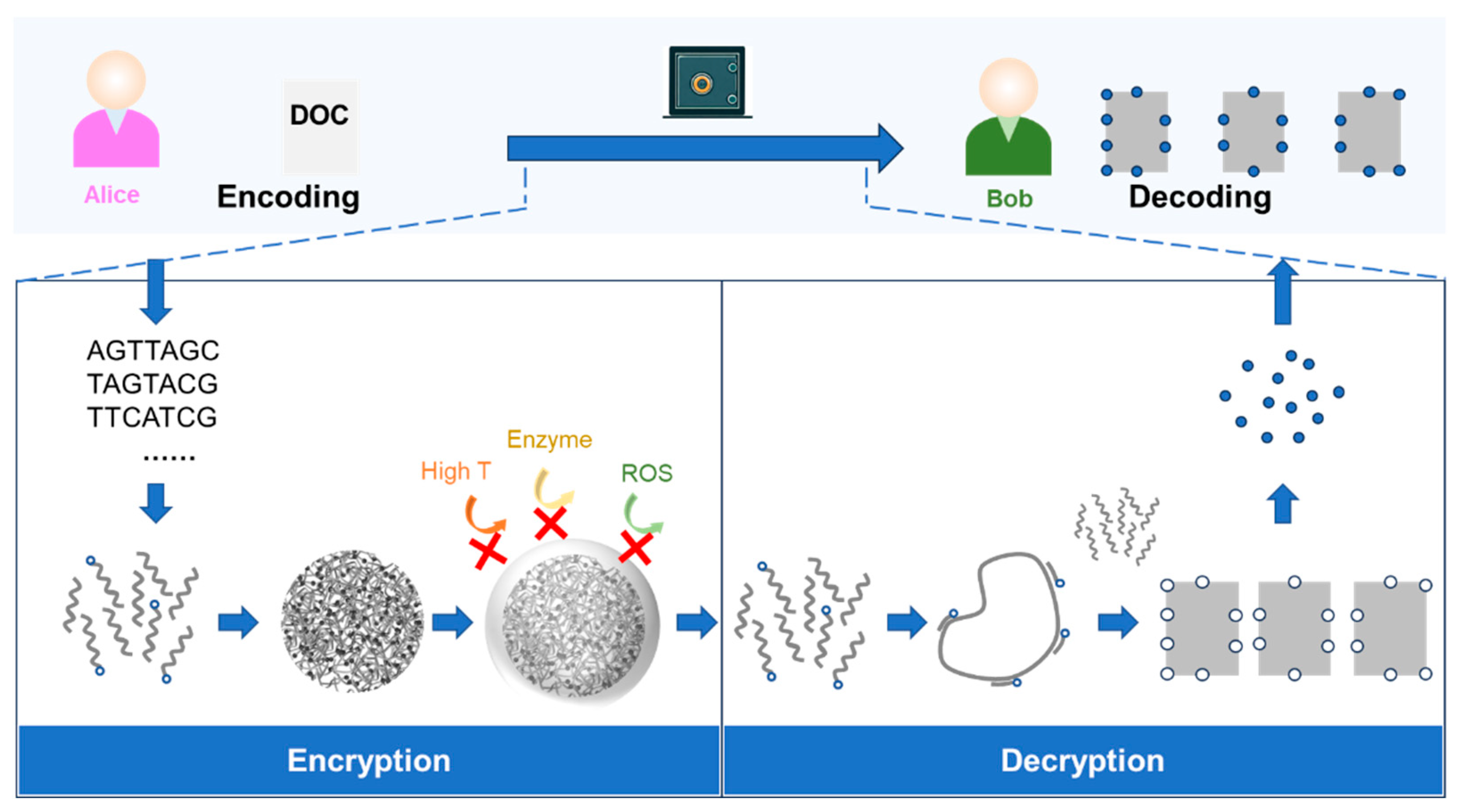
4.1. The Principles of Encrypted Communication
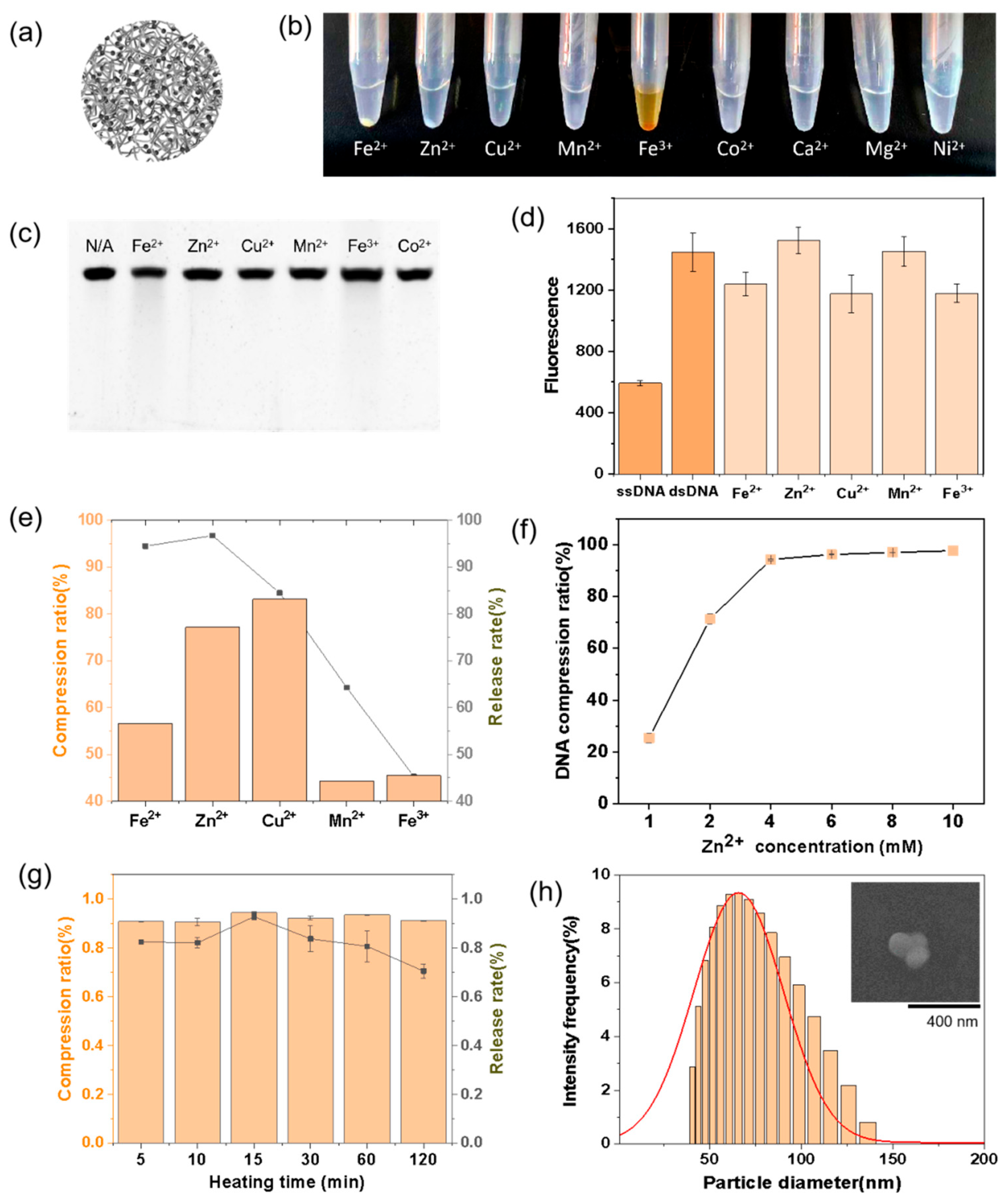
4.2. Preparation of DNA Nanospheres
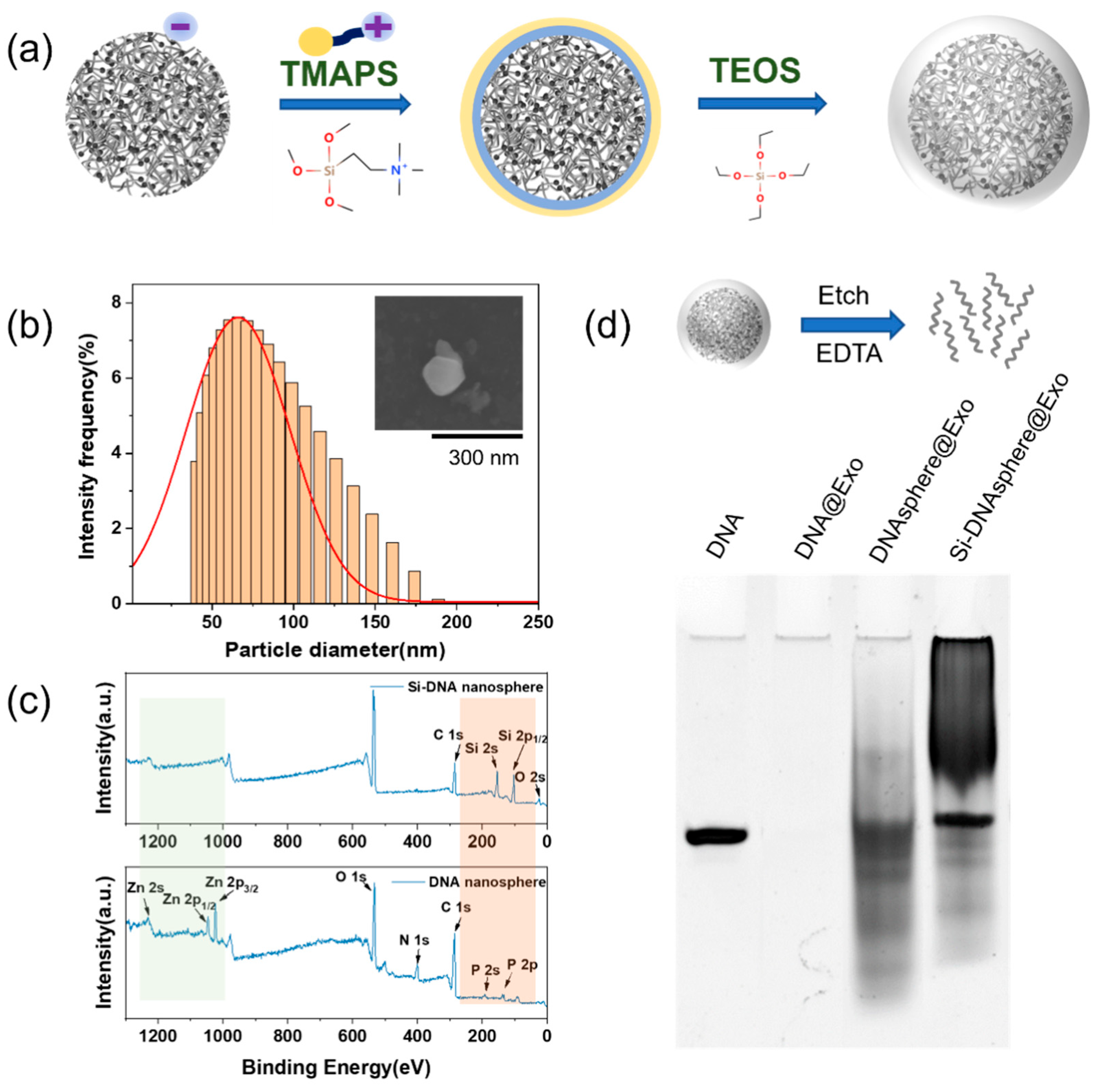
4.3. Preparation of Si-DNA Nanospheres
4.4. Reading of Encrypted Information in the Si-DNA Nanosphere
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, X.; Du, Y.; Fan, T.; Guo, J.; Ren, J.; Wu, R.; Zheng, T. Threat analysis for space information network based on network security attributes: A review. Complex Intell. Syst. 2023, 9, 3429–3468. [Google Scholar] [CrossRef]
- Sarkar, T.; Selvakumar, K.; Motiei, L.; Margulies, D. Message in a molecule. Nat. Commun. 2016, 7, 11374. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Bae, H.J.; Kim, J.; Shin, S.; Choi, S.-E.; Lee, S.H.; Kwon, S.; Park, W. Lithographically Encoded Polymer Microtaggant Using High-Capacity and Error-Correctable QR Code for Anti-Counterfeiting of Drugs. Adv. Mater. 2012, 24, 5924–5929. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, X.; Yang, Q.; Li, Y.; Li, W.; Li, H.; Chen, S.; Li, M.; Song, Y. Patterned photonic crystals for hiding information. J. Mater. Chem. C. 2017, 5, 4621–4628. [Google Scholar] [CrossRef]
- Diffie, W.; Hellman, M.E. Exhaustive Crypt-Analysis of Nbs Data Encryption Standard. Computer 1977, 10, 74–84. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Park, S.J.; Lee, C.S.; Hwang, S.; Shin, Y.B.; Ha, T.H.; Kim, M. Long-Term Stability and Integrity of Plasmid-Based DNA Data Storage. Polymers 2018, 10, 28. [Google Scholar] [CrossRef]
- Church, G.M.; Gao, Y.; Kosuri, S. Next-Generation Digital Information Storage in DNA. Science 2012, 337, 1628. [Google Scholar] [CrossRef] [PubMed]
- Goldman, N.; Bertone, P.; Chen, S.; Dessimoz, C.; LeProust, E.M.; Sipos, B.; Birney, E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 2013, 494, 77–80. [Google Scholar] [CrossRef]
- Clelland, C.T.; Risca, V.; Bancroft, C. Hiding messages in DNA microdots. Nature 1999, 399, 533–534. [Google Scholar] [CrossRef]
- Bancroft, C.; Bowler, T.; Bloom, B.; Clelland, C.T. Long-Term Storage of Information in DNA. Science 2001, 293, 1763–1765. [Google Scholar] [CrossRef]
- Wong, P.C.; Wong, K.K.; Foote, H. Organic data memory using the DNA approach. Commun. ACM 2003, 46, 95–98. [Google Scholar] [CrossRef]
- Ailenberg, M.; Rotstein, O. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques 2009, 47, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.P. Long-term data storage in DNA. Trends Biotechnol. 2001, 19, 247–250. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, F.; Chao, J.; Xie, M.; Liu, H.; Pan, M.; Kopperger, E.; Liu, X.; Li, Q.; Shi, J.; et al. DNA origami cryptography for secure communication. Nat. Commun. 2019, 10, 5469. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.V.; Tørring, T.; Rotaru, A.; Jacobsen, M.F.; Ravnsbæk, J.B.; Subramani, R.; Mamdouh, W.; Kjems, J.; Mokhir, A.; Besenbacher, F.; et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 2010, 5, 200–203. [Google Scholar] [CrossRef]
- Fang, W.; Xie, M.; Hou, X.; Liu, X.; Zuo, X.; Chao, J.; Wang, L.; Fan, C.; Liu, H.; Wang, L. DNA Origami Radiometers for Measuring Ultraviolet Exposure. J. Am. Chem. Soc. 2020, 142, 8782–8789. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Nyberg, B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 1974, 13, 3405–3410. [Google Scholar] [CrossRef]
- Koag, M.-C.; Jung, H.; Kou, Y.; Lee, S. Bypass of the Major Alkylative DNA Lesion by Human DNA Polymerase η. Molecules 2019, 24, 3928. [Google Scholar] [CrossRef] [PubMed]
- Knips, A.; Zacharias, M. Both DNA global deformation and repair enzyme contacts mediate flipping of thymine dimer damage. Sci. Rep. 2017, 7, 41324. [Google Scholar] [CrossRef]
- Athanasiadou, D.; Carneiro, K.M.M. DNA nanostructures as templates for biomineralization. Nat. Rev. Chem. 2021, 5, 93–108. [Google Scholar] [CrossRef]
- Grass, R.N.; Heckel, R.; Puddu, M.; Paunescu, D.; Stark, W.J. Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. 2015, 54, 2552–2555. [Google Scholar] [CrossRef] [PubMed]
- Paunescu, D.; Fuhrer, R.; Grass, R.N. Protection and deprotection of DNA--high-temperature stability of nucleic acid barcodes for polymer labeling. Angew. Chem. Int. Ed. 2013, 52, 4269–4272. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.D.; Kohll, A.X.; Nguyen, B.H.; Koch, J.; Heckel, R.; Stark, W.J.; Ceze, L.; Strauss, K.; Grass, R.N. Combining Data Longevity with High Storage Capacity—Layer-by-Layer DNA Encapsulated in Magnetic Nanoparticles. Adv. Funct. Mater. 2019, 29, 1901672. [Google Scholar] [CrossRef]
- Nguyen, M.-K.; Nguyen, V.H.; Natarajan, A.K.; Huang, Y.; Ryssy, J.; Shen, B.; Kuzyk, A. Ultrathin Silica Coating of DNA Origami Nanostructures. Chem. Mater. 2020, 32, 6657–6665. [Google Scholar] [CrossRef]
- Sigel, R.K.; Sigel, H. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc. Chem. Res. 2010, 43, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, C.; Di, Z.; Li, H.; Zhang, J.; Xue, W.; Zhao, M.; Zhang, K.; Zhao, Y.; Li, L. Engineering Multifunctional DNA Hybrid Nanospheres through Coordination-Driven Self-Assembly. Angew. Chem. Int. Ed. 2019, 58, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhu, L.; Yang, M.; Xu, W. Synthesis of Amorphous/Crystalline Hetero-Phase Nanozymes With Peroxidase-Like Activity by Coordination-Driven Self-Assembly for Biosensors. Small 2023, 19, 2204782. [Google Scholar] [CrossRef]
- Liu, B.; Hu, F.; Zhang, J.; Wang, C.; Li, L. A Biomimetic Coordination Nanoplatform for Controlled Encapsulation and Delivery of Drug-Gene Combinations. Angew. Chem. Int. Ed. 2019, 58, 8804–8808. [Google Scholar] [CrossRef]
- Han, Q.; Zhang, X.; Jia, Y.; Guo, S.; Zhu, J.; Luo, S.; Na, N.; Ouyang, J. Synthesis and Characteristics of Self-Assembled Multifunctional Ln(3+) -DNA Hybrid Coordination Polymers. Chemistry 2022, 28, 202200281. [Google Scholar] [CrossRef]
- Lu, C.; Xu, Y.; Huang, P.J.; Zandieh, M.; Wang, Y.; Zheng, J.; Liu, J. Protection of DNA by metal ions at 95 degrees C: From lower critical solution temperature (LCST) behavior to coordination-driven self-assembly. Nanoscale 2022, 14, 14613–14622. [Google Scholar] [CrossRef]
- Lee, W.K.; Kwon, K.; Choi, Y.; Lee, J.S. Dynamic metallization of spherical DNA via conformational transition into gold nanostructures with controlled sizes and shapes. J. Colloid Interface Sci. 2021, 594, 160–172. [Google Scholar] [CrossRef]
- Du, Z.; Zhu, L.; Wang, P.; Lan, X.; Lin, S.; Xu, W. Coordination-Driven One-Step Rapid Self-Assembly Synthesis of Dual-Functional Ag@Pt Nanozyme. Small 2023, 19, 2301048. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Shen, X.; Sun, F.; Na, N.; Ouyang, J. Metal-DNA coordination based bioinspired hybrid nanospheres for in situ amplification and sensing of microRNA. J. Mater. Chem. B 2020, 8, 11074–11081. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Yu, J.; Lu, H.; Wei, Z.; Chao, Z.; Wang, Z.; Wu, W.; Jiang, H.; Tian, L. Mn-DNA coordination of nanoparticles for efficient chemodynamic therapy. Chem. Commun. 2021, 57, 1734–1737. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Liu, B.; Zhou, Z.; Li, W.; Zhao, J.; Li, L.; Zhao, Y. Engineering hierarchical metal-organic@metal-DNA heterostructures for combinational tumor treatment. Nano Res. 2023. [Google Scholar] [CrossRef]
- Fu, J.; Liu, M.; Liu, Y.; Woodbury, N.W.; Yan, H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 2012, 134, 5516–5519. [Google Scholar] [CrossRef]
- Antkowiak, P.L.; Koch, J.; Nguyen, B.H.; Stark, W.J.; Strauss, K.; Ceze, L.; Grass, R.N. Integrating DNA Encapsulates and Digital Microfluidics for Automated Data Storage in DNA. Small 2022, 18, 2107381. [Google Scholar] [CrossRef]
- Ramos-Valle, A.; Marín-Caba, L.; Hevia, L.G.; Correa-Duarte, M.A.; Fanarraga, M.L. One-pot synthesis of compact DNA silica particles for gene delivery and extraordinary DNA preservation. Mater. Today 2023, 18, 100357. [Google Scholar] [CrossRef]
- Sreedhara, A.; Cowan, J.A. Catalytic hydrolysis of DNA by metal ions and complexes. J. Biol. Inorg. Chem. 2001, 6, 337–347. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Cai, Z.; Wang, J.; Liu, H. Condensed DNA Nanosphere for DNA Origami Cryptography. Chemistry 2023, 5, 2406-2417. https://doi.org/10.3390/chemistry5040159
Gao R, Cai Z, Wang J, Liu H. Condensed DNA Nanosphere for DNA Origami Cryptography. Chemistry. 2023; 5(4):2406-2417. https://doi.org/10.3390/chemistry5040159
Chicago/Turabian StyleGao, Rui, Zhuang Cai, Jianbang Wang, and Huajie Liu. 2023. "Condensed DNA Nanosphere for DNA Origami Cryptography" Chemistry 5, no. 4: 2406-2417. https://doi.org/10.3390/chemistry5040159
APA StyleGao, R., Cai, Z., Wang, J., & Liu, H. (2023). Condensed DNA Nanosphere for DNA Origami Cryptography. Chemistry, 5(4), 2406-2417. https://doi.org/10.3390/chemistry5040159




