Investigating Grape Seed Extract as a Natural Antibacterial Agent for Water Disinfection in Saudi Arabia: A Pilot Chemical, Phytochemical, Heavy-Metal, Mineral, and CB-Dock Study Employing Water and Urine Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sample Collection
2.2. Water Sample Collection
2.3. Urine Sample Collection and Preparation
2.4. Gas Chromatography–Mass Spectrometry Analysis
2.5. Phytochemical Determination
2.6. Metal Analyses
2.7. Bacterial Population Analysis
2.8. Docking Analysis
2.9. Antibacterial Activity
2.10. Statistical Analysis
3. Results
3.1. GC-MS and Analyses of GSE
3.2. Phytochemical Components and Metal Contents of GSE
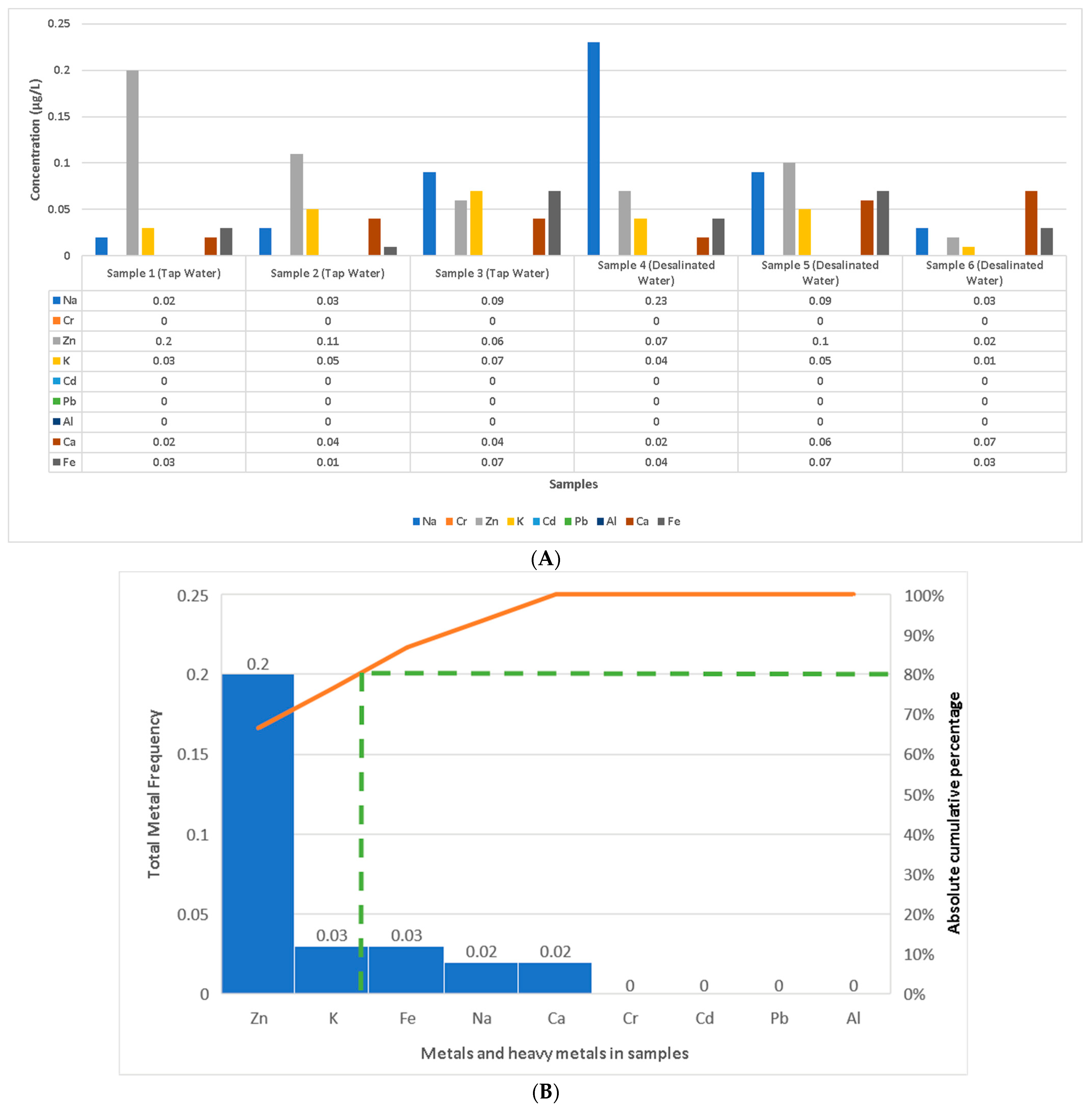
3.3. Metal Analysis in Water Samples
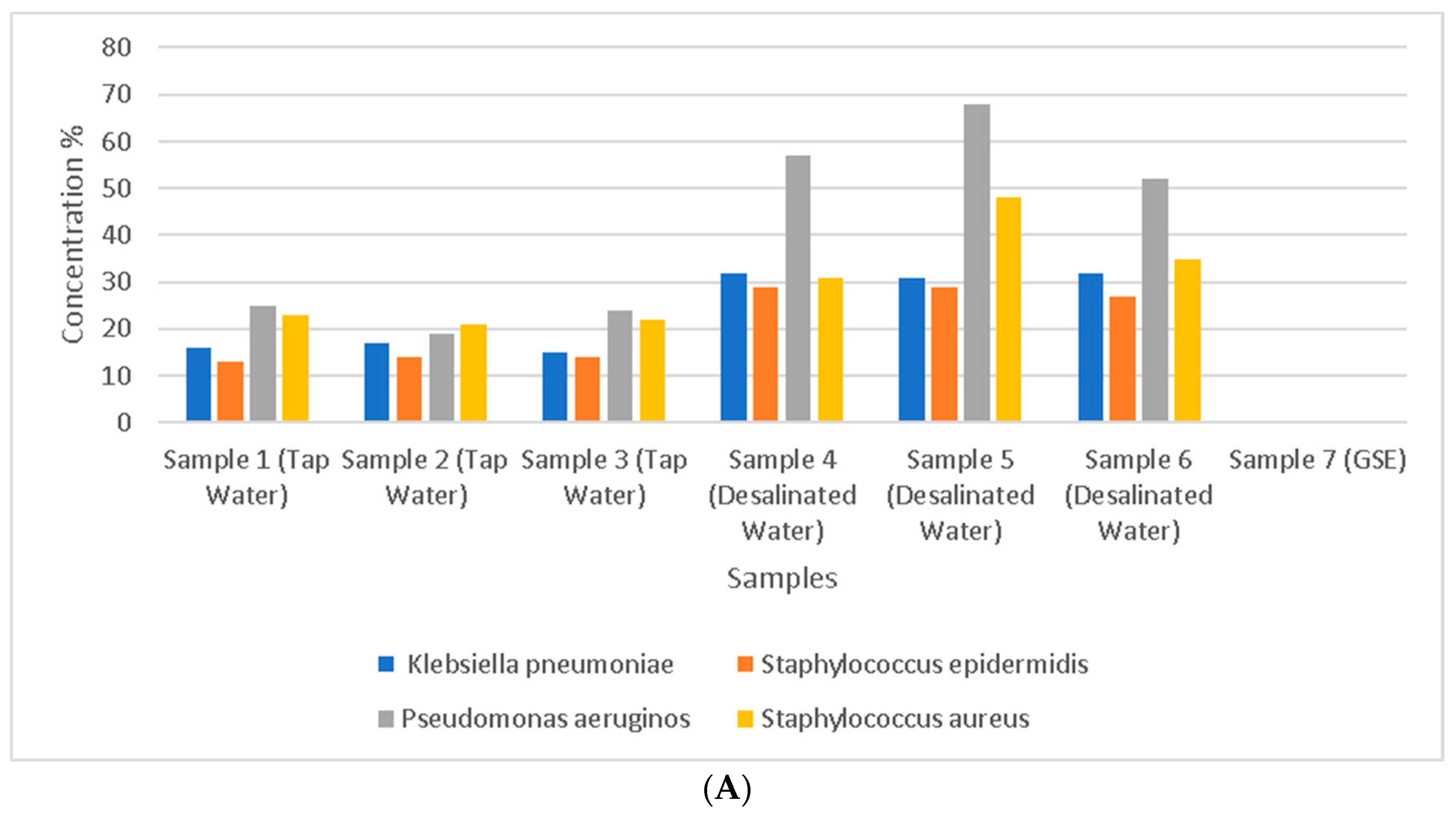
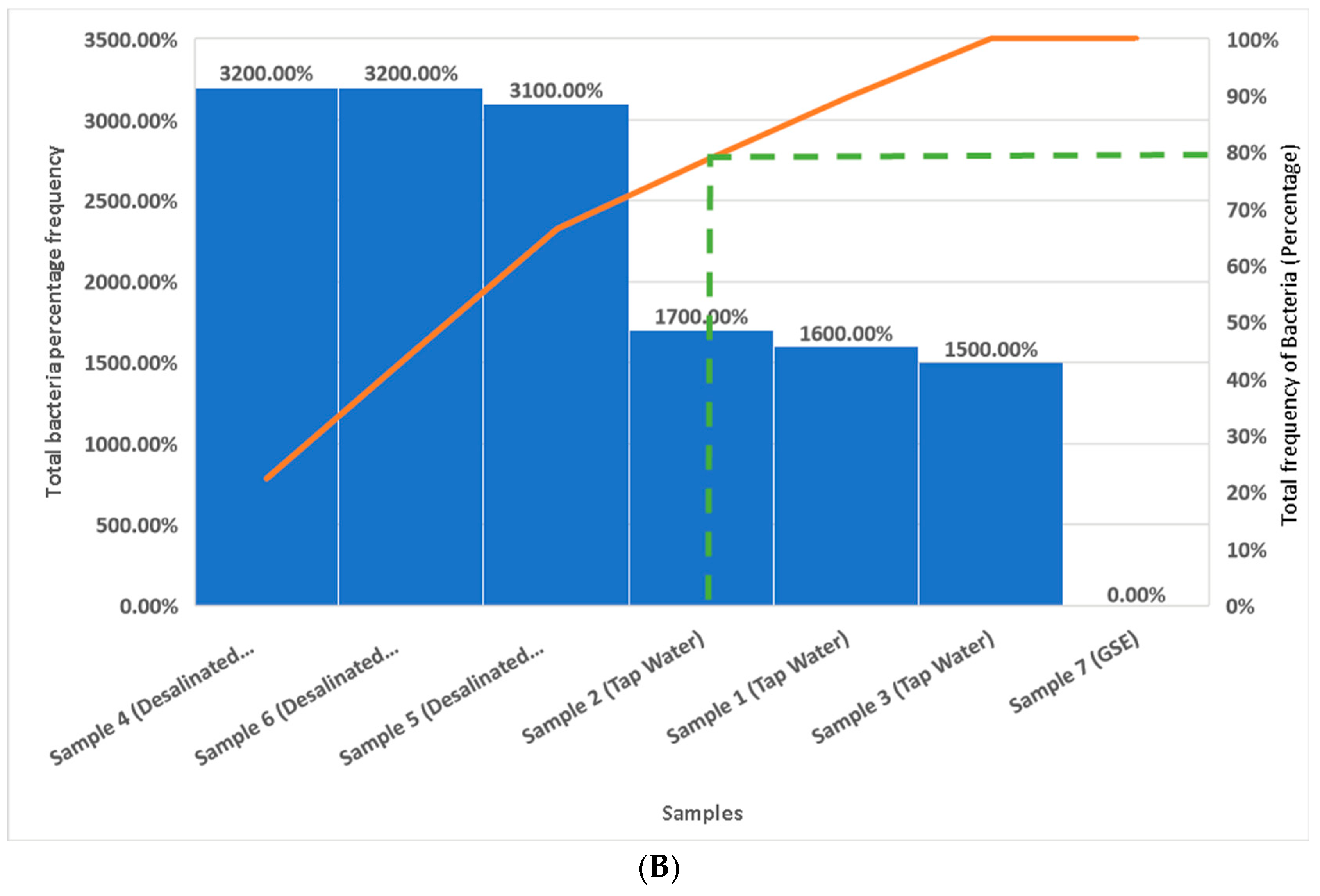
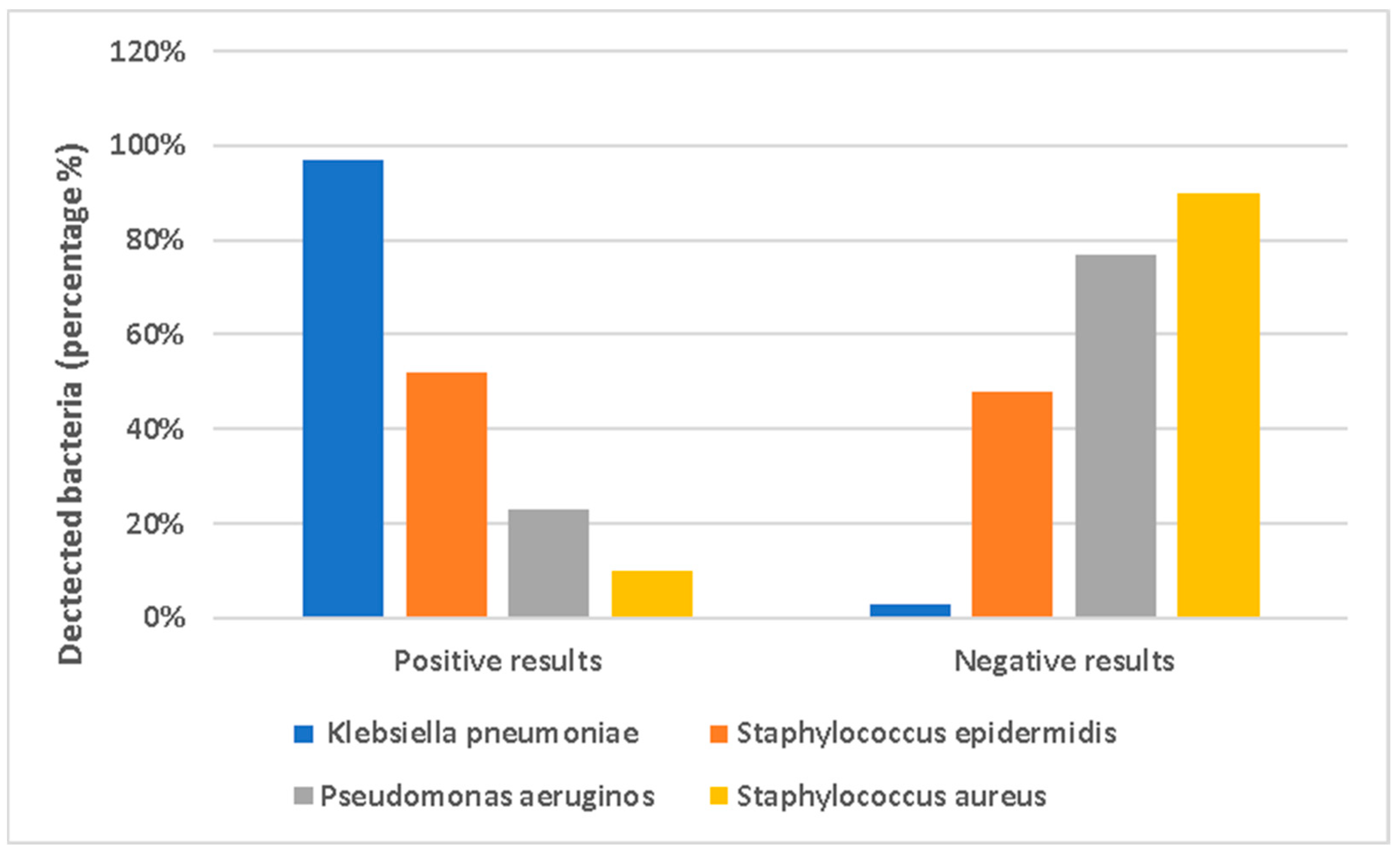
3.4. Analysis of Bacterial Species in Water and GSE Samples
3.5. Screening of Urine Biological Samples for Heavy Metals, Other Elements, and Bacteria
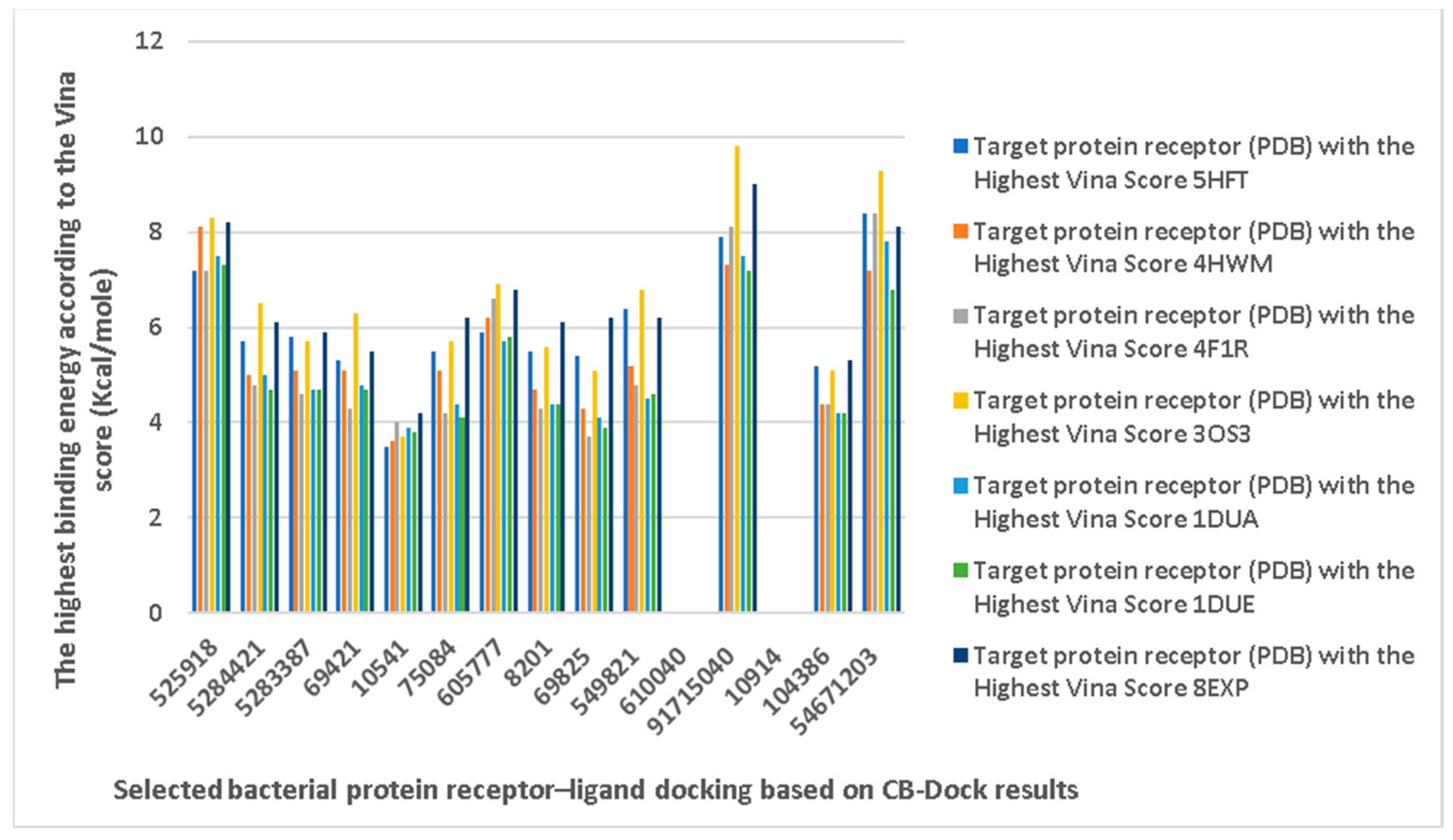
3.6. CB-Dock Analysis
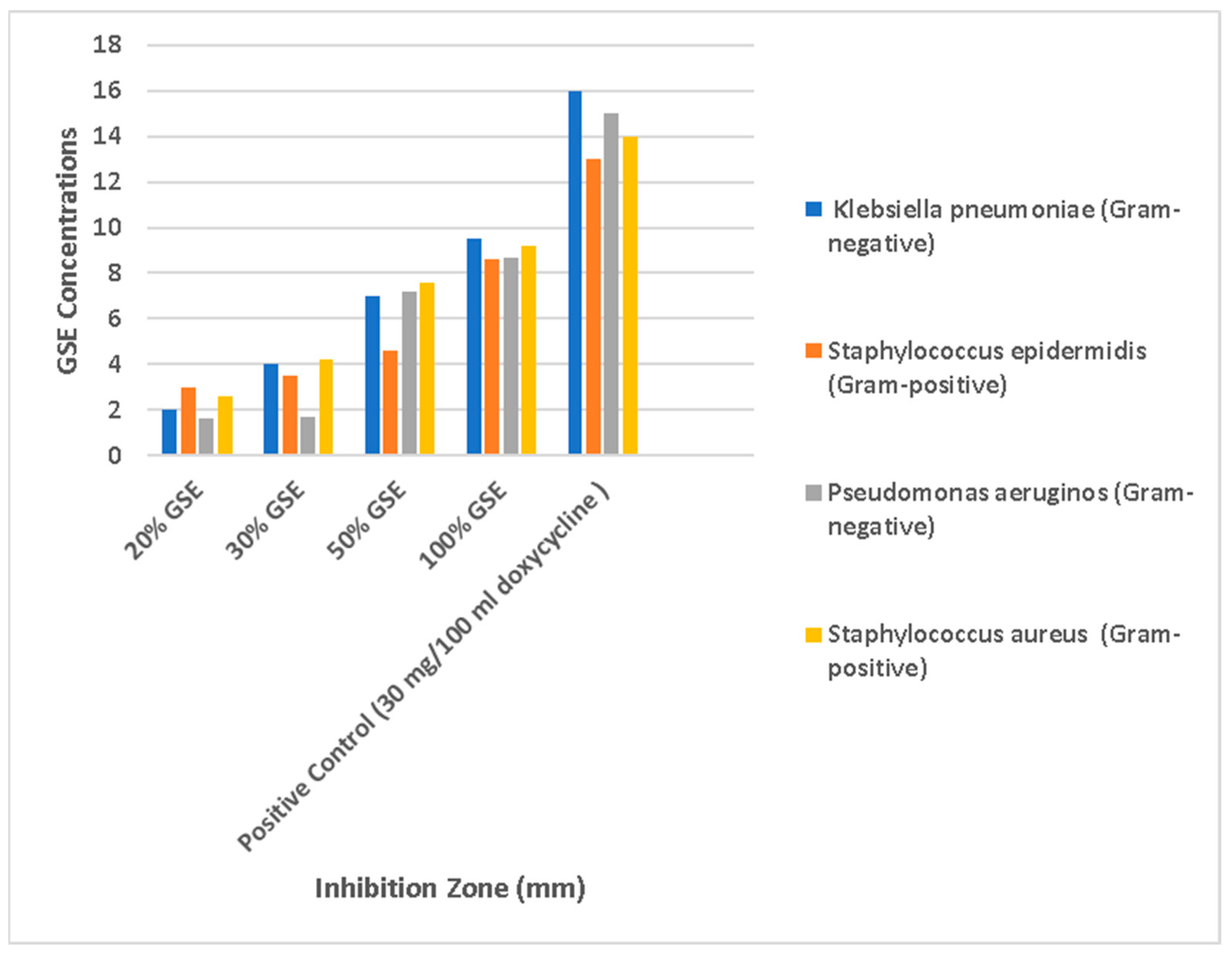
3.7. Antibacterial Effects at Different Concentrations of GSE
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Sayed, A.A.H.; Al-Blehed, M.S. Bacterial Isolate from Arabian Gulf Coast Soils in Saudi Arabia Able to Degrade Arab Crude Oil. J. King Saud Univ. Eng. Sci. 1999, 11, 251–260. [Google Scholar] [CrossRef]
- Abdalwahab, M.A.; Dawood, S.A.; Abouhosh, E. Evaluation of Microbial or Bacterial Quality of the Drinking Water of Duba Province North Saudi Arabia. Asian J. Agric. Food Sci. 2017, 5, 13–18. Available online: https://ajouronline.com/index.php/AJAFS/article/view/4447#google_vignette (accessed on 12 January 2023).
- AlOtaibi, E.L.S. Bacteriological assessment of urban water sources in Khamis Mushait Governorate, southwestern Saudi Arabia. Int. J. Health Geogr. 2009, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Saati, A.; Faidah, H. Environmental Prevalence of Pathogens in Different Drinking Water Resources in Makkah City (Kingdom of Saudi Arabia). Curr. World Environ. J. 2013, 8, 37–53. [Google Scholar] [CrossRef][Green Version]
- Al-Wohaib, F.A.; Al-Sharif, I.; Al-Zain, H.; Murad, D.; Al-Harbi, L.; Al-Mozaini, M. Evaluation of two molecular detection platforms for gastroenteritis pathogens in treated sewage water in the Eastern province of Saudi Arabia. Sci. Rep. 2022, 12, 21744. [Google Scholar] [CrossRef]
- Alqahtani, J.M.; Asaad, A.M.; Ahmed, E.M.; Qureshi, M.A. Drinking water quality and public health in Southwestern Saudi Arabia: The need for a national monitoring program. J. Fam. Community Med. 2015, 22, 19–24. [Google Scholar] [CrossRef]
- Alaa el-Din, M.N.; Madany, I.M.; Al-Tayaran, A.; Al-Jubair, A.H.; Gomaa, A. Trends in water quality of some wells in Saudi Arabia, 1984–1989. Sci. Total Environ. 1994, 143, 173–181. [Google Scholar] [CrossRef]
- Pritchard, M.; Mkandawire, T.; O’Neill, J.G. Biological, Chemical and Physical Drinking Water Quality from Shallow Wells in Malawi: Case Study of Blantyre, Chiradzulu and Mulanje. Phys. Chem. Earth Parts A/B/C 2007, 32, 1167–1177. [Google Scholar] [CrossRef]
- Agus, E.; Voutchkov, N.; Sedlak, D. Disinfection by-products and their potential impact on the quality of water produced by desalination systems: A literature review. Desalination 2009, 237, 214–237. [Google Scholar] [CrossRef]
- Kim, D.; Amy, G.L.; Karanfil, T. Disinfection by-product formation during seawater desalination: A review. Water Research 2015, 81, 343–355. [Google Scholar] [CrossRef]
- Fatima, F.; Siddiqui, S.; Khan, W.A. Nanoparticles as Novel Emerging Therapeutic Antibacterial Agents in the Antibiotics Resistant Era. Biol. Trace Elem. Res. 2021, 199, 2552–2564. [Google Scholar] [CrossRef]
- Xin, X.; Qi, C.; Xu, L.; Gao, Q.; Liu, X. Green synthesis of silver nanoparticles and their antibacterial effects. Front. Chem. Eng. 2022, 4, 941240. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, M.; Cheng, J.; Wan, Y.; Zhu, L.; Gao, Z.; Jiang, L. Antibiotic Alternatives: Multifunctional Ultra-Small Metal Nanoclusters for Bacterial Infectious Therapy Application. Molecules 2024, 29, 3117. [Google Scholar] [CrossRef] [PubMed]
- Crisan, C.M.; Mocan, T.; Manolea, M.; Lasca, L.I.; Tăbăran, F.-A.; Mocan, L. Review on Silver Nanoparticles as a Novel Class of Antibacterial Solutions. Appl. Sci. 2021, 11, 1120. [Google Scholar] [CrossRef]
- Al Bahri, M.; Calvo, L.; Gilarranz, M.A.; Rodriguez, J.J. Activated carbon from grape seeds upon chemical activation with phosphoric acid: Application to the adsorption of diuron from water. Chem. Eng. J. 2012, 203, 348–356. [Google Scholar] [CrossRef]
- Hamouda, A.F.; Farawilla, T.-L.M.; Attafi, I.M.; Khardali, I.A.; Attafi, M.A.; Oraiby, M.E.; Abualsail, F.M. Screening Pilot Study of Fruit Seed Compositions by GC-MS and Their Potential Scenario Anti ACE2 and 2rh1 Receptors as a Recycling Possibility in the Coronavirus Pandemic. J. Clin. Med. Res. 2021, 2, 1–65. [Google Scholar] [CrossRef]
- Raota, C.S.; Cerbaro, A.F.; Salvador, M.; Delamare, A.P.L.; Echeverrigaray, S.; Crespo, J.d.S.; da Silva, T.B.; Giovanela, M. Green synthesis of silver nanoparticles using an extract of Ives cultivar (Vitis labrusca) pomace: Characterization and application in wastewater disinfection. J. Environ. Chem. Eng. 2019, 7, 103383. [Google Scholar] [CrossRef]
- Munther Al Agha, A.G.; Muslih Al Khafaji, N.J.; Al Azawi, A.K.S. Isolation and Identification of Klebsiella pneumoniae using API-20E analytical system and conventional PCR assay. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 203–210. [Google Scholar] [CrossRef]
- Rice, E.E.W.; Baird, R.B.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater. 2017. Available online: https://yabesh.ir/wp-content/uploads/2018/02/Standard-Methods-23rd-Perv.pdf (accessed on 18 February 2023).
- Al-Abdalall, A.H.A. Isolation and identification of microbes associated with mobile phones in Dammam in eastern Saudi Arabia. J. Fam. Community Med. 2010, 17, 11–14. [Google Scholar] [CrossRef]
- O’hara, C.M. Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin. Microbiol. Rev. 2005, 18, 147–162. [Google Scholar] [CrossRef]
- Hamouda, A.F.; Felemban, S. A Bio-Indicator Pilot Study Screening Selected Heavy Metals in Female Hair, Nails, and Serum from Lifestyle Cosmetic, Canned Food, and Manufactured Drink Choices. Molecules 2023, 28, 5582. [Google Scholar] [CrossRef]
- Hamouda, A.F.; Felemban, S. Biochemical Pilot Study on Effects of Pomegranate Seed Oil Extract and Cosmetic Cream on Neurologically Mediated Skin Inflammation in Animals and Humans: A Comparative Observational Study. Molecules 2023, 28, 903. [Google Scholar] [CrossRef]
- Bolajoko, E.; Onyeaghala, A.A.; Akinosun, O.; Attah, A.; Moody, J.O.; Khine, A.A. Comparative proximate, phytochemical and mineral contents of freeze-dried and aqueous extracts of vernonia amygdalina del. leaves and garcinia kola heckel seeds. Int. J. Herbs Pharmacol. Res. 2020, 9, 2–13. [Google Scholar]
- Felemban, S.; Hamouda, A.F. A Pilot Study of the Effects of Ajwa Date Seed Extract in a Diabetic Animal with Parallel Observations on Human Subjects. J. Pharm. Res. Int. 2022, 34, 23–33. [Google Scholar] [CrossRef]
- Hamouda, A.F.; Hassan, A.A.M.; Khardali, I.A.; Attafi, I.M.; Oraiby, M.E.; Attafi, M.A.; Mahmoud, M.H.; Salem, S.F.A.; Sahli, K.A.Y.; Hamdi, F.Y.; et al. A Screening Pilot Study on the Relation between Body Mass Index, and Heavy Metal and Mineral Levels in College Students. Electron. J. Biol. 2019, 15, 79–89. [Google Scholar] [CrossRef]
- Aamir, M.; Singh, V.K.; Dubey, M.K.; Meena, M.; Kashyap, S.P.; Katari, S.K.; Upadhyay, R.S.; Umamaheswari, A.; Singh, S. In silico Prediction, Characterization, Molecular Docking, and Dynamic Studies on Fungal SDRs as Novel Targets for Searching Potential Fungicides Against Fusarium Wilt in Tomato. Front. Pharmacol. 2018, 9, 1038. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Huang, Y.; Wang, C.; Chen, F.; Yang, L.; Ling, L.; Che, Z.; Chen, X. Recent developments in molecular docking technology applied in food science: A review. Int. J. Food Sci. Technol. 2020, 55, 33–45. [Google Scholar] [CrossRef]
- Cattaneo, A.; Macchi, F.; Plazzotta, G.; Veronica, B.; Bocchio-Chiavetto, L.; Riva, M.A.; Pariante, C.M. Inflammation and neuronal plasticity: A link between childhood trauma and depression pathogenesis. Front. Cell. Neurosci. 2015, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Grimm, M.; Dai, W.-T.; Hou, M.-C.; Xiao, Z.-X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L. Improved protein-ligand binding affinity prediction by using a curvature-dependent surface-area model. Bioinformatics 2014, 30, 1674–1680. [Google Scholar] [CrossRef]
- Mohamed, A.A.; Ali, S.I.; EL-Baz, F.K.; Hegazy, A.K.; Kord, M.A. Chemical composition of essential oil and in vitro antioxidant and antimicrobial activities of crude extracts of Commiphora myrrha resin. Ind. Crops Prod. 2014, 57, 10–16. [Google Scholar] [CrossRef]
- El-Shazly, H.A.M.; Mohamed, E.S.A.; Abdelatif, S.H. Effect of Pomegranate Seed Oil on Certain Food Spoilage Microorganisms and its Role in Shelf Life of Cream. J. Food Dairy Sci. 2017, 8, 173–178. [Google Scholar] [CrossRef]
- Hamouda, A.F.; Felemban, S. A Pilot Study of the Amelioration of Avocado Seed Oil in Obese Female Rats Induced by Carbon Tetrachloride and Alloxan Monohydrate. J. Drug Alcohol Res. 2022, 11, 236177. [Google Scholar] [CrossRef]
- Witkowska, D.; Słowik, J.; Chilicka, K. Heavy Metals and Human Health: Possible Exposure Pathways and the Competition for Protein Binding Sites. Molecules 2021, 26, 6060. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Martin-Loeches, I.; Garcia-Vidal, C.; San Jose, A.; Torres, A. Microbial Etiology of Pneumonia: Epidemiology, Diagnosis and Resistance Patterns. Int. J. Mol. Sci. 2016, 17, 2120. [Google Scholar] [CrossRef]
- Ginsburg, A.S.; Izadnegahdar, R.; Klugman, K.P. World Pneumonia Day 2016: Pulse oximetry and oxygen. Lancet Glob. Heal. 2016, 4, e893–e894. [Google Scholar] [CrossRef]
- Huang, S.S.; Singh, R.; McKinnell, J.A.; Park, S.; Gombosev, A.; Eells, S.J.; Gillen, D.L.; Kim, D.; Rashid, S.; Macias-Gil, R.; et al. Decolonization to Reduce Postdischarge Infection Risk among MRSA Carriers. N. Engl. J. Med. 2019, 380, 638–650. [Google Scholar] [CrossRef]
- Bratzler, D.W.; Dellinger, E.P.; Olsen, K.M.; Perl, T.M.; Auwaerter, P.G.; Bolon, M.K.; Fish, D.N.; Napolitano, L.M.; Sawyer, R.G.; Slain, D.; et al. Clinical practice guidelines for antimicrobial prophylaxis in surgery. Surg. Infect. 2013, 14, 73–156. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Prospero-Garcia, O.; Siuzdak, G.; Gilula, N.B.; Henriksen, S.J.; Boger, D.L.; Lerner, R.A. Chemical characterization of a family of brain lipids that induce sleep. Science 1995, 268, 1506–1509. [Google Scholar] [CrossRef]
- Thomas, A.; A Stevenson, L.; Wease, K.N.; Price, M.R.; Baillie, G.; A Ross, R.; Pertwee, R.G. Evidence that the plant cannabinoid Delta9-tetrahydrocannabivarin is a cannabinoid CB1 and CB2 receptor antagonist. Br. J. Pharmacol. 2005, 146, 917–926. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.A.; Mullish, B.H.; Pechlivanis, A.; Liu, Z.; Brignardello, J.; Kao, D.; Holmes, E.; Li, J.V.; Clarke, T.B.; Thursz, M.R.; et al. Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota. Gastroenterology 2018, 155, 1495–1507.e15. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Pramanik, A.; Agrawal, P.K. Evaluation of bioactive secondary metabolites from endophytic fungus Pestalotiopsis neglecta BAB-5510 isolated from leaves of Cupressus torulosa D.Don. 3 Biotech 2016, 6, 210. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Ban, X.; He, J.; Tong, J.; Tian, J.; Wang, Y. Comparative analysis of essential oil components and antioxidant activity of extracts of Nelumbo nucifera from various areas of China. J. Agric. Food Chem. 2010, 58, 441–448. [Google Scholar] [CrossRef]
- Khan, R.; Ali, S.; Mumtaz, S.; Kanwal, L.; Nauroze, T. Ameliorating and pharmacological intervention potential of grape seed extract against lead- and cadmium-induced toxicity. Int. J. Environ. Sci. Technol. 2021, 19, 10441–10456. [Google Scholar] [CrossRef]
- Marhamati, F.; Mahdavian, M.; Bazgir, S. Corrosion mitigation of mild steel in hydrochloric acid solution using grape seed extract. Sci. Rep. 2021, 11, 18374. [Google Scholar] [CrossRef]
- Gupta, M.; Dey, S.; Marbaniang, D.; Pal, P.; Ray, S.; Mazumder, B. Grape seed extract: Having a potential health benefits. J. Food Sci. Technol. 2020, 57, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Hamouda, A.F. Approach to Cope with Cancer Treatment: I Will Rise Again—Our Voices Must Be Heard, 1st ed.; B P International: London, UK, 2024; pp. 1–105. ISBN 978-81-976377-8-0. [Google Scholar] [CrossRef]
| No. | Name | Base Peak | RT (min) | Chromatogram |
|---|---|---|---|---|
| 1. | 9-Octadecenamide, (Z)- | 59 | 12.351–12.426 | 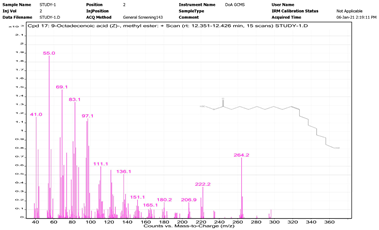 |
| 2. | Stigmastan-3,5-diene | 396.4 | 18.213–18.272 | 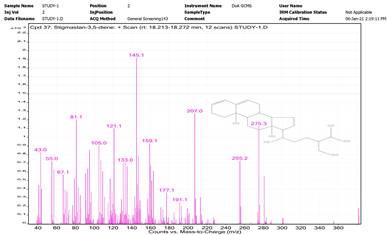 |
| 3. | 9,12-Octadecadienoic acid (Z,Z)-, methyl ester | 81.1 | 12.308–12.351 | 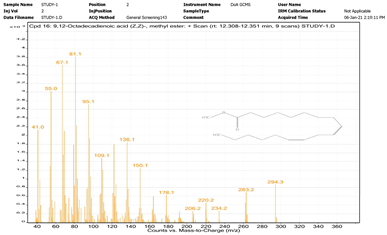 |
| 4. | Oleamide | 59 | 15.121–15.218 | 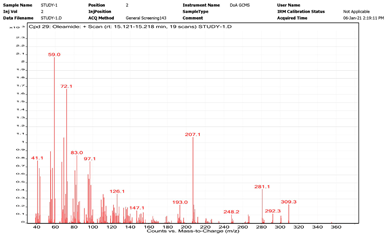 |
| 5. | Hexadecanamide | 59 | 12.928–12.961 | 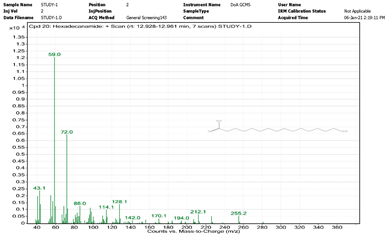 |
| 6. | Phosphoric acid, trimethyl ester | 110 | 3.151–3.296 | 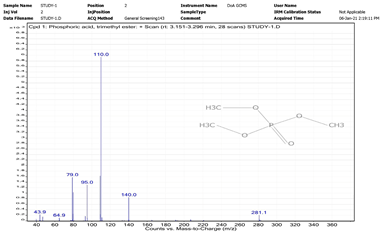 |
| 7. | Dodecyl acrylate | 55 | 9.564–9.591 |  |
| 8. | Pentanoic acid, 5-hydroxy-, 2,4-di-t-butylphenyl esters | 191.2 | 8.227–8.297 | 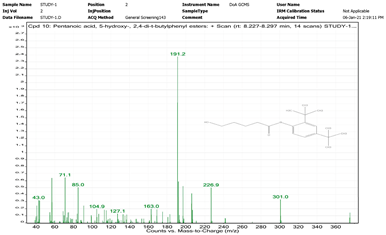 |
| 9. | Methyl stearate | 74 | 12.485–12.511 | 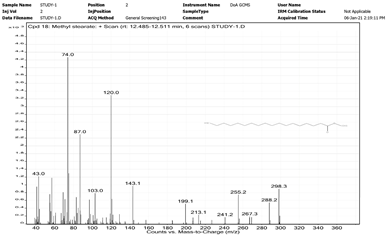 |
| 10. | 1-Pentadecyne | 55 | 12.720–12.789 | 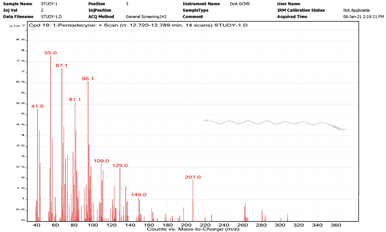 |
| 11. | 11,13-Dimethyl-12-tetradecen-1-ol acetate | 207 | 14.645–14.688 | 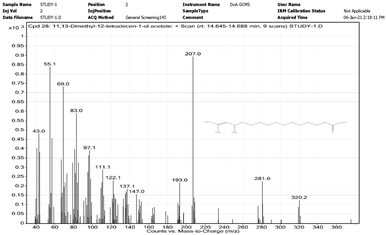 |
| 12. | 4-Methyl-2,4-bis(4′-trimethylsilyloxyphenyl)pentene-1 | 207 | 21.919–22.198 | 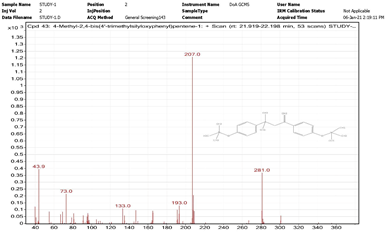 |
| 13. | 1,3-Benzenediol, o-(4-methylbenzoyl)-o′-(2-methoxybenzoyl)- | 135.1 | 5.644–5.697 | 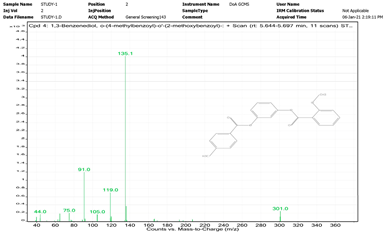 |
| 14. | Cyclotrisiloxane, hexamethyl- | 207 | 17.411–17.469 | 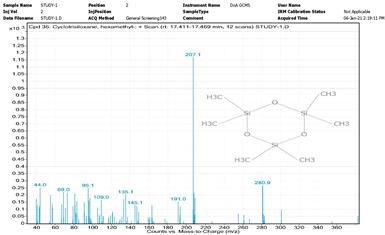 |
| 15. | Isooctyl 3-mercaptopropionate | 57 | 11.891–11.923 | 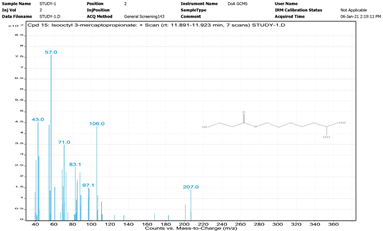 |
| Ligand | Target Protein Receptor (PDB) | ||
|---|---|---|---|
| 1. | CID 525918 Stigmastan-3,5-diene | Klebsiella pneumoniae | |
5HFT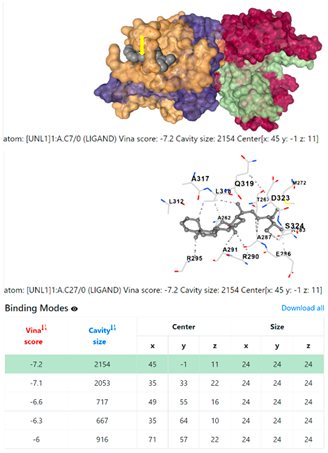 | 4HWM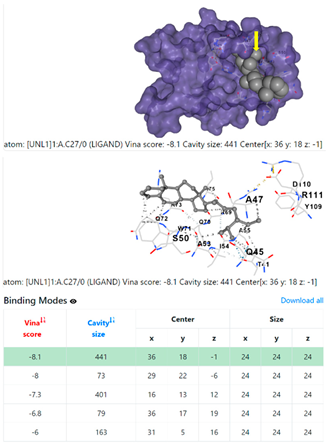 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R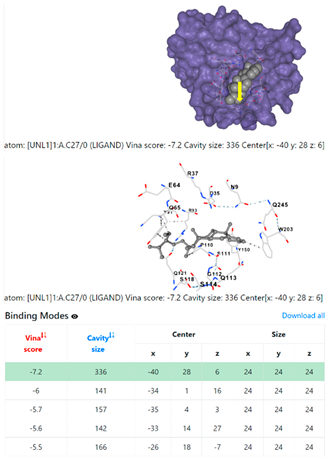 | 3OS3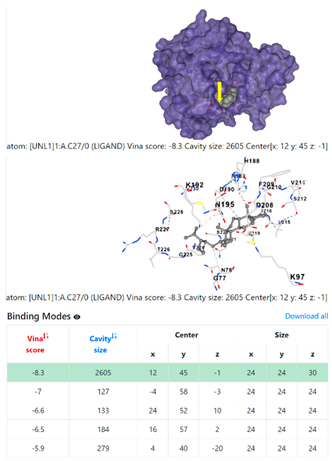 | ||
| Staphylococcus aureus | |||
1DUA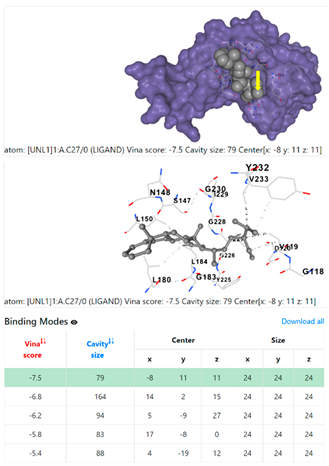 | 1DUE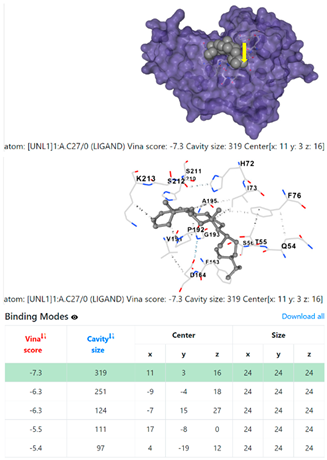 | ||
8EXP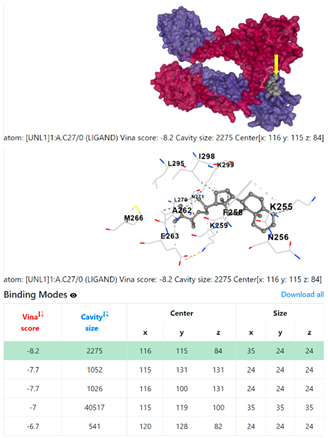 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 2. | CID 5284421 9,12-Octadecadienoic acid (Z,Z)-, methyl ester | Klebsiella pneumoniae | |
5HFT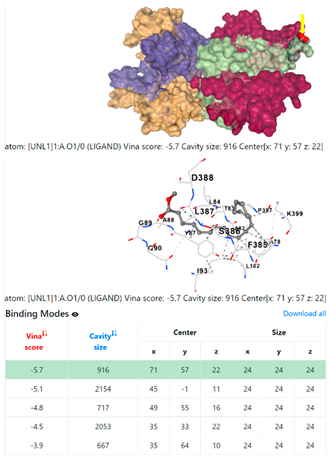 | 4HWM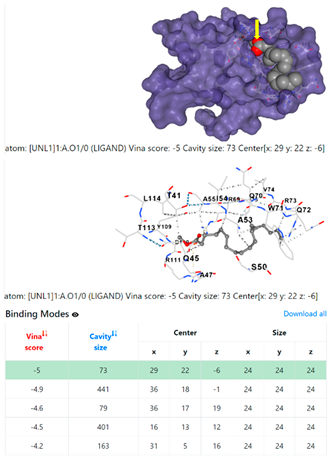 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R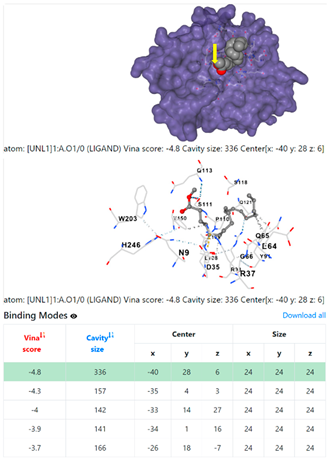 | 3OS3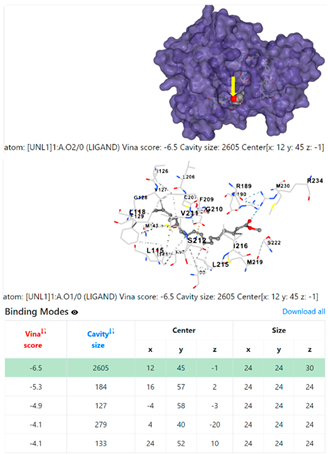 | ||
| Staphylococcus aureus | |||
1DUA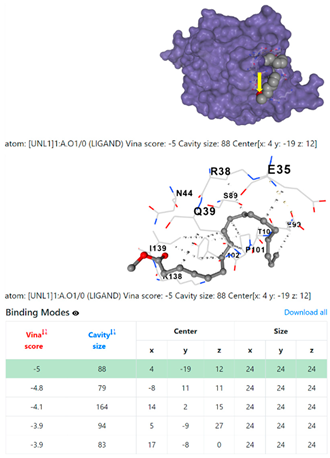 | 1DUE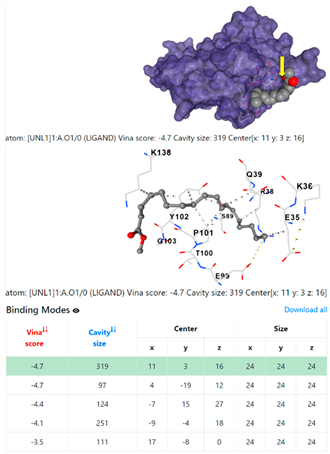 | ||
8EXP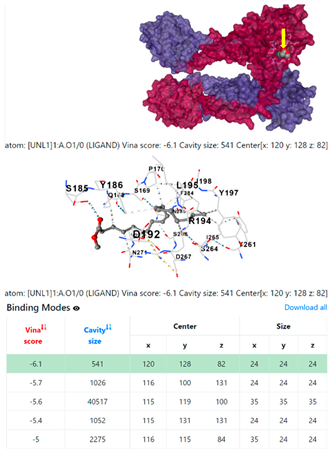 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 3. | CID 5283387 Oleamide | Klebsiella pneumoniae | |
5HFT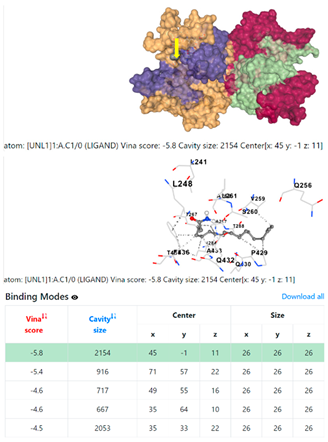 | 4HWM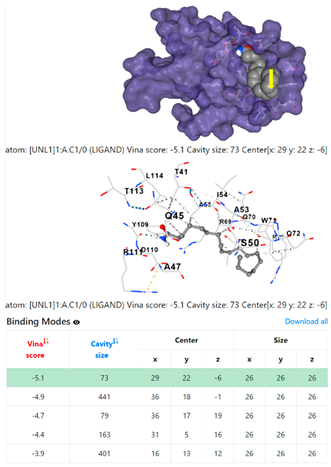 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R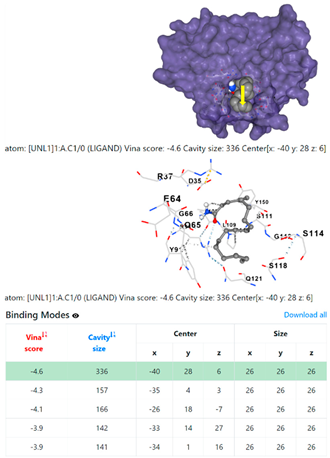 | 3OS3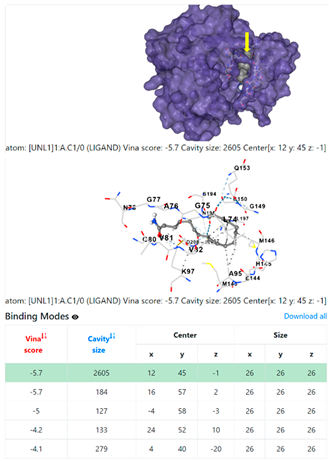 | ||
| Staphylococcus aureus | |||
1DUA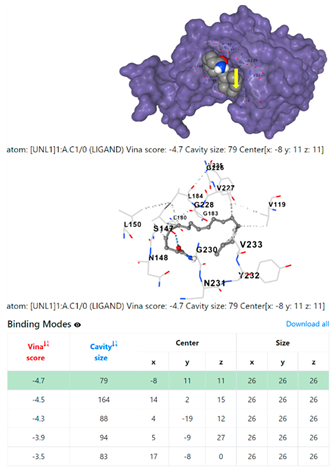 | 1DUE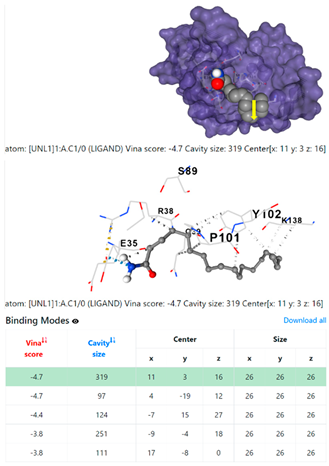 | ||
8EXP | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 4. | CID 69421 Hexadecanamide | Klebsiella pneumoniae | |
5HFT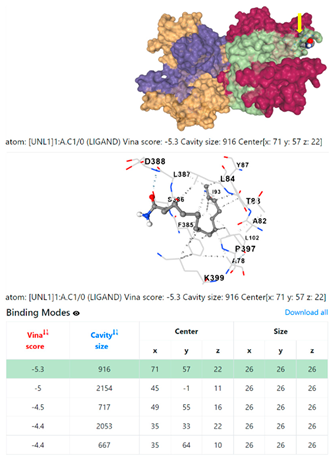 | 4HWM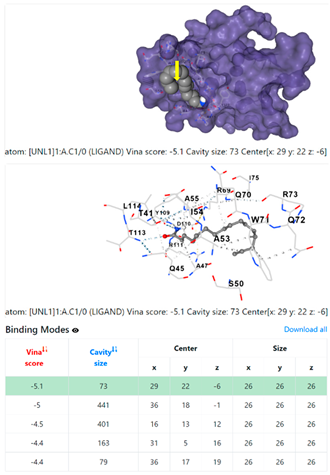 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R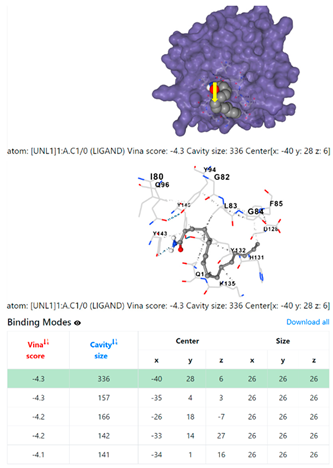 | 3OS3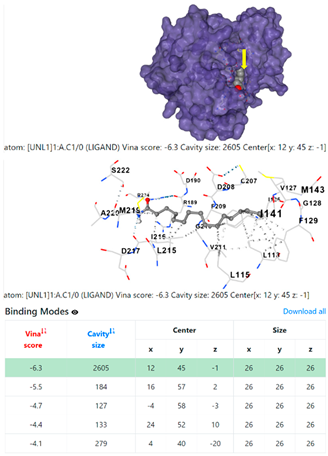 | ||
| Staphylococcus aureus | |||
1DUA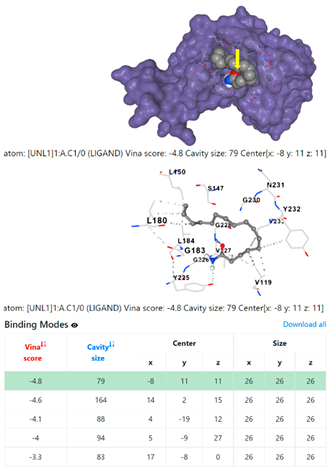 | 1DUE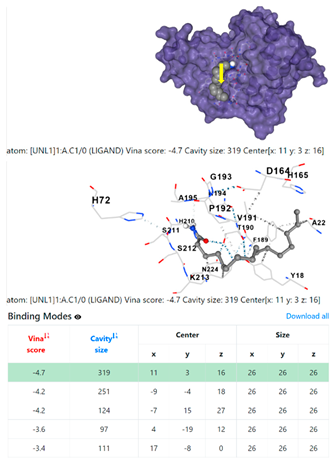 | ||
8EXP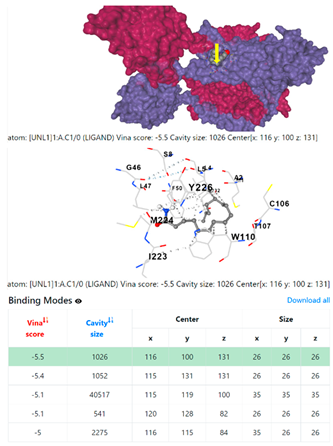 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 5. | CID 10541 Phosphoric acid, trimethyl ester | Klebsiella pneumoniae | |
5HFT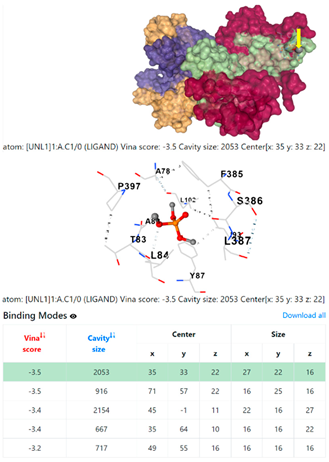 | 4HWM | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R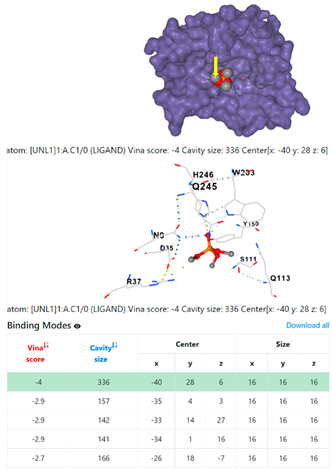 | 3OS3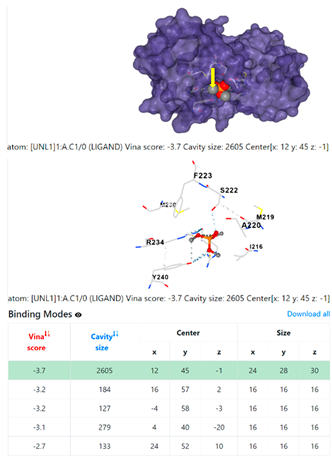 | ||
| Staphylococcus aureus | |||
1DUA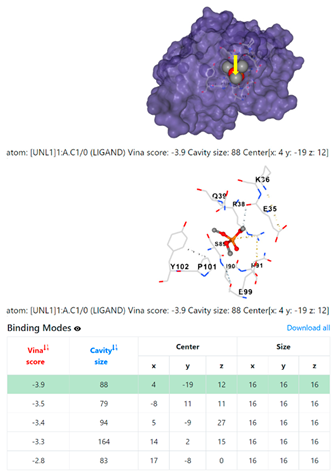 | 1DUE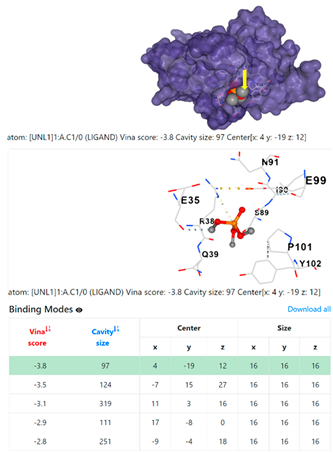 | ||
8EXP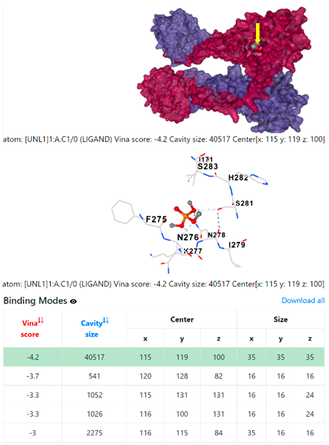 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 6. | CID 75084 Dodecyl acrylate | Klebsiella pneumoniae | |
5HFT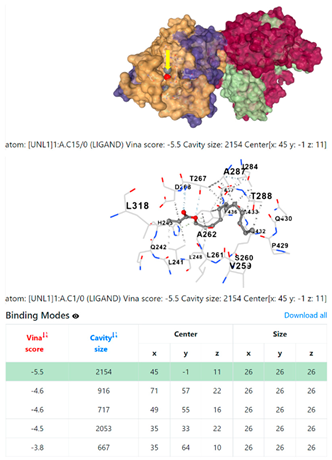 | 4HWM | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R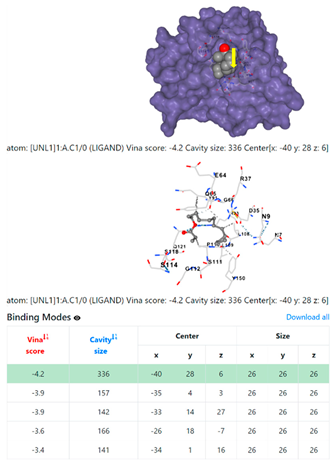 | 3OS3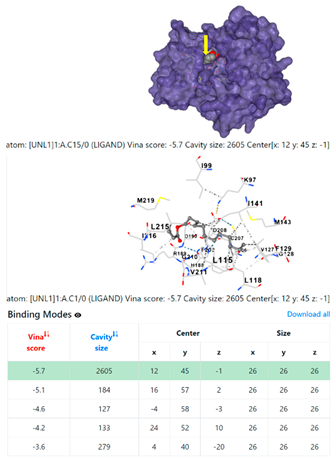 | ||
| Staphylococcus aureus | |||
1DUA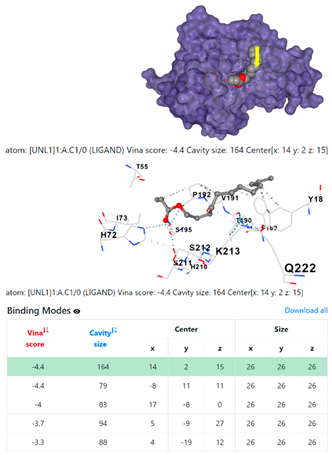 | 1DUE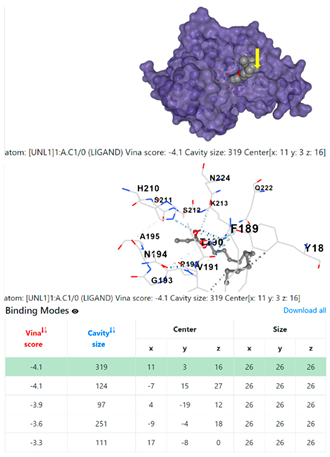 | ||
8EXP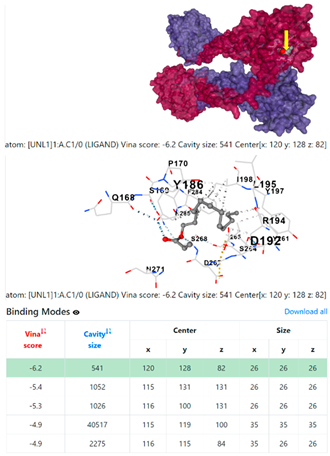 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 7. | CID 605777 Pentanoic acid, 5-hydroxy-, 2,4-di-t-butylphenyl esters | Klebsiella pneumoniae | |
5HFT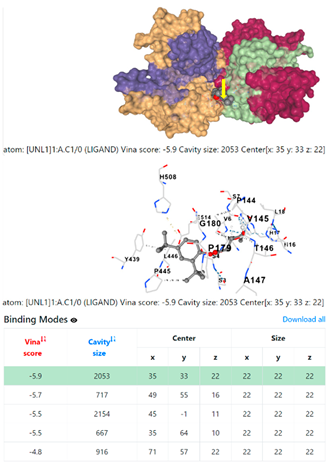 | 4HWM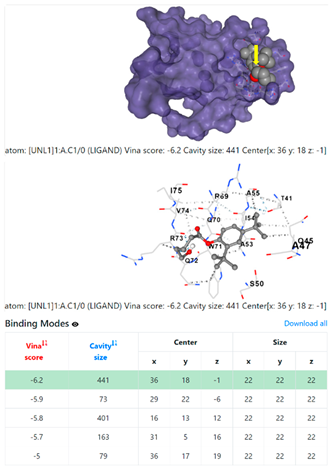 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R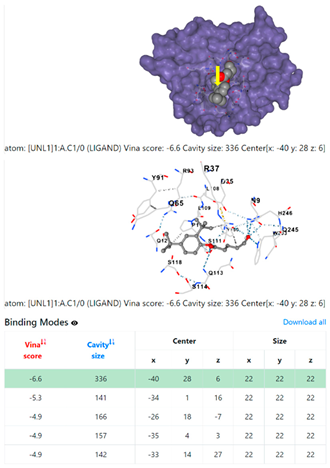 | 3OS3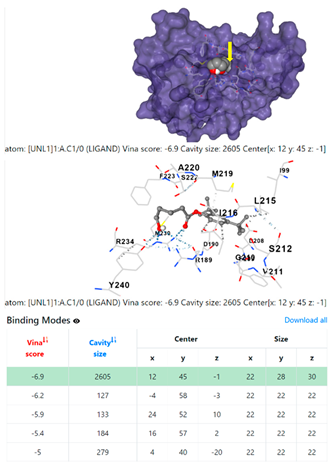 | ||
| Staphylococcus aureus | |||
1DUA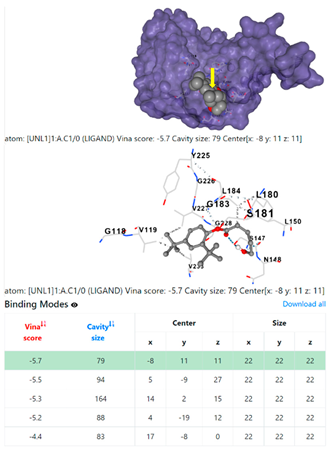 | 1DUE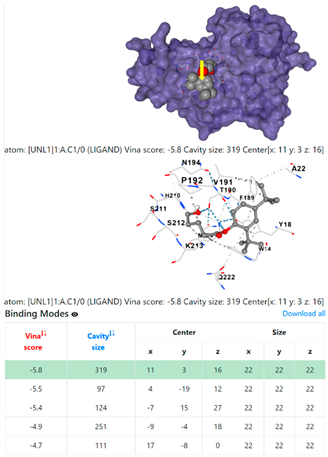 | ||
8EXP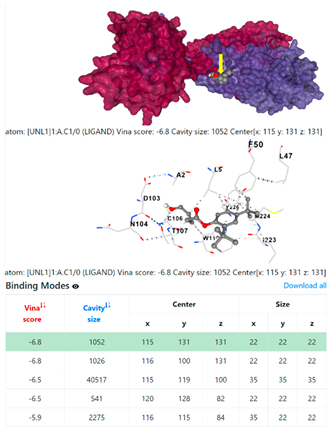 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 8. | CID 8201 Methyl stearate | Klebsiella pneumoniae | |
5HFT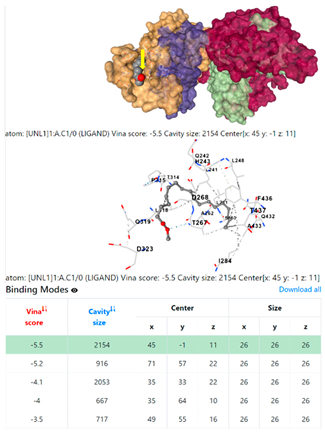 | 4HWM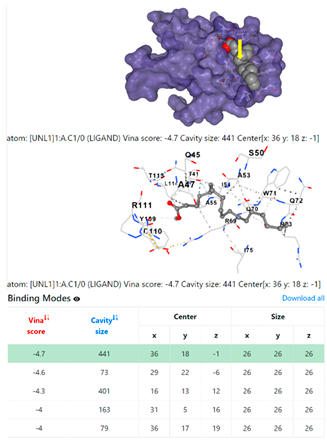 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R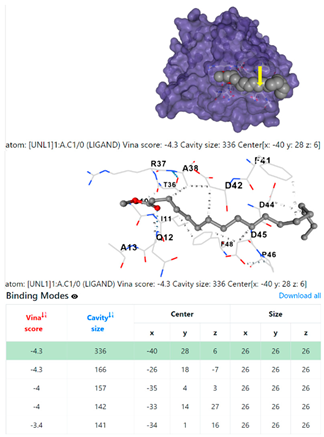 | 3OS3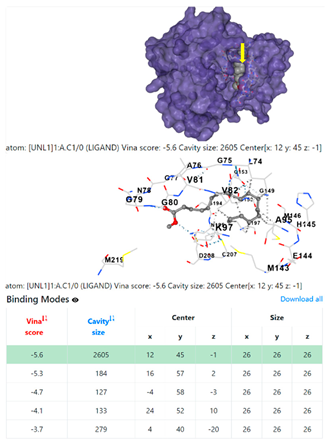 | ||
| Staphylococcus aureus | |||
1DUA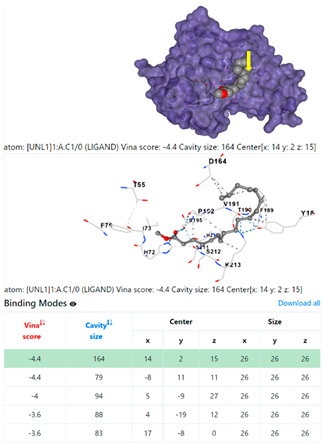 | 1DUE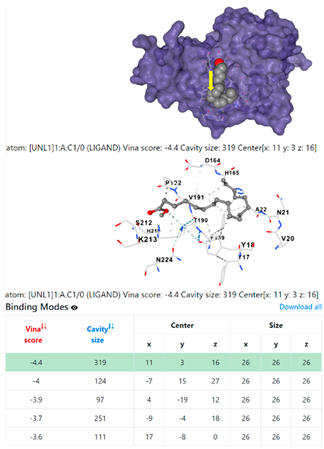 | ||
8EXP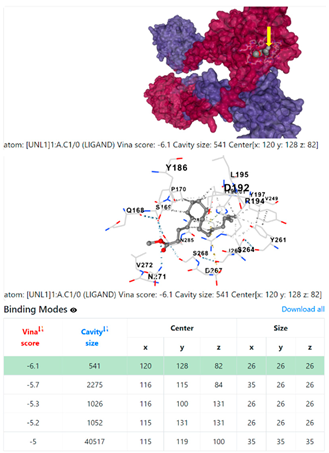 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 9. | CID 69825 1-Pentadecyne | Klebsiella pneumoniae | |
5HFT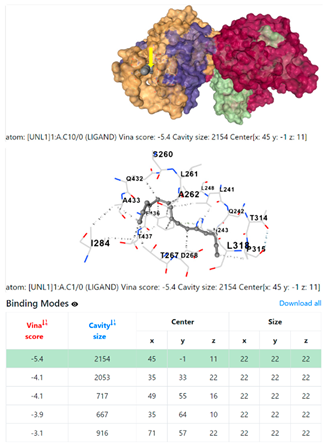 | 4HWM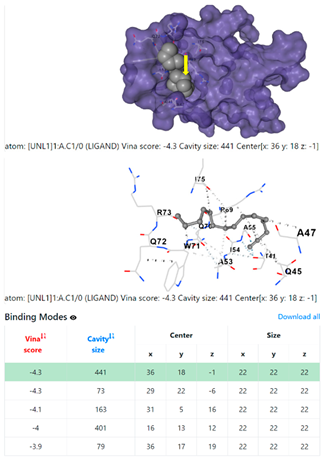 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R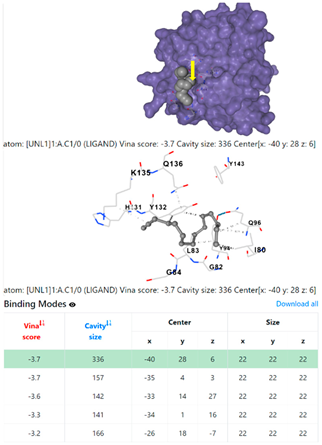 | 3OS3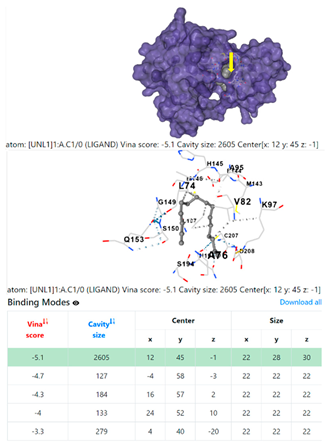 | ||
| Staphylococcus aureus | |||
1DUA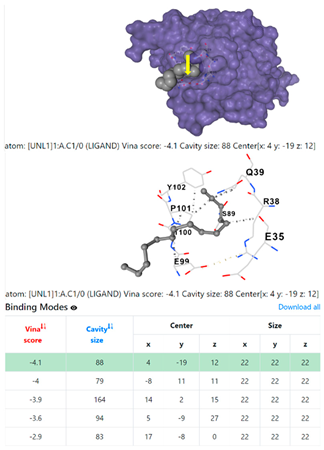 | 1DUE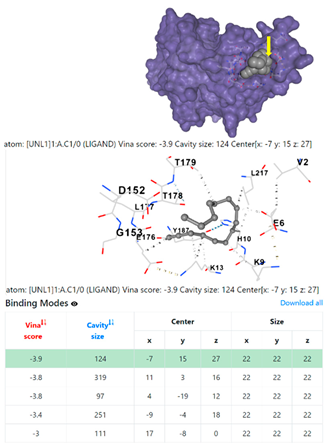 | ||
8EXP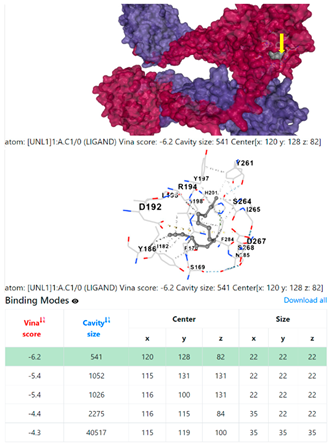 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 10. | CID 549821 11,13-Dimethyl-12-tetradecen-1-ol acetate | Klebsiella pneumoniae | |
5HFT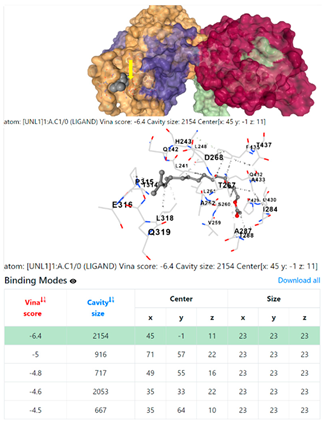 | 4HWM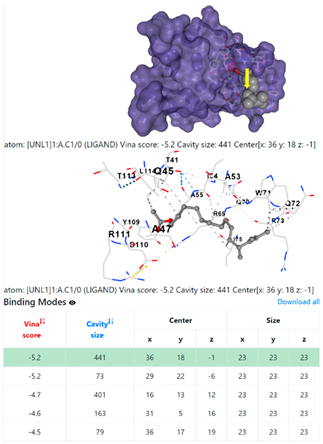 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R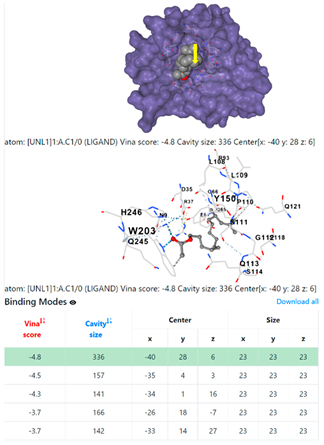 | 3OS3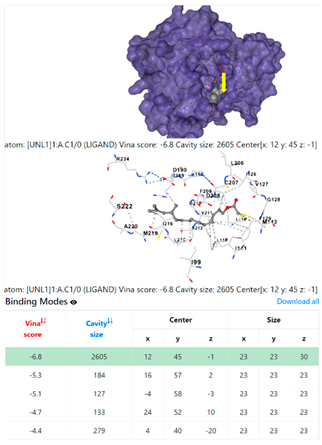 | ||
| Staphylococcus aureus | |||
1DUA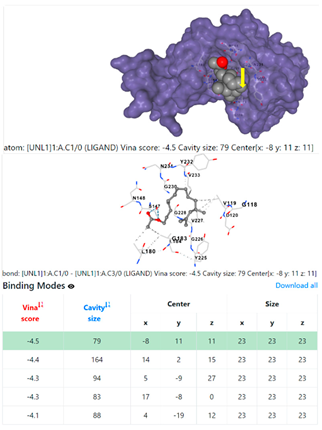 | 1DUE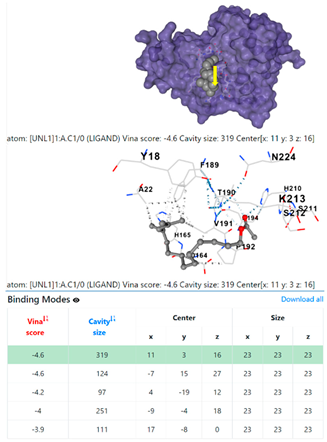 | ||
8EXP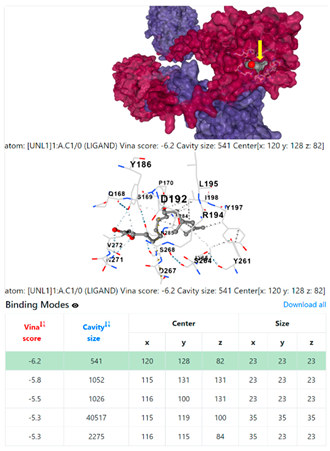 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 11. | CID 91715040 1,3-Benzenediol, o-(4-methylbenzoyl)-o′-(2-methoxybenzoyl)- | Klebsiella pneumoniae | |
5HFT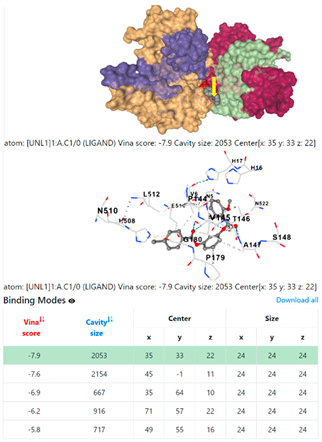 | 4HWM | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R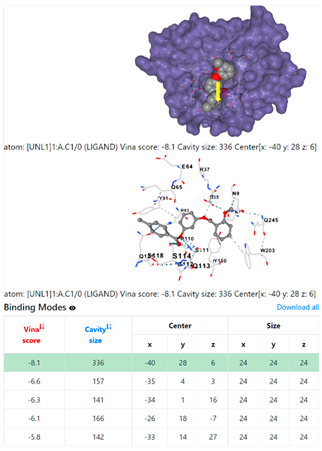 | 3OS3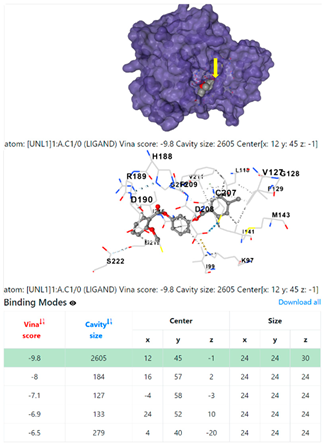 | ||
| Staphylococcus aureus | |||
1DUA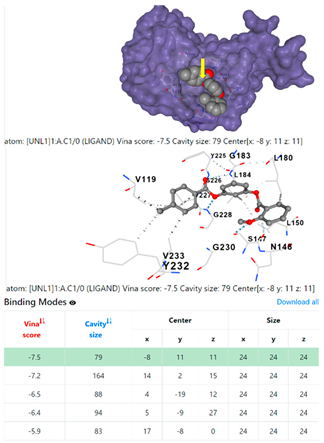 | 1DUE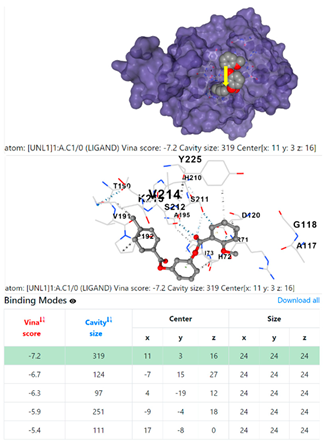 | ||
8EXP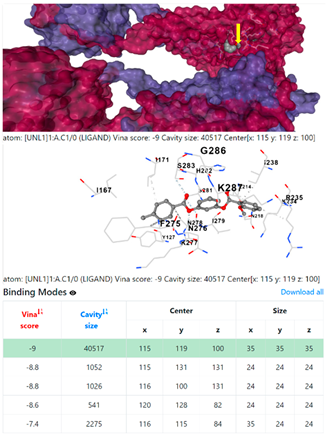 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 12. | CID 104386 Isooctyl 3-mercaptopropionate | Klebsiella pneumoniae | |
5HFT | 4HWM | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R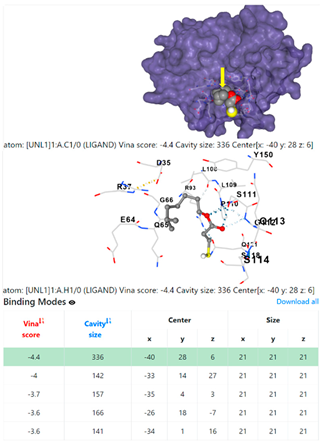 | 3OS3 | ||
| Staphylococcus aureus | |||
1DUA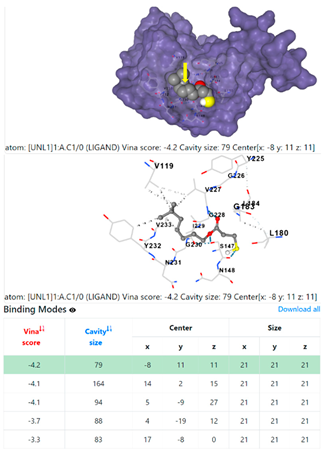 | 1DUE | ||
8EXP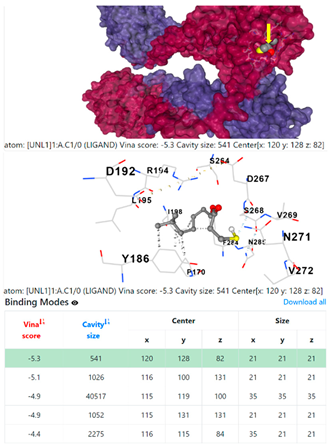 | |||
| Ligand | Target Protein Receptor (PDB) | ||
| 13. | CID 54671203 Standard antibiotic (doxycycline) | Klebsiella pneumoniae | |
5HFT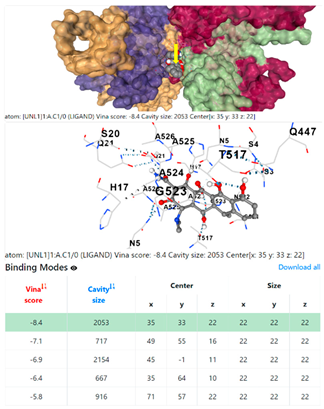 | 4HWM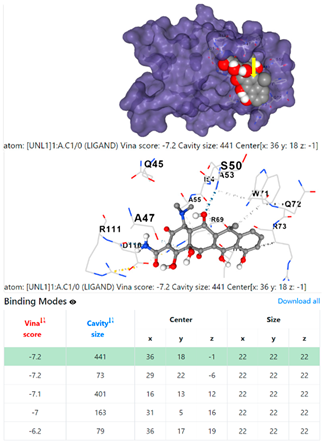 | ||
| Pseudomonas aeruginosa | 3OS prokaryotic ribosoma (binding unit of antibiotic) | ||
4F1R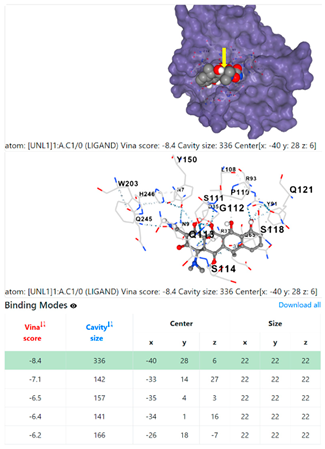 | 3OS3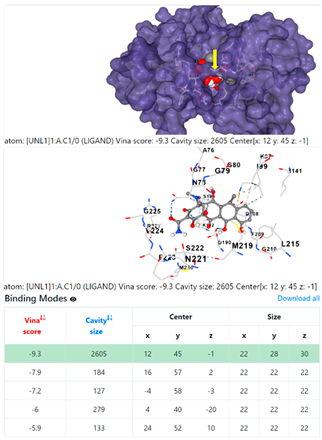 | ||
| Staphylococcus aureus | |||
1DUA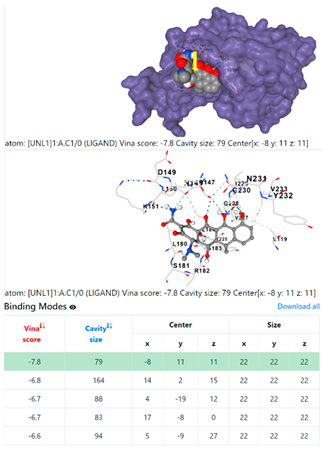 | 1DUE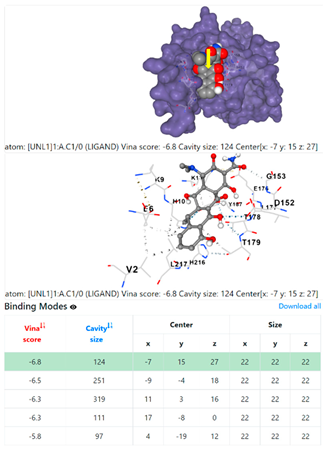 | ||
8EXP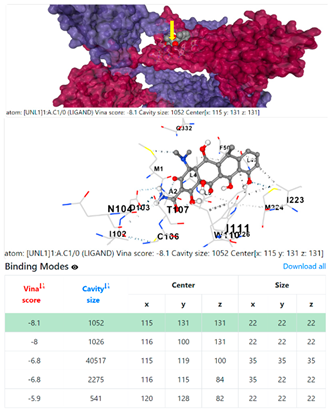 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felemban, S.; Hamouda, A.F. Investigating Grape Seed Extract as a Natural Antibacterial Agent for Water Disinfection in Saudi Arabia: A Pilot Chemical, Phytochemical, Heavy-Metal, Mineral, and CB-Dock Study Employing Water and Urine Samples. Chemistry 2024, 6, 852-898. https://doi.org/10.3390/chemistry6050051
Felemban S, Hamouda AF. Investigating Grape Seed Extract as a Natural Antibacterial Agent for Water Disinfection in Saudi Arabia: A Pilot Chemical, Phytochemical, Heavy-Metal, Mineral, and CB-Dock Study Employing Water and Urine Samples. Chemistry. 2024; 6(5):852-898. https://doi.org/10.3390/chemistry6050051
Chicago/Turabian StyleFelemban, Shifa, and Asmaa Fathi Hamouda. 2024. "Investigating Grape Seed Extract as a Natural Antibacterial Agent for Water Disinfection in Saudi Arabia: A Pilot Chemical, Phytochemical, Heavy-Metal, Mineral, and CB-Dock Study Employing Water and Urine Samples" Chemistry 6, no. 5: 852-898. https://doi.org/10.3390/chemistry6050051
APA StyleFelemban, S., & Hamouda, A. F. (2024). Investigating Grape Seed Extract as a Natural Antibacterial Agent for Water Disinfection in Saudi Arabia: A Pilot Chemical, Phytochemical, Heavy-Metal, Mineral, and CB-Dock Study Employing Water and Urine Samples. Chemistry, 6(5), 852-898. https://doi.org/10.3390/chemistry6050051








