Molecular Phylogenetics of Trapezia Crabs in the Central Mexican Pacific
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sampling and Taxonomic Identification
2.3. DNA Extraction, Amplification and Sequencing
3. Results
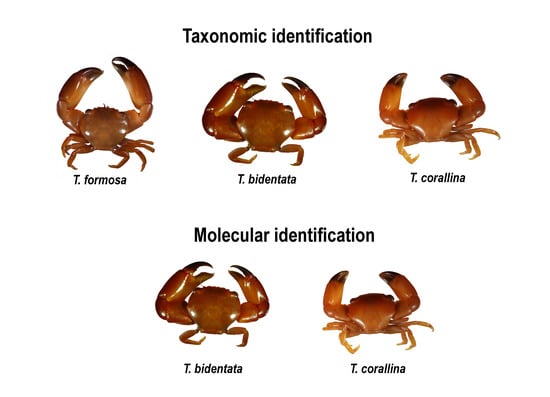
3.1. Morphologic Identification
3.2. Molecular Identification
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Richter, C.; Wunsch, M.; Rasheed, M.; Koetter, I.; Badran, M.I. Endoscopic exploration of Red Sea coral reefs reveals dense populations of cavity dwelling sponges. Nature 2001, 413, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Stella, J.S.; Pratchett, M.; Hutchings, P.; Jones, G. Coral-associated invertebrates: Diversity, ecological importance and vulnerability to disturbance. Oceanogr. Mar. Biol. 2011, 49, 43–104. [Google Scholar]
- Gagnon, J.M.; Beaudin, L.; Silverberg, N.; Mauviel, A. Mesocosm and in situ observations of the burrowing shrimp Calocaris templemani (Decapoda: Thalassinidea) and its bioturbation activities in soft sediments of the Laurentian Trough. Mar. Biol. 2013, 160, 2687–2697. [Google Scholar] [CrossRef]
- Rice, M.M.; Ezzat, L.; Burkepile, D.E. Corallivory in the Anthropocene: Interactive effects of anthropogenic stressors and corallivory on coral reefs. Front. Mar. Sci. 2018, 5, 525. [Google Scholar] [CrossRef] [Green Version]
- Cortés, J. Marine biodiversity baseline for Área de Conservación Guanacaste, Costa Rica: Published records. ZooKeys 2017, 652, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.N.; Woodhead, P.M. Ecological studies of the coral predator Acanthaster planci in the South Pacific. Mar. Biol. 1970, 6, 12–17. [Google Scholar] [CrossRef]
- Garth, J.S. The Crustacea Decapoda (Brachyura and Anomura) of Eniwetok Atoll, Marshall Islands, with special reference to the obligate commensals of branching corals. Micronesica 1964, 1, 137–144. [Google Scholar]
- Stimson, J. Stimulation of fat-body production in the polyps of the coral Pocillopora damicornis by the presence of mutualistic crabs of the genus Trapezia. Mar. Biol. 1990, 106, 211–218. [Google Scholar] [CrossRef]
- Glynn, P.W. Fine-scale interspecific interactions on coral reefs: Functional roles of small and cryptic metazoans. Smithson. Contr. Mar. Sci. 2013, 39, 229–248. [Google Scholar]
- Patton, W.K. Community structure among the animals inhabiting the coral Pocillopora damicornis at Heron Island Australia. In Symbiosis in the Sea, 1st ed.; Vernberg, W., Ed.; Univ. South Carolina Press: Columbia, SC, USA, 1974; pp. 219–243. [Google Scholar]
- Garth, J.S. Decapod crustaceans inhabiting reef-building corals of Ceylon and the Maldive Islands. J. Mar. Biol. Assoc. India. 1974, 15, 195–212. [Google Scholar]
- Nyenzi, B.; Lefale, P.F. El Nino southern oscillation (ENSO) and global warming. Adv. Geosci. 2006, 6, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Glynn, P.W.; Alvarado, J.J.; Banks, S.; Cortés, J.; Feingold, J.S.; Jiménez, C.; Maragos, J.E.; Martínez, P.; Maté, J.L.; Moanga, D.A.; et al. Eastern Pacific Coral Reef Provinces, Coral Community Structure and Composition: An Overview. In Coral Reefs of the Eastern Tropical Pacific, 1st ed.; Glynn, P., Manzello, D., Enochs, I., Eds.; Springer: Dordrecht, The Netherlands, 2017; Volume 8, pp. 107–176. [Google Scholar] [CrossRef] [Green Version]
- Richmond, R.H. Energetics, competency, and long-distance dispersal of planula larvae of the coral Pocillopora damicornis. Mar. Biol. 1987, 93, 527–533. [Google Scholar] [CrossRef]
- Toth, L.T.; Aronson, R.B.; Vollmer, S.V.; Hobbs, J.W.; Urrego, D.H.; Cheng, H.; Enochs, I.C.; Combosch, D.J.; van Woesik, R.; Macintyre, I.G. ENSO drove 2500-year collapse of eastern Pacific coral reefs. Science 2012, 337, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, P. Eastern Pacific species of Trapezia (Crustacea, Brachyura: Trapeziidae), sibling species symbiotic with reef corals. Bull. Mar. Sci. 1996, 58, 531–554. [Google Scholar]
- Reyes-Bonilla, H.; Calderón Aguilera, L.E.; Cruz-Piñón, G.; Medina-Rosas, P.; López-Perez, R.A.; Herrero-Perezrul, M.D.; Leyte-Morales, G.E.; Cupul-Magaña, A.L.; Carriquiry-Beltran, J.D. Atlas de Corales Pétreos (Anthozoa: Scleractinia) del Pacífico Mexicano, 1st ed.; Sociedad Mexicana de Arrecifes Coralinos AC, CICESE, CONABIO, CONACYT, DBM/UABCS, CUC/UdeG, Umar: Mexico City, México, 2005; p. 129. [Google Scholar]
- CONANP (Comisión Nacional de Áreas Naturales Protegidas). Programa de Conservación Y Manejo; Parque Nacional Islas Marietas: Mexico City, México, 2007. [Google Scholar]
- Wyrtki, K. Surface currents of the eastern tropical Pacific Ocean. Bull. Inter. Am. Trop. Tuna Commn. 1965, 9, 268–305. [Google Scholar]
- Roden, G.I.; Groves, G.W. Recent oceanographic investigations in the Gulf of California. J. Mar. Res. 1959, 18, 10–35. [Google Scholar]
- Griffiths, R.C. Physical, chemical and biological oceanography at the entrance to the Gulf of California, spring of 1960. US Fish. Wild. Serv. Spec. Sci. Rep. Fish. 1968, 573, 1–47. [Google Scholar]
- Fiedler, P.C. Seasonal climatologies and variability of eastern tropical Pacific surface waters. NOAA Tech. Rep. 1992, 109, 1–65. [Google Scholar]
- Wang, C.; Fiedler, P.C. ENSO variability and the eastern tropical Pacific: A review. Prog. Oceanogr. 2006, 69, 239–266. [Google Scholar] [CrossRef]
- Castro, P.; Ng, P.K.; Ahyong, S.T. Phylogeny and systematics of the Trapeziidae Miers, 1886 (Crustacea: Brachyura), with the description of a new family. Zootaxa 2004, 643, 1–70. [Google Scholar] [CrossRef] [Green Version]
- Lai, J.C.; Ahyong, S.T.; Jeng, M.S.; Ng, P.K. Are coral-dwelling crabs monophyletic? A phylogeny of the Trapezioidea (Crustacea: Decapoda: Brachyura). Invertebr. Syst. 2009, 23, 402–408. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Moritz, C. Defining ‘evolutionarily significant units’ for conservation. Trends. Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- da Silva, J.M.; Creer, S.; Dos Santos, A.; Costa, A.C.; Cunha, M.R.; Costa, F.O.; Carvalho, G.R. Systematic and evolutionary insights derived from mtDNA COI barcode diversity in the Decapoda (Crustacea: Malacostraca). PLoS ONE 2011, 6, e19449. [Google Scholar] [CrossRef] [Green Version]
- Fransen, C.H.J.M.; De Grave, S. Evolution and Radiation of Shrimp-Like Decapods: An Overview. In Decapod Crustacean Phylogenetics (Crustacean Issues Book 18), 1st ed.; Martin, J.W., Crandall, K.A., Felder, D.L., Eds.; Taylor & Francis/CRC Press: Boca Raton, FL, USA, 2016; pp. 245–259. [Google Scholar]
- Moriyama, E.N.; Powell, J.R. Synonymous substitution rates in Drosophila: Mitochondrial versus nuclear genes. J. Mol. Evol. 1997, 45, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Niemiller, M.L.; Near, T.J.; Fitzpatrick, B.M. Delimiting species using multilocus data: Diagnosing cryptic diversity in the Southern cavefish, Typhlichthys subterraneus (Teleostei: Amblyopsidae). Evolution 2012, 66, 846–866. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, J.W. Behavioral homology and phylogeny. Annu. Rev. Ecol. Evol. Syst. 1992, 23, 361–381. [Google Scholar] [CrossRef]
- Miller, J.S.; Wenzel, J.W. Ecological characters and phylogeny. Annu. Rev. Entomol. 1995, 40, 389–415. [Google Scholar] [CrossRef]
- Castro, P. Notes on symbiotic decapod crustaceans from Gorgona Island, Colombia, with a revision of the eastern Pacific species of Trapezia (Brachyura, Xanthidae), symbionts of scleractinian corals. An. Inst. Inv. Mar. Punta Betín 1982, 12, 9–17. [Google Scholar] [CrossRef]
- Castro, P. Shallow-water Trapeziidae and Tetraliidae (Crustacea: Brachyura) of the Philippines (Panglao 2004 Expedition), New Guinea, and Vanuatu (Santo 2006 Expedition). Raffles Bull. Zool. 2009, 20, 271–281. [Google Scholar]
- Castro, P. Systematic status and geographic distribution of Trapezia formosa Smith, 1869 (Crustacea, Brachyura, Trapeziidae), a symbiont of reef corals. Zoosystema 1998, 2, 177–181. [Google Scholar]
- Cabezas, P.; Macpherson, E.; Machordom, A. Taxonomic revision of the genus Paramunida Baba, 1988 (Crustacea: Decapoda: Galatheidae): A morphological and molecular approach. Zootaxa 2010, 1, 1–60. [Google Scholar] [CrossRef] [Green Version]
- Glynn, P.W. Some physical and biological determinants of coral community structure in the eastern Pacific. Ecol. Monogr. 1976, 4, 431–456. [Google Scholar] [CrossRef]
- Jiménez, C.; Cortés, J. Growth of seven species of scleractinian corals in an upwelling environment of the eastern Pacific (Golfo de Papagayo, Costa Rica). Bull. Mar. Sci. 2003, 1, 187–198. [Google Scholar]
- Reyes-Bonilla, H. Coral reefs of the Pacific coast of Mexico. In Latin American Coral Reefs, 1st ed.; Cortés, J., Ed.; Elsevier Science: Amsterdam, The Netherlands; London, UK, 2003; pp. 331–349. [Google Scholar] [CrossRef]
- Rodríguez-Troncoso, A.P.; Carpizo-Ituarte, E.; Cupul-Magaña, A.L. Physiological response to high temperature in the Tropical Eastern Pacific coral Pocillopora verrucosa. Mar. Ecol. 2016, 37, 1168–1176. [Google Scholar] [CrossRef]
- Martínez-Castillo, V.; Rodríguez-Troncoso, A.P.; Santiago-Valentín, J.D.; Cupul-Magaña, A.L. The influence of urban pressures on coral physiology on marginal coral reefs of the Mexican Pacific. Coral Reef. 2020, 39, 625–637. [Google Scholar] [CrossRef]
- Knudsen, J.W. Trapezia and Tetralia (Decapoda, Brachyura, Xanthidae) as obligate ectoparasites of pocilloporid and acroporid corals. Pac. Sci. 1967, 21, 51–57. [Google Scholar]




| Species | Carapace Width | Carapace Length | Cheliped Width | Cheliped Length | ||||
|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | Males | Females | Males | Females | |
| Trapezia formosa | 6.10 ± 0.99 | 5.90 ± 1.31 | 5.06 ± 0.64 | 5.19 ± 0.78 | 2.54 ± 0.50 | 2.85 ± 0.87 | 6.41 ± 1.27 | 6.09 ± 0.52 |
| Trapezia bidentata | 6.95 ± 0.78 | 8.93 ± 2.43 | 5.86 ± 1.76 | 6.32 ± 1.91 | 3.26 ± 0.19 | 3.15 ± 1.52 | 7.69 ± 1.83 | 6.156 ± 0.921 |
| Trapezia corallina | 9.03 ± 1.56 | N/A | 8.17 ± 3.01 | N/A | 4.19 ± 0.52 | N/A | 9.78 ± 2.33 | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canizales-Flores, H.M.; Rodríguez-Troncoso, A.P.; Bautista-Guerrero, E.; Cupul-Magaña, A.L. Molecular Phylogenetics of Trapezia Crabs in the Central Mexican Pacific. Oceans 2020, 1, 156-164. https://doi.org/10.3390/oceans1030011
Canizales-Flores HM, Rodríguez-Troncoso AP, Bautista-Guerrero E, Cupul-Magaña AL. Molecular Phylogenetics of Trapezia Crabs in the Central Mexican Pacific. Oceans. 2020; 1(3):156-164. https://doi.org/10.3390/oceans1030011
Chicago/Turabian StyleCanizales-Flores, Hazel M., Alma P. Rodríguez-Troncoso, Eric Bautista-Guerrero, and Amílcar L. Cupul-Magaña. 2020. "Molecular Phylogenetics of Trapezia Crabs in the Central Mexican Pacific" Oceans 1, no. 3: 156-164. https://doi.org/10.3390/oceans1030011
APA StyleCanizales-Flores, H. M., Rodríguez-Troncoso, A. P., Bautista-Guerrero, E., & Cupul-Magaña, A. L. (2020). Molecular Phylogenetics of Trapezia Crabs in the Central Mexican Pacific. Oceans, 1(3), 156-164. https://doi.org/10.3390/oceans1030011