Environmentally-Driven Variation in the Physiology of a New Caledonian Reef Coral
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of Research Expedition and Analytical Goals
2.2. Night Coral Sampling
2.3. Response Variables (RV)
2.4. Multivariate Data Analysis
2.5. Univariate Data Analysis
3. Results
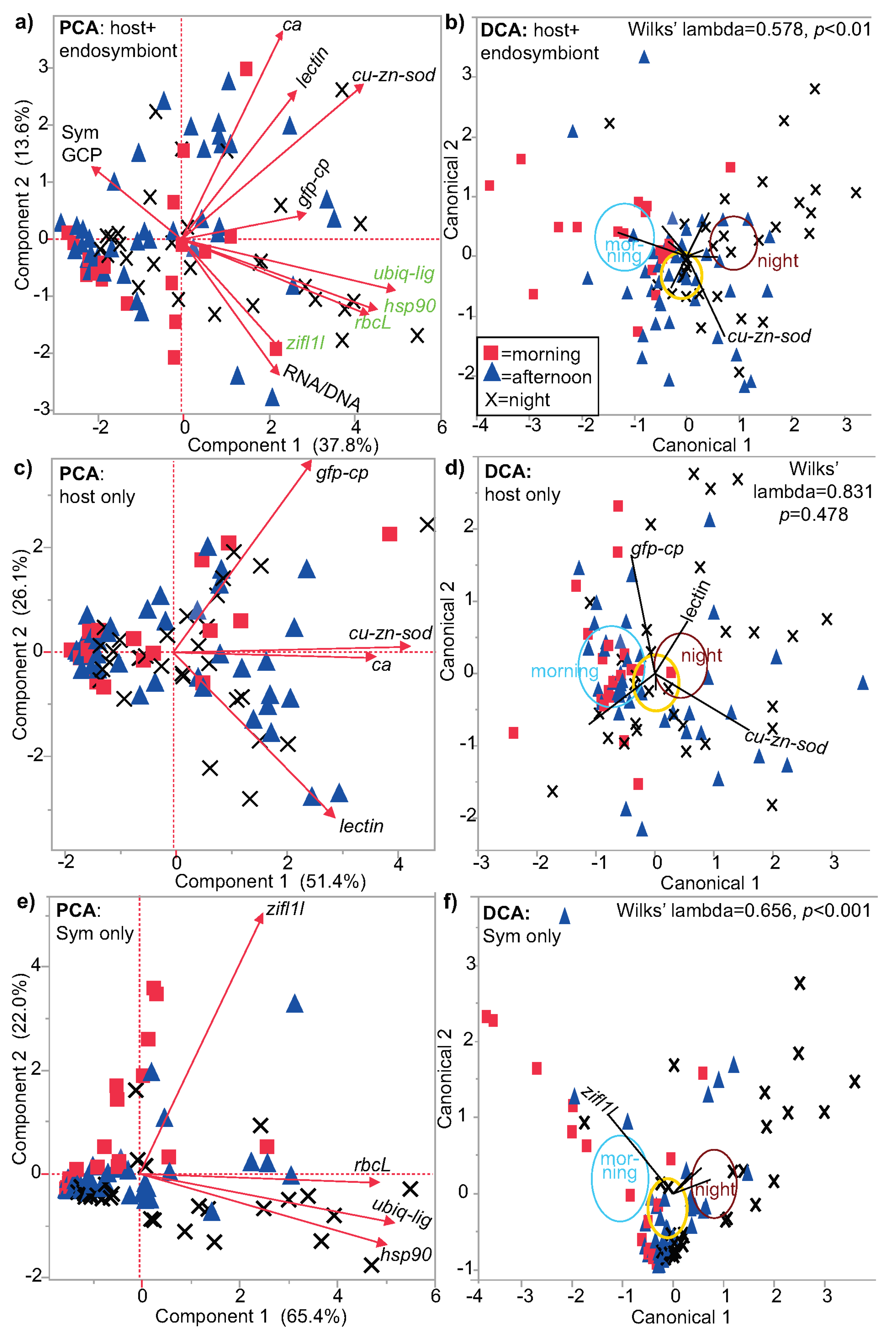
3.1. Multivariate Analysis
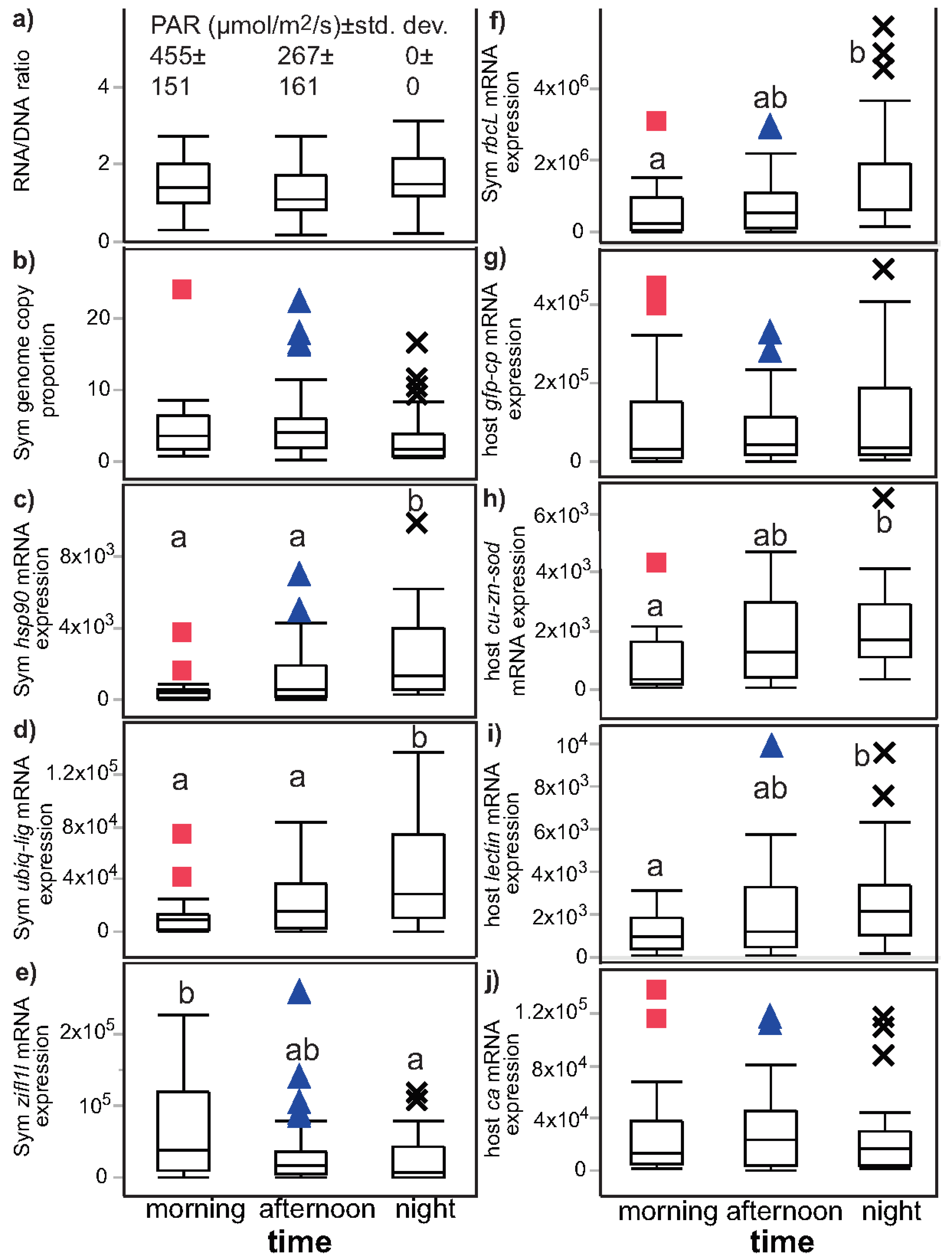
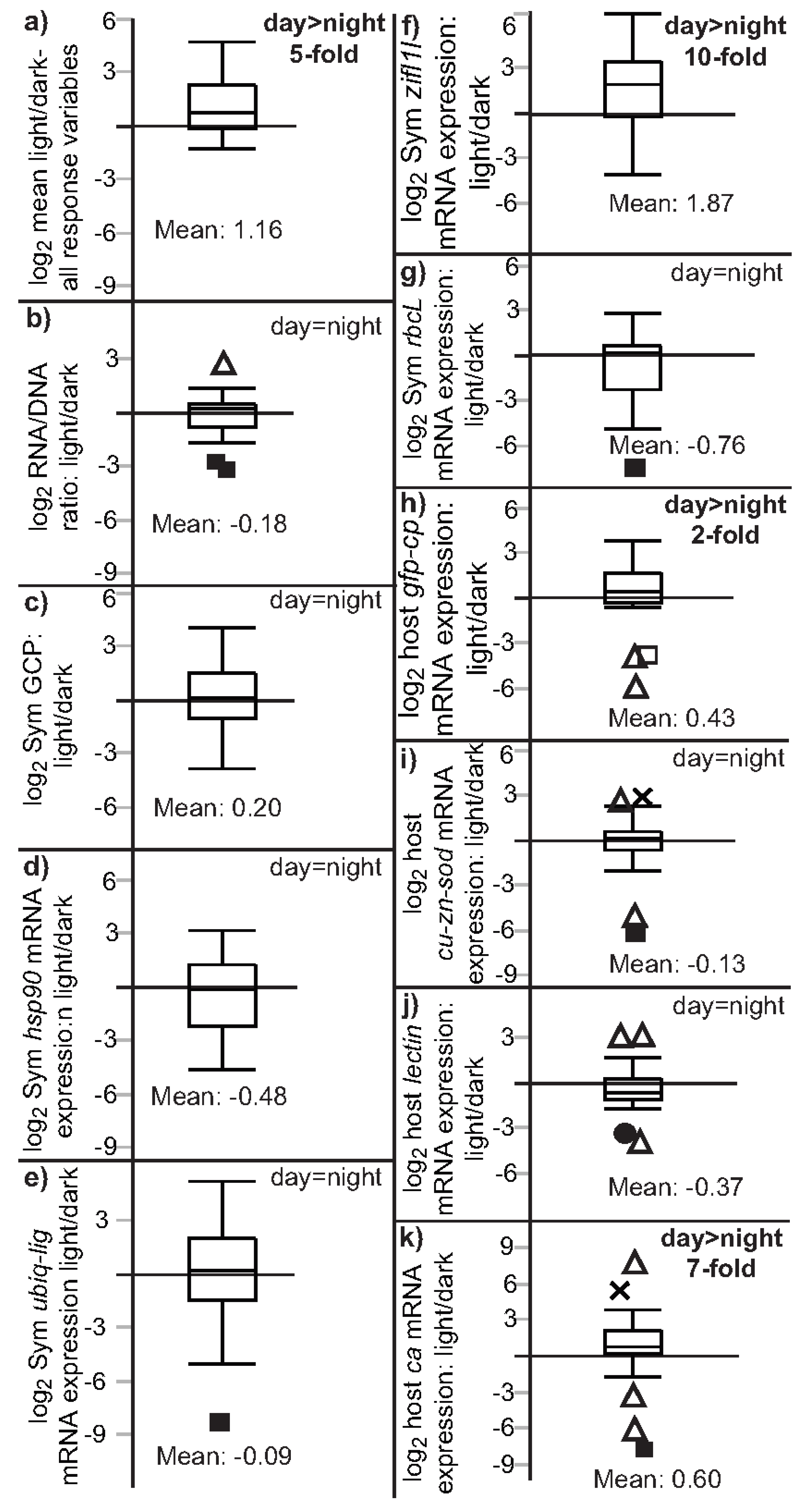
3.2. Univariate Effects of Time and Light on Coral Response Variables
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoegh-Guldberg, O.; Poloczanska, E.S.; Skirving, W.; Dove, S. Coral reef ecosystems under climate change and ocean acidification. Front. Mar. Sci. 2017, 4, 158. [Google Scholar] [CrossRef] [Green Version]
- Hughes, T.P.; Barnes, M.L.; Bellwood, D.R.; Cinner, J.E.; Cumming, G.S.; Jackson, J.B.C.; Kleypas, J.; van de Leemput, I.A.; Lough, J.M.; Morrison, T.H.; et al. Coral reefs in the anthropocene. Nature 2017, 546, 82–90. [Google Scholar] [CrossRef]
- Sheppard, C.; Sheppard, A.; Mogg, A.; Bayley, D.; Dempsey, A.C.; Roche, R.; Turner, J.; Purkis, S. Coral bleaching and mortality in the Chagos Archipelago. Atoll Res. Bull. 2017, 613, 1–26. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Chen, M.; Meng, P.J.; Lin, H.J.; Chen, C.S.; Liu, P.J. The physiological response of the reef coral Pocillopora damicornis to elevated temperature: Results from coral reef mesocosm experiments in Southern Taiwan. Mar. Environ. Res. 2013, 86, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cruz-García, R.; Rodríguez-Troncoso, A.P.; Rodríguez-Zaragoza, F.A.; Cupul-Magaña, A.L.; Mayfield, A.B. Ephemeral effects of El Niño southern oscillation events on an eastern tropical Pacific coral community. Mar. Freshw. Res. 2020, 71, 1259–1268. [Google Scholar] [CrossRef]
- Gintert, B.E.; Manzello, D.P.; Enochs, I.C.; Kolodziej, G.; Carlton, R.D.; Gleason, A.C.R.; Gracias, N. Marked annual coral bleaching resilience of an inshore patch reef in the Florida Keys: A nugget of hope, aberrance, or last man standing? Coral Reefs 2018, 37, 533–547. [Google Scholar] [CrossRef] [Green Version]
- Darling, E.S.; McClanahan, T.R.; Maina, J.; Gurney, G.G.; Graham, N.A.J.; Januchowski-Hartley, F.; Cinner, J.E.; Mora, C.; Hicks, C.C.; Maire, E.; et al. Social–environmental drivers inform strategic management of coral reefs in the Anthropocene. Nat. Ecol. Evol. 2019, 3, 1341–1350. [Google Scholar] [CrossRef]
- IPCC. Impacts, adaptation and vulnerability. In Fourth Assessment Report on the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Mayfield, A.B.; Chen, C.S.; Dempsey, A.C.; Bruckner, A.W. The molecular ecophysiology of closely related pocilloporids from the South Pacific: A case study from the Austral and Cook Islands. Platax 2016, 13, 1–25. [Google Scholar]
- Andrefouet, S.; Cabioch, G.; Flamand, B.; Pelletier, B. A reappraisal of the diversity of geomorphological and genetic processes of New Caledonian coral reefs: A synthesis from optical remote sensing, coring and acoustic multibeam observations. Coral Reefs 2009, 28, 691–707. [Google Scholar] [CrossRef]
- Cunning, R.; Bay, R.A.; Gillette, P.; Baker, A.; Traylor-Knowles, N. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci. Rep. 2018, 8, 16134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayfield, A.B.; Chen, C.S.; Dempsey, A.C. The molecular ecophysiology of closely related pocilloporid corals of New Caledonia. Platax 2017, 14, 1–45. [Google Scholar]
- Barshis, D.J.; Stillman, J.H.; Gates, R.D.; Toonen, R.J.; Smith, L.W.; Birkeland, C. Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: Does host genotype limit phenotypic plasticity? Mol. Ecol. 2010, 19, 1705–1720. [Google Scholar] [CrossRef]
- Putnam, H.M.; Edmunds, P.J.; Fan, T.Y. Effect of a fluctuating thermal regime on adult and larval reef corals. Invertebr. Biol. 2010, 129, 199–209. [Google Scholar] [CrossRef]
- Edmunds, P.J.; Cumbo, V.; Fan, T.Y. Effects of temperature on the respiration of brooded larvae from tropical reef corals. J. Exp. Biol. 2011, 214, 2783–2790. [Google Scholar] [CrossRef] [Green Version]
- Schmidt-Roach, S.; Miller, K.J.; Lundgren, P.; Andreakis, N. With eyes wide open: A revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zool. J. Linnaean Soc. 2014, 170, 1–33. [Google Scholar] [CrossRef]
- Putnam, H.M.; Barott, K.; Ainsworth, T.D.; Gates, R.D. The vulnerability and resilience of reef-building corals. Curr. Biol. 2017, 27, R528–R540. [Google Scholar] [CrossRef] [Green Version]
- van Hooidonk, R.; Maynard, J.; Tamelander, J.; Gove, J.; Ahmadia, G.; Raymundo, L.; Williams, G.; Heron, S.F.; Planes, S. Local-scale projections of coral reef futures and implications of the Paris Agreement. Sci. Rep. 2016, 6, 39666. [Google Scholar] [CrossRef]
- UNEP. Projections of Future Coral Bleaching Conditions Using IPCC CMIP6 Models: Climate Policy Implications, Management Applications, and Regional Seas Summaries; United Nations Environment Programme: Nairobi, Kenya, 2020.
- Saul, S.; Purkis, S. Semi-automated object-based classification of coral reef habitat using discrete choice models. Remote Sens. 2015, 7, 15894–15916. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.B.; Hsiao, Y.Y.; Fan, T.Y.; Chen, C.S.; Gates, R.D. Evaluating the temporal stability of stress-activated protein kinase and cytoskeleton gene expression in the Pacific corals Pocillopora damicornis and Seriatopora hystrix. J. Exp. Mar. Biol. Ecol. 2010, 395, 215–222. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Hirst, M.B.; Gates, R.D. Gene expression normalization in a dual compartment system: A real-time PCR protocol for symbiotic anthozoans. Mol. Ecol. Res. 2009, 9, 462–470. [Google Scholar] [CrossRef]
- Putnam, H.M.; Mayfield, A.B.; Fan, T.Y.; Chen, C.S.; Gates, R.D. The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature and pCO2. Mar. Biol. 2013, 160, 2157–2173. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Fan, T.Y.; Chen, C.S. Real-time PCR-based gene expression analysis in the model reef-building coral Pocillopora damicornis: Insight from a salinity stress study. Platax 2013, 10, 1–29. [Google Scholar]
- Mayfield, A.B.; Chen, C.S.; Liu, P.J. Decreased green fluorescent protein-like chromoprotein gene expression in specimens of the reef-building coral Pocillopora damicornis undergoing high temperature-induced bleaching. Platax 2014, 11, 1–23. [Google Scholar]
- Mayfield, A.B.; Wang, Y.B.; Chen, C.S.; Chen, S.H.; Lin, C.Y. Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures. Mol. Ecol. 2014, 23, 5816–5830. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.N.U.; Kang, H.J.; Weis, V.M.; Mayfield, A.B.; Fang, L.S.; Chen, C.S. Diel rhythmicity of lipid body formation in a coral-Symbiodinium endosymbiosis. Coral Reefs 2012, 31, 521–534. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Hsiao, Y.Y.; Fan, T.Y.; Chen, C.S. Temporal variation in RNA/DNA and protein/DNA ratios in four anthozoan-dinoflagellate endosymbioses of the Indo-Pacific: Implications for molecular diagnostics. Platax 2012, 9, 1–24. [Google Scholar]
- Wall, C.B.; Ritson-Williams, R.; Popp, B.N.; Gates, R.D. Spatial variation in the biochemical and isotopic composition of corals during bleaching and recovery. Limnol. Oceanogr. 2019, 64, 2011–2028. [Google Scholar] [CrossRef]
- Hoadley, K.D.; Szmant, A.M.; Pyott, S.J. Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PLoS ONE 2011, 6, e19755. [Google Scholar] [CrossRef] [Green Version]
- Levy, O.; Kaniewska, P.; Alon, S.; Eisenberg, E.; Karako-Lampert, S.; Bay, L.K.; Reef, R.; Miller, D.J.; Hoegh-Guldberg, O. Complex diel cycles of gene expression in coral-algal symbiosis. Science 2011, 331, 175. [Google Scholar] [CrossRef]
- Bertucci, A.; Forêt, S.; Ball, E.E.; Miller, D.J. Transcriptomic differences between day and night in Acropora millepora provide new insights into metabolite exchange and light-enhanced calcification in corals. Mol. Ecol. 2015, 24, 4489–4504. [Google Scholar] [CrossRef]
- Hemond, E.M.; Vollmer, S.V. Diurnal and nocturnal transcriptomic variation in the Caribbean staghorn coral, Acropora cervicornis. Mol. Ecol. 2015, 24, 4460–4473. [Google Scholar] [CrossRef] [PubMed]
- Gates, R.D. Seawater temperature and sublethal coral bleaching in Jamaica. Coral Reefs 1990, 8, 193–197. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Gates, R.D. Osmoregulation in anthozoan-dinoflagellate symbiosis. Comp. Biochem. Physiol. 2007, 147, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.D.; Williams, G.J.; Johnson, M.D.; Radice, V.Z.; Zgliczynski, B.J.; Kelly, E.L.A.; Rohwer, F.L.; Sandin, S.A.; Smith, J.E. Gradients in primary production predict trophic strategies of mixotrophic corals across spatial scales. Curr. Biol. 2018, 28, 3355–3363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Seneca, F.O.; Traylor-Knowles, N.; Palumbi, S.R. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. USA 2013, 110, 1387–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayfield, A.B.; Chen, Y.J.; Lu, C.Y.; Chen, C.S. Proteins responsive to variable temperature exposure in the reef-building coral Seriatopora hystrix. In Coral Reefs: Ecosystems, Environmental Impact and Current Threats; Ortiz, S., Ed.; NOVA Publishing: New York, NY, USA, 2016; pp. 1–72. [Google Scholar]
- Mayfield, A.B.; Wang, Y.B.; Chen, C.S.; Chen, S.H.; Lin, C.Y. Dual-compartmental transcriptomic+proteomic analysis of a marine endosymbiosis exposed to environmental change. Mol. Ecol. 2016, 25, 5944–5958. [Google Scholar] [CrossRef]
- Peng, S.E.; Chen, W.N.U.; Chen, H.K.; Lu, C.Y.; Mayfield, A.B.; Chen, C.S. Lipid bodies in coral-dinoflagellate endosymbiosis: Ultrastructural and proteomic analyses. Proteomics 2011, 17, 3540–3555. [Google Scholar] [CrossRef] [PubMed]
- Levy, O.; Achituv, Y.; Schnider, K.; Dubinsky, Z.; Gorbunov, M. Diurnal hysteresis in coral photosynthesis. Mar. Ecol. Prog. Ser. 2004, 268, 105–117. [Google Scholar] [CrossRef]
- Moya, A.; Tambutté, S.; Lotto, S.; Allemand, D.; Zoccola, D. Carbonic anhydrase in the scleractinian coral Stylophora pistillata: Characterization, localization, and role in biomineralization. J. Biol. Chem. 2008, 283, 25475–25484. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.B.; Hsiao, Y.Y.; Chen, H.K.; Chen, C.S. Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Mar. Biotech. 2014, 16, 371–384. [Google Scholar] [CrossRef]
- Chen, H.K.; Song, S.N.; Wang, L.H.; Mayfield, A.B.; Chen, Y.J.; Chen, W.N.U.; Chen, C.S. A compartmental comparison of major lipid species in a coral-Symbiodinium endosymbiosis: Evidence that the coral host regulates lipogenesis of its cytosolic lipid bodies. PLoS ONE 2015, 10, e0132519. [Google Scholar] [CrossRef] [PubMed]
- Lesser, M.P. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 1997, 16, 187–192. [Google Scholar] [CrossRef]
- Lesser, M.P. Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu. Rev. Physiol. 2006, 68, 253–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrito, T.R.; Teixeira, M.C.; Duarte, A.A.; Duque, P.; Sa Correia, I. Heterologous expression of a Tpo1 homolog from Arabidopsis thaliana confers resistance to the herbicide 2,4-D and other chemical stresses in yeast. Appl. Microbiol. Biotechnol. 2009, 84, 927–936. [Google Scholar] [CrossRef] [PubMed]

| EP (n = 13) | Number and Description of Categories/Bins |
|---|---|
| Region | 2: south vs. north |
| Island/atoll | 6: see Figures 1 and 2 of Mayfield et al. [12] a |
| Reef exposure | 3: protected, intermediate, exposed |
| Reef zone | 4: backreef, forereef, lagoon, channel b |
| Reef type | 4: patch reef, fringing reef, barrier reef, atoll |
| Time | 3: <10:00, 10:00–14:00, >18:00 |
| Light (PAR) | 2: >0 (light) vs. 0 (dark) µmol/photons m2/s |
| Depth | 6: 5–10, 10–15, 15–20, 20–25, 25–30, >30 m |
| Coral cover | 3: 15–30, 30–45%, undetermined |
| Temperature | 5: 23–24, 24–25, 25–26, >26 °C, undetermined |
| Salinity | 4: 34.9, 35.1, 35.2, 35.3 |
| Colony color | 5: normal, pale, very pale, green, purple |
| Symbiodiniaceae assemblage | 2: Cladocopium only vs. Symbiodinium + Cladocopium |
| RV (n = 12) | Function/Notes |
| Maximum colony length (cm) | colony size |
| Planar surface area (cm2) | colony size |
| RNA/DNA ratio | proxy for total gene expression level |
| Symbiodiniaceae genome copy proportion | proxy for endosymbiont density |
| Symbiodiniaceae rbcL mRNA expression | ribulose-1,5-bisphosphate carboxylase/oxygenase (photosynthesis) |
| Symbiodiniaceae ubiq-lig mRNA expression | ubiquitin ligase (stress response) |
| Symbiodiniaceae hsp90 mRNA expression | heat shock protein 90 (stress response) |
| Symbiodiniaceae zifl1l mRNA expression | zinc-induced facilitator-like 1-like (unknown/osmoregulation) |
| Host coral ca mRNA expression | carbonic anhydrase (metabolism) |
| Host coral lectin mRNA expression | cell adhesion |
| Host coral cu-zn-sod mRNA expression | copper-zinc superoxide dismutase (stress response) |
| Host coral gfp-like cp mRNA expression | green fluorescent protein-like chromoprotein (light regulation) |
| Environmental Parameter | PERMANOVA | PLS-% Variation Explained | PLS-% Misclassified | Predictor Score |
|---|---|---|---|---|
| light | p < 0.001 | 7.0 | 26 | 81 |
| time | p < 0.01 | 7.4 | 40 | 67 |
| coral cover | p < 0.01 | 9.0 | 45 | 64 |
| temperature | p < 0.01 | 11.6 | 48 | 64 |
| colony color | p < 0.01 | 7.3 | 51 | 56 |
| region | p = 0.01 | 4.0 | 24 | 80 |
| reef type | p < 0.045 | 9.9 | 40 | 70 |
| reef exposure | p < 0.045 | 4.4 | 36 | 68 |
| island/atoll | p < 0.045 | 9.6 | 52 | 58 |
| depth | p < 0.045 | 7.7 | 51 | 57 |
| salinity | NS | 2.8 | 33 | 70 |
| Sym assemblage | NS | 1.1 | 35 | 66 |
| reef zone | NS | 7.4 | 49 | 58 |
| Effect type | Time (df = 2) | Light (df = 1) | Light minus dark | Light/dark |
| Sample size | 89 | 89 | 26 | 26 |
| Type of test | Wilcoxon test | Wilcoxon test | Matched pairs (signed-rank) | Signed-rank vs. 1 |
| Figure | Figure 2 | Figure 3 | Not plotted | Figure 4 and Figure S1 |
| Response variable | ||||
| RNA/DNA | NS (p = 0.055) | night > day (p = 0.012) a | NS (p = 0.853) | NS (p = 0.824) |
| Sym GCP | NS (p = 0.044) | day > night (p = 0.010) | NS (p = 0.261) | NS (p = 0.108) |
| Sym hsp90 | night > day (p < 0.001) a | night > day (p < 0.001) a | NS (p = 0.683) | NS (p = 0.890) |
| Sym ubiq-lig | night > day (p = 0.002) | night > day (p = 0.001) | NS (p = 0.612) | NS (p = 0.351) |
| Sym zifl1l | day > night (p = 0.019) a | NS (p = 0.222) b | day > night (p = 0.001) | day > night (p < 0.001) |
| Sym rbcL | night > day (p = 0.008) | night > day (p = 0.002) | NS (p = 0.971) | NS (p = 0.931) |
| host ca | NS (p = 0.724) | NS (p = 0.585) | day > night (p = 0.012) | day > night (p = 0.003) |
| host lectin | night > day (p = 0.018) | night > day (p = 0.014) | NS (p = 0.053) | NS (p = 0.831) |
| host cu-zn-sod | night > day (p = 0.006) | night > day (p = 0.006) | NS (p = 0.170) | NS (p = 0.107) |
| host gfp-cp | NS (p = 0.874) | NS (p = 0.863) | day > night (p = 0.007) | day > night (p = 0.009) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayfield, A.B.; Dempsey, A.C. Environmentally-Driven Variation in the Physiology of a New Caledonian Reef Coral. Oceans 2022, 3, 15-29. https://doi.org/10.3390/oceans3010002
Mayfield AB, Dempsey AC. Environmentally-Driven Variation in the Physiology of a New Caledonian Reef Coral. Oceans. 2022; 3(1):15-29. https://doi.org/10.3390/oceans3010002
Chicago/Turabian StyleMayfield, Anderson B., and Alexandra C. Dempsey. 2022. "Environmentally-Driven Variation in the Physiology of a New Caledonian Reef Coral" Oceans 3, no. 1: 15-29. https://doi.org/10.3390/oceans3010002
APA StyleMayfield, A. B., & Dempsey, A. C. (2022). Environmentally-Driven Variation in the Physiology of a New Caledonian Reef Coral. Oceans, 3(1), 15-29. https://doi.org/10.3390/oceans3010002







