Signal “Off-On” Biosensor Based on Fluorescence Resonance Energy Transfer (FRET) for Detection of Sorghum Mosaic Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Apparatus
2.2. Synthesis of CdTe QD
2.3. Synthesis of AuNP
2.4. Conjugation of SrMV Ab and CdTe QD
2.5. Conjugation of SrMV CP and AuNP
2.6. Determination of Quenching Efficiency
2.7. Quantitative Detection of Purified CP
3. Results
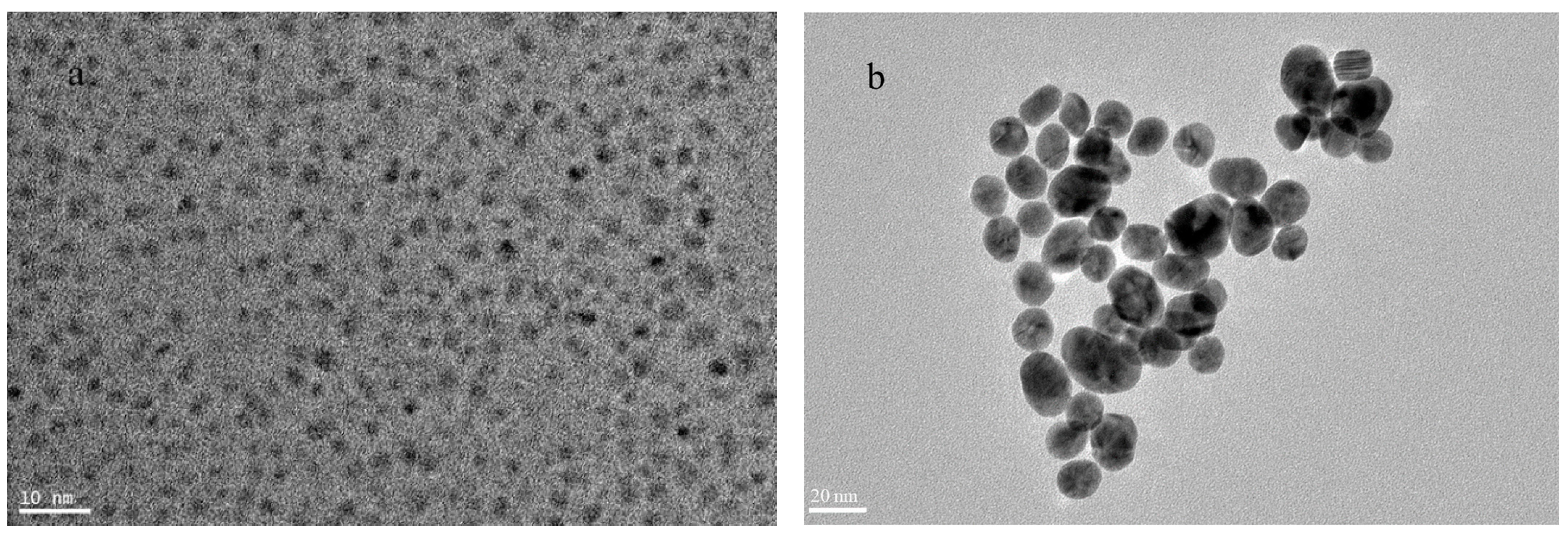
3.1. Morphological and Spectral Characterizations of CdTe QD and AuNP
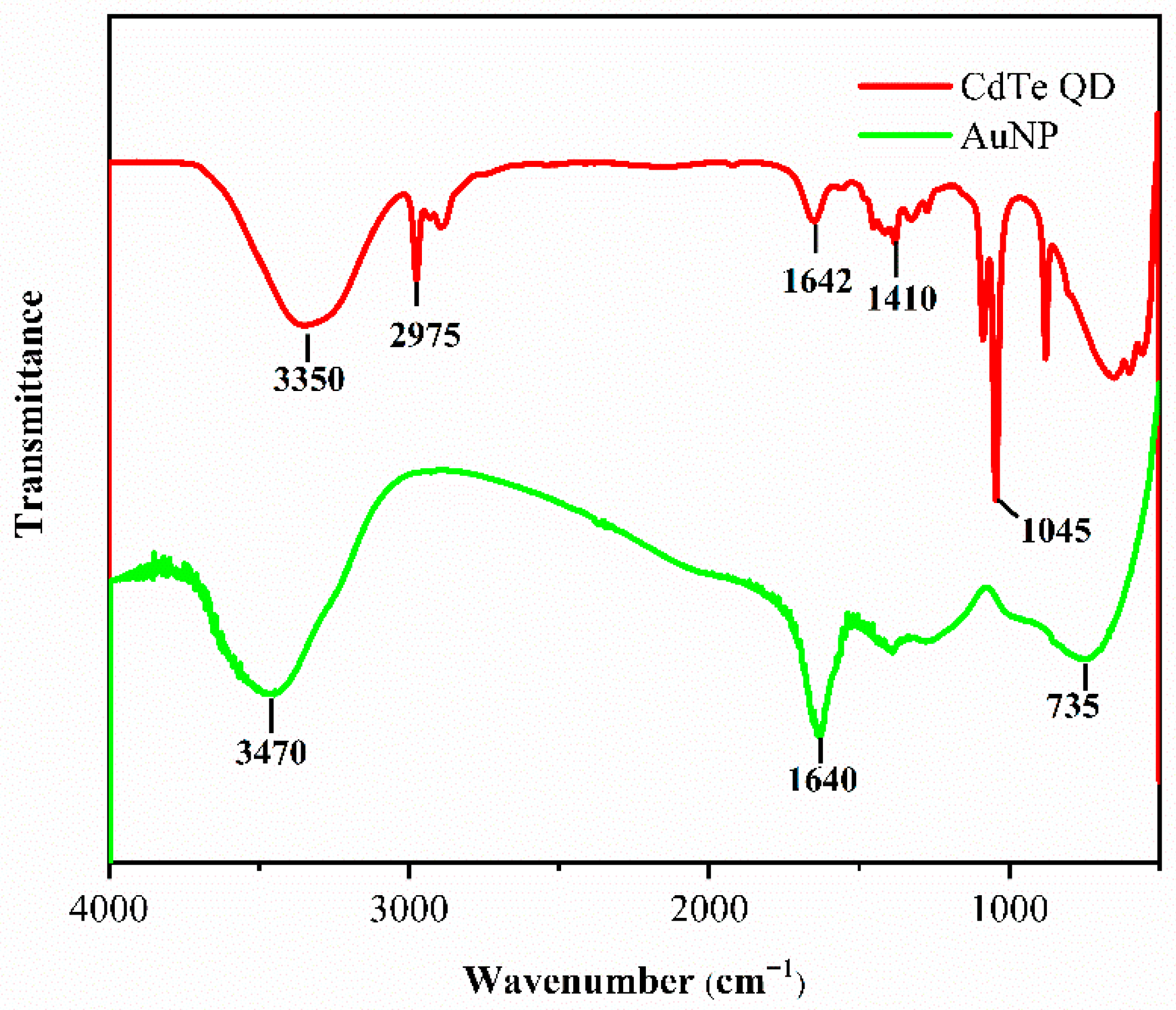
3.2. Characterization of Bioconjugate
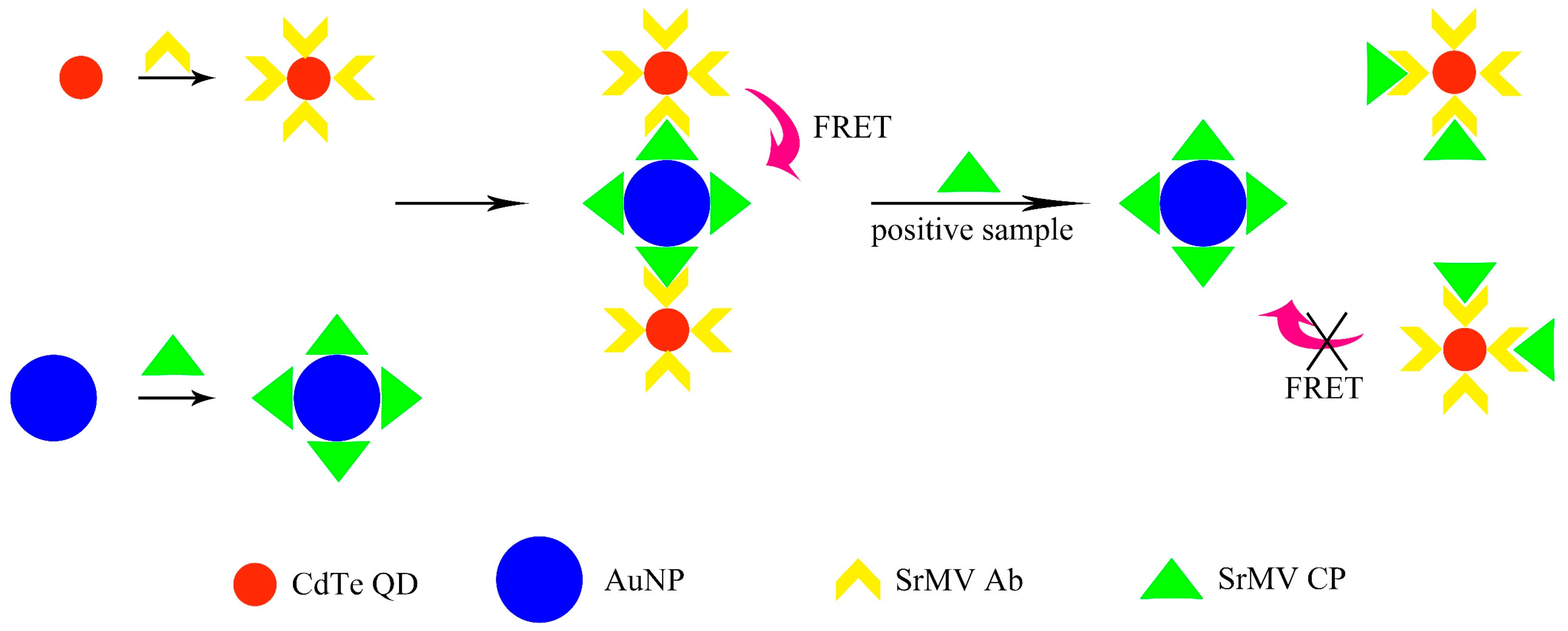
3.3. Principle Validation
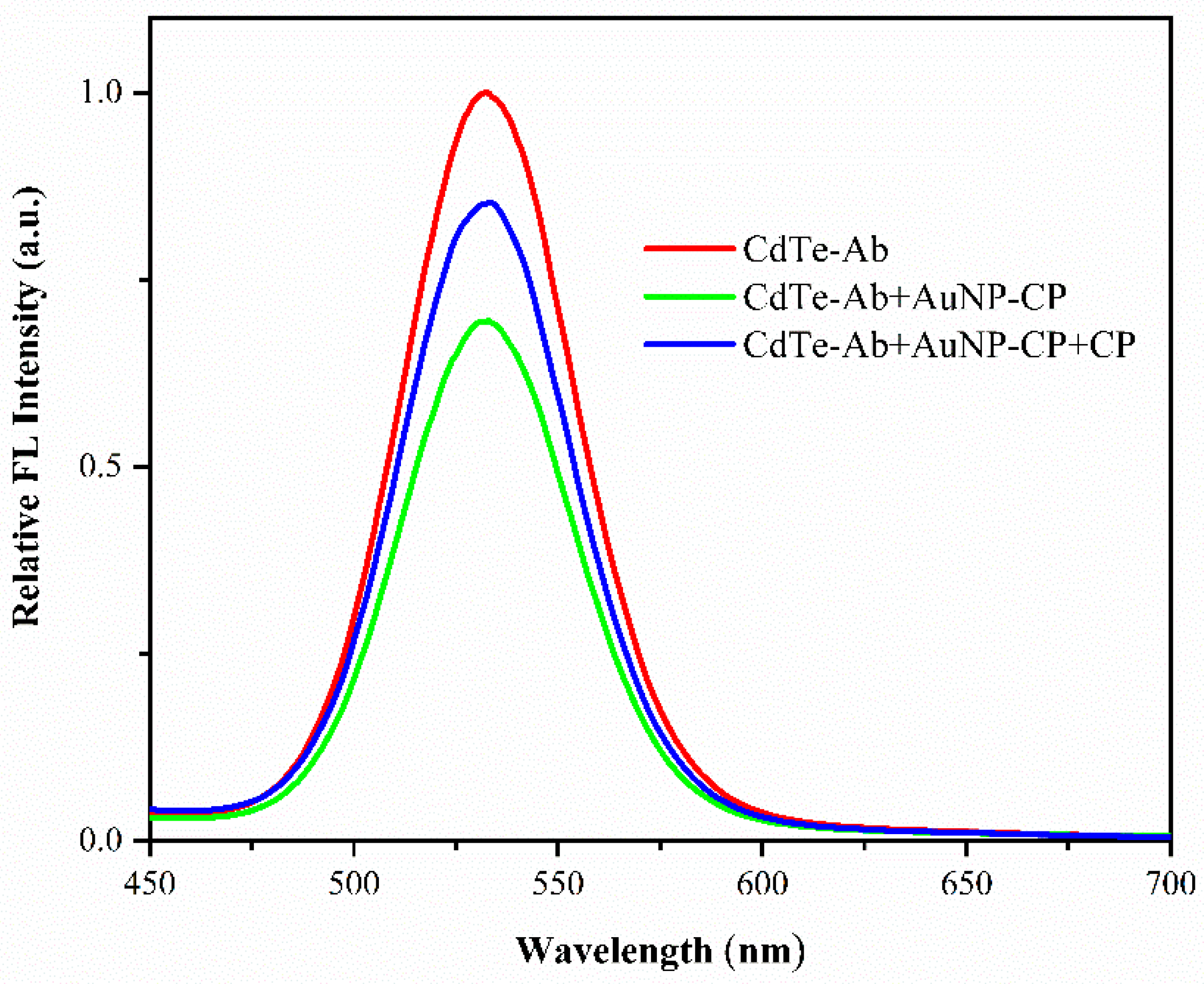
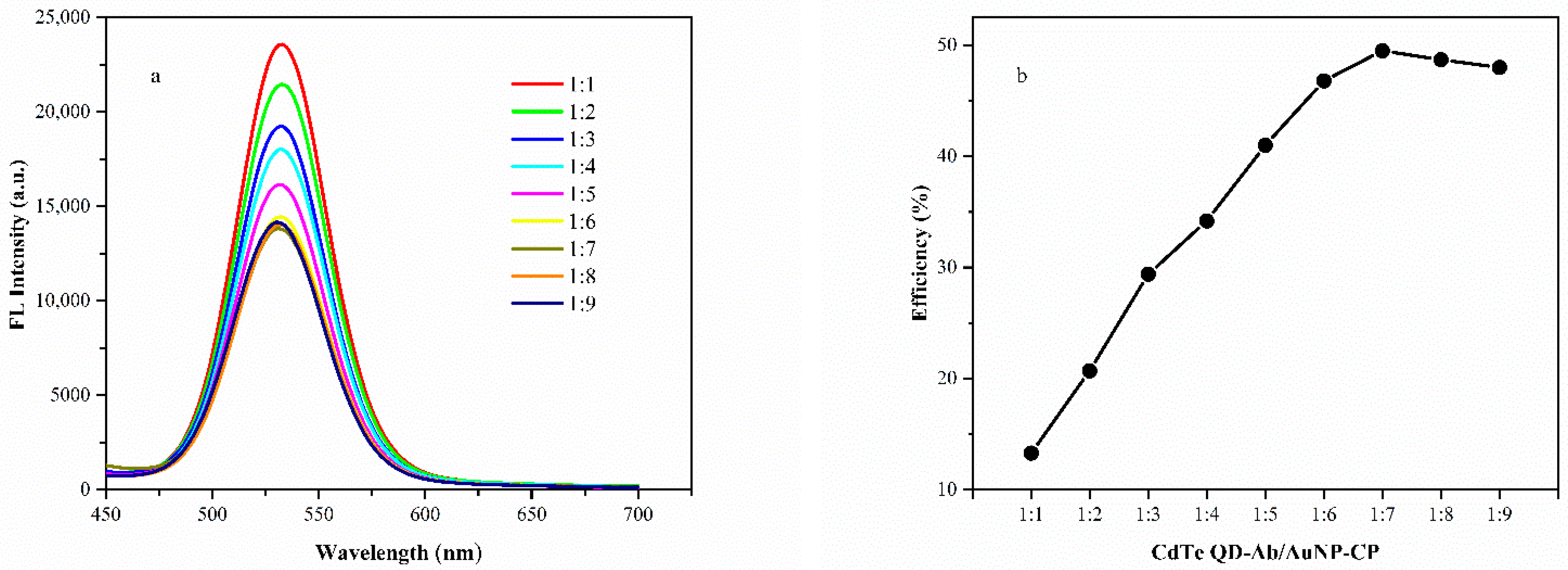
3.4. FRET Efficiency
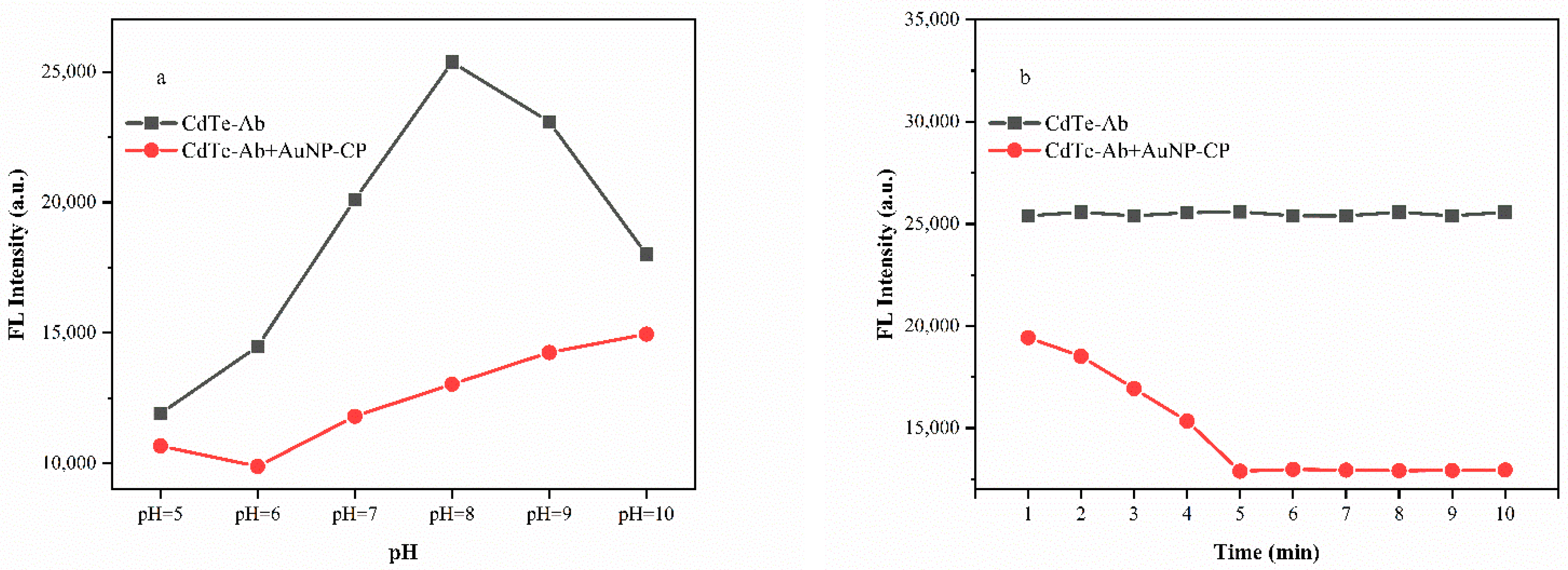
3.5. Optimization for the Biosensor
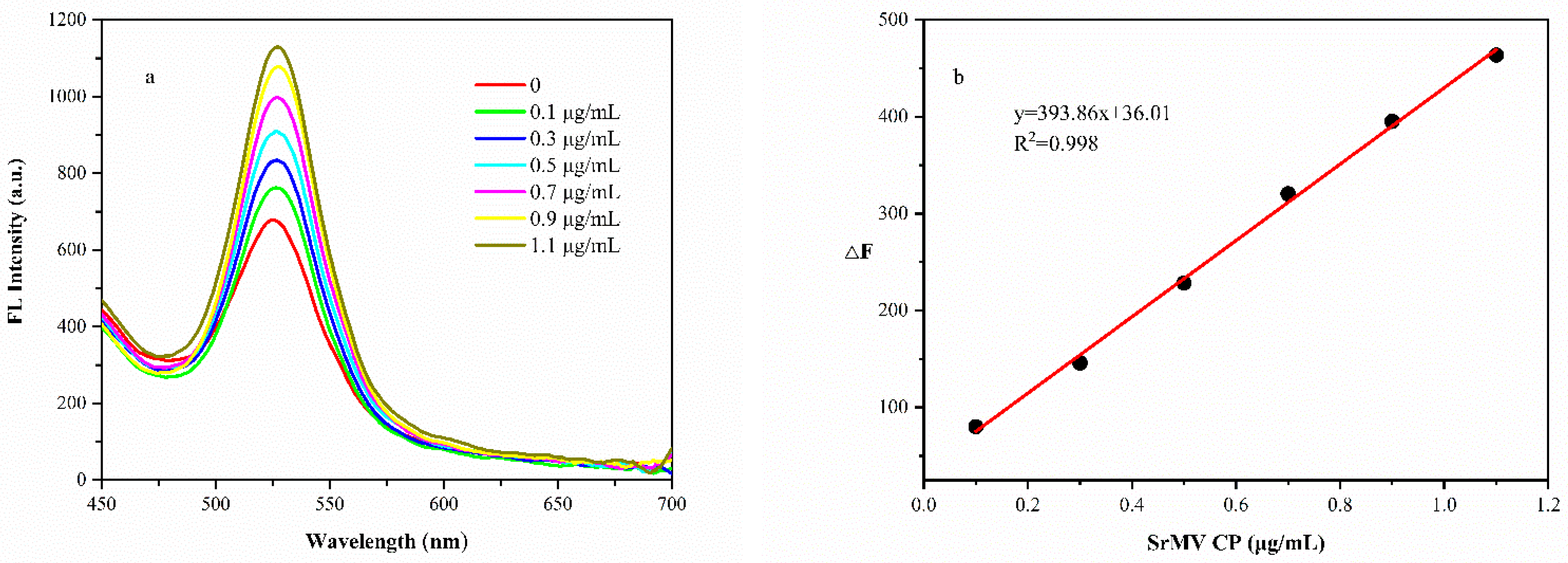
3.6. Feasibility Verification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, W.F.; Shan, H.L.; Zhang, R.Y.; Wang, X.Y.; Yang, K.; Luo, Z.M.; Yin, J.; Cang, X.Y.; Li, J.; Huang, Y.K. Identification of resistance to Sugarcane streak mosaic virus (SCSMV) and Sorghum mosaic virus (SrMV) in new elite sugarcane varieties/clones in China. Crop Prot. 2018, 110, 77–82. [Google Scholar] [CrossRef]
- Wang, X.Y.; Li, W.F.; Huang, Y.K.; Zhang, R.Y.; Shan, H.L.; Yin, J.; Luo, Z.M. Molecular detection and phylogenetic analysis of viruses causing mosaic symptoms in new sugarcane varieties in China. Eur. J. Plant Pathol. 2017, 148, 931–940. [Google Scholar] [CrossRef]
- Chen, H.; Lin, Y.F.; Ali, N.; Lv, W.Z.; Shen, Y.N.; Chen, B.S.; Wen, R.H. Comparison of IC-RT-PCR, Dot-ELISA and Indirect-ELISA for the detection of Sorghum mosaic virus in field-grown sugarcane plants. Sugar Tech. 2019, 22, 122–129. [Google Scholar] [CrossRef]
- Ling, H.; Huang, N.; Xu, L.; Peng, Q.; Liu, F.; Yang, Y.; Que, Y. Suitable Reference Genes/miRNAs for qRT-PCR Normalization of Expression Analysis in Sugarcane Under Sorghum mosaic virus infection. Sugar Tech. 2019, 21, 780–793. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2014, 35, 1–25. [Google Scholar] [CrossRef]
- Keizerweerd, A.T.; Chandra, A.; Grisham, M.P. Development of a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of Sugarcane mosaic virus and Sorghum mosaic virus in sugarcane. J. Virol. Methods 2015, 212, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Song, F.; Xiong, X.; Peng, X. Fluorescent Nanosensors Based on Fluorescence Resonance Energy Transfer (FRET). Ind. Eng. Chem. Res. 2013, 52, 11228–11245. [Google Scholar] [CrossRef]
- Imani, M.; Mohajeri, N.; Rastegar, M.; Zarghami, N. Recent advances in FRET-Based biosensors for biomedical applications. Anal. Biochem. 2021, 630, 114323. [Google Scholar] [CrossRef]
- Cardoso Dos Santos, M.; Algar, W.R.; Medintz, I.L.; Hildebrandt, N. Quantum dots for Förster Resonance Energy Transfer (FRET). TrAC Trends Anal. Chem. 2020, 125, 115819. [Google Scholar] [CrossRef]
- Yu, L.; Andriola, A. Quantitative gold nanoparticle analysis methods: A review. Talanta 2010, 82, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, J.; Li, H.; Zhou, J.; Zhang, H.; Fu, L. A sensitive immunosensor based on FRET between gold nanoparticles and InP/ZnS quantum dots for arginine kinase detection. Food Chem. 2021, 354, 129536. [Google Scholar] [CrossRef]
- Deng, J.; Lu, Q.; Hou, Y.; Liu, M.; Li, H.; Zhang, Y.; Yao, S. Nanosensor composed of nitrogen-doped carbon dots and gold nanoparticles for highly selective detection of cysteine with multiple signals. Anal. Chem. 2015, 87, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.P.; Pan, Y.; Zhang, H.; Zhang, Z.M.; Li, M.J.; Yi, C.Q.; Yang, M.S. A dual-mode nanosensor based on carbon quantum dots and gold nanoparticles for discriminative detection of glutathione in human plasma. Biosens. Bioelectron. 2014, 56, 39–45. [Google Scholar] [CrossRef]
- Sabela, M.; Balme, S.; Bechelany, M.; Janot, J.-M.; Bisetty, K. A Review of Gold and Silver Nanoparticle-Based Colorimetric Sensing Assays. Adv. Eng. Mater. 2017, 19, 1700270. [Google Scholar] [CrossRef]
- Shojaei, T.R.; Salleh, M.A.M.; Sijam, K.; Rahim, R.A.; Mohsenifar, A.; Safarnejad, R.; Tabatabaei, M. Fluorometric immunoassay for detecting the plant virus Citrus tristeza using carbon nanoparticles acting as quenchers and antibodies labeled with CdTe quantum dots. Microchim. Acta 2016, 183, 2277–2287. [Google Scholar] [CrossRef]
- Sabzehparvar, F.; Cherati, T.R.; Mohsenifar, A.; Shojaei, T.R.; Tabatabaei, M. Immobilization of gold nanoparticles with rhodamine to enhance the fluorescence resonance energy transfer between quantum dots and rhodamine; new method for downstream sensing of infectious bursal disease virus. Spectrochim. Acta A 2019, 212, 173–179. [Google Scholar] [CrossRef]
- Cheng, S.; Liao, L.S.; Sun, J.; Ye, Y.Y.; Yang, J.X.; Cao, C.F.; Lv, J.Q.; Fang, L.R.; Wu, F.; Lin, Y.X.; et al. A new immunoassay of serum antibodies against Peste des petits ruminants virus using quantum dots and a lateral-flow test strip. Anal. Bioanal. Chem. 2017, 409, 133–141. [Google Scholar] [CrossRef]
- Shen, W.; Gao, Z. Quantum dots and duplex-specific nuclease enabled ultrasensitive detection and serotyping of Dengue viruses in one step in a single tube. Biosens. Bioelectron. 2015, 65, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Fang, J.; Guo, Y.C.; Tao, Y.; Han, X.L.; Hua, Y.X.; Wang, J.J.; Li, L.Y.; Jian, Y.L.; Xie, G. A target-triggered biosensing platform for detection of HBV DNA based on DNA walker and CHA. Anal. Biochem. 2018, 554, 16–22. [Google Scholar] [CrossRef]
- Hong, S.; Lee, C. The Current Status and Future Outlook of Quantum Dot-Based Biosensors for Plant Virus Detection. Plant Pathol. J. 2018, 34, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Khater, M.; Escosura-Muniz, A.d.l.; Merkoci, A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Shanehsaz, M.; Mohsenifar, A.; Hasannia, S.; Pirooznia, N.; Samaei, Y.; Shamsipur, M. Detection of Helicobacter pylori with a nanobiosensor based on fluorescence resonance energy transfer using CdTe quantum dots. Microchim. Acta 2012, 180, 195–202. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, D.; Tang, Y.; Wang, Y.; Yan, F.; Li, Z.; Wang, J.; Zhou, H.S. Highly sensitive and selective colorimetric detection of cartap residue in agricultural products. Talanta 2012, 101, 382–387. [Google Scholar] [CrossRef]
- Yu, W.W.; Qu, L.H.; Guo, W.Z.; Peng, X.G. Experimental Determination of the Extinction Coefficient of CdTe, CdSe, and CdS Nanocrystals. Chem. Mater. 2003, 15, 2854–2860. [Google Scholar] [CrossRef]
- Yeh, K.C.; Chiang, Y.; Chang, S.W. Full Atomistic Simulation of Cross-Linked Gold Nanoparticle Assemblies. Multiscale Sci. Eng. 2020, 2, 242–251. [Google Scholar] [CrossRef]
- Stanisavljevic, M.; Krizkova, S.; Vaculovicova, M.; Kizek, R.; Adam, V. Quantum dots-fluorescence resonance energy transfer-based nanosensors and their application. Biosens. Bioelectron. 2015, 74, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Shojaei, T.R.; Salleh, M.A.; Sijam, K.; Rahim, R.A.; Mohsenifar, A.; Safarnejad, R.; Tabatabaei, M. Detection of Citrus tristeza virus by using fluorescence resonance energy transfer-based biosensor. Spectrochim. Acta A 2016, 169, 216–222. [Google Scholar] [CrossRef]
- Ruiz, G.; Tripathi, K.; Okyem, S.; Driskell, J.D. pH Impacts the Orientation of Antibody Adsorbed onto Gold Nanoparticles. Bioconjug. Chem. 2019, 30, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Zekavati, R.; Safi, S.; Hashemi, S.J.; Rahmani-Cherati, T.; Tabatabaei, M.; Mohsenifar, A.; Bayat, M. Highly sensitive FRET-based fluorescence immunoassay for aflatoxin B1 using cadmium telluride quantum dots. Microchim. Acta 2013, 180, 1217–1223. [Google Scholar] [CrossRef]
- Wang, S.; Mamedova, N.; Kotov, N.A.; Wei, C.; Studer, J.J.N.L. Antigen/Antibody Immunocomplex from CdTe Nanoparticle Bioconjugates. Nano Lett. 2002, 2, 817–822. [Google Scholar] [CrossRef]


| Sample | Added (μg) | Measured (μg) | Recovery (%) | RSD (n = 3, %) |
|---|---|---|---|---|
| 1 | 1 | 1.07 ± 0.09 | 107.4 | 8.45 |
| 2 | 5 | 4.91 ± 0.34 | 98.2 | 6.92 |
| 3 | 10 | 9.77 ± 0.62 | 97.7 | 6.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.; Du, P.; Wen, R.; Chen, B.; He, X. Signal “Off-On” Biosensor Based on Fluorescence Resonance Energy Transfer (FRET) for Detection of Sorghum Mosaic Virus. Analytica 2022, 3, 385-393. https://doi.org/10.3390/analytica3040026
Han Z, Du P, Wen R, Chen B, He X. Signal “Off-On” Biosensor Based on Fluorescence Resonance Energy Transfer (FRET) for Detection of Sorghum Mosaic Virus. Analytica. 2022; 3(4):385-393. https://doi.org/10.3390/analytica3040026
Chicago/Turabian StyleHan, Zhenlong, Pengfei Du, Ronghui Wen, Baoshan Chen, and Xipu He. 2022. "Signal “Off-On” Biosensor Based on Fluorescence Resonance Energy Transfer (FRET) for Detection of Sorghum Mosaic Virus" Analytica 3, no. 4: 385-393. https://doi.org/10.3390/analytica3040026
APA StyleHan, Z., Du, P., Wen, R., Chen, B., & He, X. (2022). Signal “Off-On” Biosensor Based on Fluorescence Resonance Energy Transfer (FRET) for Detection of Sorghum Mosaic Virus. Analytica, 3(4), 385-393. https://doi.org/10.3390/analytica3040026







