Conservation Innovations and Future Directions for the Study of Rhinoceros Gut Microbiome
Abstract
1. Introduction
2. The Animal Gut Microbiome under Human Management and Implications for In-Situ/Ex-Situ Conservation
3. The Rhinoceros Gut Microbiome
3.1. White Rhinoceros
3.2. Black Rhinoceros
3.3. Greater One-Horned Rhinoceros
3.4. Sumatran Rhinoceros
4. Common Pitfalls within Rhinoceros Gut Microbiome Studies
5. Addressing Pitfalls and Future Directions
5.1. Addressing Non-Standardized Sequencing and Bioinformatic Methodology Pitfalls through Collaboration
5.2. Addressing Individual- and Population-Related Pitfalls through the Establishment of Reference Datasets across Managed Facilities and Wild Populations
5.3. Future Avenue of Research: Address Reproductive Dysfunction
5.4. Future Avenue of Research: Fill Species Gaps in the Literature
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Rex Gaskins, H.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Ruan, R.; McLaughlin, R.W.; Hao, Y.; Zheng, J.; Wang, D. Fecal bacterial composition of the endangered Yangtze finless porpoises living under captive and semi-natural conditions. Curr. Microbiol. 2016, 72, 306–314. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, V.J.; Song, S.J.; Delsuc, F.; Prest, T.L.; Oliverio, A.M.; Korpita, T.M.; Alexiev, A.; Amato, K.R.; Metcalf, J.L.; Kowalewski, M.; et al. The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 2017, 57, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fu, H.; Zhou, Y.; Yan, M.; Chen, D.; Yang, M.; Xiao, S.; Chen, C.; Huang, L. Identification of the gut microbiota biomarkers associated with heat cycle and failure to enter oestrus in gilts. Microb. Biotechnol. 2021, 14, 1316–1330. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, M.A.; Orguilia, L.; Palacio, M.I.; Malpartida, A.; Mayol, S.; Mor, G.; Gutiérrez, G. Potential biomarkers of infertility associated with microbiome imbalances. Am. J. Reprod. Immunol. 2021, 86, e13438. [Google Scholar] [CrossRef]
- Carthey, A.J.R.; Blumstein, D.T.; Gallagher, R.V.; Tetu, S.G.; Gillings, M.R. Conserving the Holobiont. Funct. Ecol. 2020, 34, 764–776. [Google Scholar] [CrossRef]
- International Union for Conservation of Nature (IUCN). The IUCN Red List of Threatened Species, Version 2022-1; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2022. [Google Scholar]
- Ellis, S.; Talukdar, B. Rhinoceros sondaicus. In The IUCN Red List of Threatened Species; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2020; p. e.T19495A18493900. [Google Scholar] [CrossRef]
- Ellis, S.; Talukdar, B. Dicerorhinus sumatrensis. In The IUCN Red List of Threatened Species; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2020; p. e.T6553A18493355. [Google Scholar] [CrossRef]
- Emslie, R. Ceratotherium simum. In The IUCN Red List of Threatened Species; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2020; p. e.T4185A45813880. [Google Scholar] [CrossRef]
- Ellis, S.; Talukdar, B. Rhinoceros unicornis. In The IUCN Red List of Threatened Species; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2019; p. e.T19496A18494149. [Google Scholar] [CrossRef]
- Dennis, P.M.; Funk, J.A.; Rajala-Schultz, P.J.; Blumer, E.S.; Miller, R.E.; Wittum, T.E.; Saville, W.J.A. A review of some of the health issues of captive black rhinoceroses (Diceros bicornis). J. Zoo Wildl. Med. 2007, 38, 509–517. [Google Scholar] [CrossRef]
- Hildebrandt, T.B.; Hermes, R.; Walzer, C.; Sós, E.; Molnar, V.; Mezösi, L.; Schnorrenberg, A.; Silinski, S.; Streich, J.; Schwarzenberger, F.; et al. Artificial insemination in the anoestrous and the postpartum white rhinoceros using GnRH analogue to induce ovulation. Theriogenology 2007, 67, 1473–1484. [Google Scholar] [CrossRef]
- Roth, T.; Miller, M.; Dierenfeld, E.; de Groot, P.; Swaisgood, R.; Stoops, M. Rhino Research Masterplan; Association of Zoos and Aquariums’ Rhino Advisory Group: Glen Rose, TX, USA, 2009. [Google Scholar]
- Metrione, L.C.; Eyres, A. Rhino Husbandry Manual; International Rhino Foundation: Fort Worth, TX, USA, 2014; pp. 1–328. [Google Scholar]
- Roth, T.L.; Switzer, A.; Watanabe-Chailland, M.; Bik, E.M.; Relman, D.A.; Romick-Rosendale, L.E.; Ollberding, N.J. Reduced gut microbiome diversity and metabolome differences in rhinoceros species at risk for iron overload disorder. Front. Microbiol. 2019, 10, 2291. [Google Scholar] [CrossRef]
- Kock, R.A.; Garnier, J. Veterinary management of three species of rhinoceroses in zoological collections. In Rhinoceros Biology and Conservation: Proceedings of an International Rhino Conference; Zoological Society of San Diego: San Diego, CA, USA, 1993; pp. 325–338. [Google Scholar]
- Pluháček, J.; Sinha, S.P.; Bartoš, L.; Šípek, P. Parity as a major factor affecting infant mortality of highly endangered Indian rhinoceros: Evidence from zoos and Dudhwa National Park, India. Biol. Conserv. 2007, 139, 457–461. [Google Scholar] [CrossRef]
- Zhang, F.M.; Wang, H.G.; Wang, M.; Cui, B.T.; Fan, Z.N.; Ji, G.Z. Fecal microbiota transplantation for severe enterocolonic fistulizing Crohn’s disease. World J. Gastroenterol. 2013, 19, 7213–7216. [Google Scholar] [CrossRef] [PubMed]
- Mardanov, A.V.; Bulygina, E.S.; Nedoluzhko, A.V.; Kadnikov, V.V.; Beletskii, A.V.; Tsygankova, S.V.; Tikhonov, A.N.; Ravin, N.V.; Prokhorchuk, E.B.; Skryabin, K.G. Molecular analysis of the intestinal microbiome composition of mammoth and woolly rhinoceros. Dokl. Biochem. Biophys. 2012, 445, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J.; Mccray, A.T. ‘Ome Sweet ‘Omics—A genealogical treasury of words. Scientist 2001, 15, 8. [Google Scholar]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The brain-gut-microbiome axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut–liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129. [Google Scholar] [CrossRef]
- Schippa, S.; Iebba, V.; Totino, V.; Santangelo, F.; Lepanto, M.; Alessandri, C.; Nuti, F.; Viola, F.; Di Nardo, G.; Cucchiara, S.; et al. A potential role of Escherichia coli pathobionts in the pathogenesis of Pediatric Inflammatory Bowel Disease. Can. J. Microbiol. 2012, 58, 426–432. [Google Scholar] [CrossRef]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef]
- Zhao, N.; Li, M.; Luo, J.; Wang, S.; Liu, S.; Wang, S.; Lyu, W.; Chen, L.; Su, W.; Ding, H.; et al. Impacts of canine distemper virus infection on the giant panda population from the perspective of gut microbiota. Sci. Rep. 2017, 7, 39954. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Worobey, M.; Kuo, C.-H.; Ndjango, J.-B.N.; Peeters, M.; Hahn, B.H.; Hugenholtz, P. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010, 8, e1000546. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.H.; Ochman, H. Rates of gut microbiome divergence in mammals. Mol. Ecol. 2018, 27, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Youngblut, N.D.; Reischer, G.H.; Walters, W.; Schuster, N.; Walzer, C.; Stalder, G.; Ley, R.E.; Farnleitner, A.H. Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat. Commun. 2019, 10, 2200. [Google Scholar] [CrossRef] [PubMed]
- Montassier, E.; Al-Ghalith, G.A.; Hillmann, B.; Viskocil, K.; Kabage, A.J.; McKinlay, C.E.; Sadowsky, M.J.; Khoruts, A.; Knights, D. CLOUD: A non-parametric detection test for microbiome outliers. Microbiome 2018, 6, 137. [Google Scholar] [CrossRef] [PubMed]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- McKenney, E.A.; Rodrigo, A.; Yoder, A.D. Patterns of gut bacterial colonization in three primate species. PLoS ONE 2015, 10, e0124618. [Google Scholar] [CrossRef]
- Gillman, S.J.; McKenney, E.A.; Lafferty, D.J.R. Wild black bears harbor simple gut microbial communities with little difference between the jejunum and colon. Sci. Rep. 2020, 10, 20779. [Google Scholar] [CrossRef] [PubMed]
- McKenney, E.A.; O’Connell, T.M.; Rodrigo, A.; Yoder, A.D. Feeding strategy shapes gut metagenomic enrichment and functional specialization in captive lemurs. Gut Microbes 2018, 9, 202–217. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.K.; Williams, C.V.; Junge, R.E.; Mahefarisoa, K.L.; Rajaonarivelo, T.; Rakotondrainibe, H.; O’Connell, T.M.; Drea, C.M. A role for gut microbiota in host niche differentiation. ISME J. 2020, 14, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Eschweiler, K.; Clayton, J.B.; Moresco, A.; McKenney, E.A.; Minter, L.J.; Suhr Van Haute, M.J.; Gasper, W.; Hayer, S.S.; Zhu, L.; Cooper, K.; et al. Host identity and geographic location significantly affect gastrointestinal microbial richness and diversity in western lowland gorillas (Gorilla gorilla gorilla) under human care. Animals 2021, 11, 3399. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, W.; Wang, L.; Hou, R.; Zhang, M.; Fei, L.; Zhang, X.; Huang, H.; Bridgewater, L.C.; Jiang, Y.; et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. mBio 2015, 6, e00022-15. [Google Scholar] [CrossRef]
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Elias, J.E.; et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 2017, 357, 802–806. [Google Scholar] [CrossRef]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci. Rep. 2018, 8, 8510. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Mshelia, E.S.; Adamu, L.; Wakil, Y.; Turaki, U.A.; Gulani, I.A.; Musa, J. The association between gut microbiome, sex, age and body condition scores of horses in Maiduguri and its environs. Microb. Pathog. 2018, 118, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Adriansjach, J.; Baum, S.T.; Lefkowitz, E.J.; Van Der Pol, W.J.; Buford, T.W.; Colman, R.J. Age-related differences in the gut microbiome of rhesus macaques. J. Gerontol. Ser. A 2020, 75, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Janiak, M.C.; Montague, M.J.; Villamil, C.I.; Stock, M.K.; Trujillo, A.E.; DePasquale, A.N.; Orkin, J.D.; Bauman Surratt, S.E.; Gonzalez, O.; Platt, M.L.; et al. Age and sex-associated variation in the multi-site microbiome of an entire social group of free-ranging rhesus macaques. Microbiome 2021, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Hehemann, J.-H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut microbiome: Profound implications for diet and disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Mushegian, A.A.; Ebert, D. Rethinking “Mutualism” in diverse host-symbiont communities. BioEssays 2016, 38, 100–108. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Sonnenburg, J.L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 2019, 17, 383–390. [Google Scholar] [CrossRef]
- Kong, F.; Zhao, J.; Han, S.; Zeng, B.; Yang, J.; Si, X.; Yang, B.; Yang, M.; Xu, H.; Li, Y. Characterization of the gut microbiota in the red panda (Ailurus fulgens). PLoS ONE 2014, 9, e87885. [Google Scholar] [CrossRef]
- Amato, K.R.; Metcalf, J.L.; Song, S.J.; Hale, V.L.; Clayton, J.; Ackermann, G.; Humphrey, G.; Niu, K.; Cui, D.; Zhao, H.; et al. Using the gut microbiota as a novel tool for examining Colobine primate GI health. Glob. Ecol. Conserv. 2016, 7, 225–237. [Google Scholar] [CrossRef]
- Clayton, J.B.; Vangay, P.; Huang, H.; Ward, T.; Hillmann, B.M.; Al-Ghalith, G.A.; Travis, D.A.; Long, H.T.; Tuan, B.V.; Minh, V.V.; et al. Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Song, S.J.; Morton, J.T.; Weiss, S.; Seguin-Orlando, A.; Joly, F.; Feh, C.; Taberlet, P.; Coissac, E.; Amir, A.; et al. Evaluating the impact of domestication and captivity on the horse gut microbiome. Sci. Rep. 2017, 7, 15497. [Google Scholar] [CrossRef]
- Gibson, K.M.; Nguyen, B.N.; Neumann, L.M.; Miller, M.; Buss, P.; Daniels, S.; Ahn, M.J.; Crandall, K.A.; Pukazhenthi, B. Gut microbiome differences between wild and captive black rhinoceros—Implications for rhino health. Sci. Rep. 2019, 9, 7570. [Google Scholar] [CrossRef] [PubMed]
- West, A.G.; Waite, D.W.; Deines, P.; Bourne, D.G.; Digby, A.; McKenzie, V.J.; Taylor, M.W. The microbiome in threatened species conservation. Biol. Conserv. 2019, 229, 85–98. [Google Scholar] [CrossRef]
- Moeller, A.H.; Shilts, M.; Li, Y.; Rudicell, R.S.; Lonsdorf, E.V.; Pusey, A.E.; Wilson, M.L.; Hahn, B.H.; Ochman, H. SIV-Induced instability of the chimpanzee gut microbiome. Cell Host Microbe 2013, 14, 340–345. [Google Scholar] [CrossRef]
- Schmidt, E.; Mykytczuk, N.; Schulte-Hostedde, A.I. Effects of the captive and wild environment on diversity of the gut microbiome of deer mice (Peromyscus maniculatus). ISME J. 2019, 13, 1293–1305. [Google Scholar] [CrossRef]
- Yao, R.; Xu, L.; Hu, T.; Chen, H.; Qi, D.; Gu, X.; Yang, X.; Yang, Z.; Zhu, L. The “wildness” of the giant panda gut microbiome and its relevance to effective translocation. Glob. Ecol. Conserv. 2019, 18, e00644. [Google Scholar] [CrossRef]
- Bian, G.; Ma, L.; Su, Y.; Zhu, W. The microbial community in the feces of the white rhinoceros (Ceratotherium simum) as determined by barcoded pyrosequencing analysis. PLoS ONE 2013, 8, e70103. [Google Scholar] [CrossRef]
- Williams, C.L.; Ybarra, A.R.; Meredith, A.N.; Durrant, B.S.; Tubbs, C.W. Gut microbiota and phytoestrogen-associated infertility in southern white rhinoceros. mBio 2019, 10, e00311-19. [Google Scholar] [CrossRef]
- Cersosimo, L.M.; Sullivan, K.E.; Valdes, E.V. Species and individual rhinoceros affect the bacterial communities, metabolites, and nutrient composition in faeces from southern black rhinoceros (Diceros bicornis minor) and southern white rhinoceros (Ceratotherium simum simum) under managed care. J. Anim. Physiol. Anim. Nutr. 2021, 106, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, M.; Kreger, M.D. Rhinoceros behaviour: Implications for captive management and conservation. Int. Zoo Yearb. 2006, 40, 150–173. [Google Scholar] [CrossRef]
- Laurie, A. Behavioural ecology of the greater one-horned rhinoceros (Rhinoceros unicornis). J. Zool. 1982, 196, 307–341. [Google Scholar] [CrossRef]
- Hazarika, B.C.; Saikia, P.K. Food habit and feeding patterns of great Indian one-horned rhinoceros (Rhinoceros unicornis) in Rajiv Gandhi Orang National Park, Assam, India. Int. Sch. Res. Not. 2012, 2012, 259695. [Google Scholar] [CrossRef]
- Burnham, C.M. Drivers of Variation in the Gut Microbiome of Southern White Rhinoceros (Ceratotherium simum simum) under Human Care. Master’s Thesis, North Carolina State University, Raleigh, NC, USA, 2021; pp. 49–87. [Google Scholar]
- Borah, P.; Dutta, R.; Barkalita, L.M.; Buragohain, L.; Deka, P.; Ali, S.; Choudhury, B.; Basumatary, P. Deciphering the fecal microbiome of Indian rhinoceros (Rhinoceros unicornis) by metagenomic approach. Asian J. Conserv. Biol. 2019, 8, 135–141. [Google Scholar]
- Kakati, P.; Paine, S.K.; Bhattacharjee, C.K.; Bhattacharyya, C.; Sharma, A.; Phukan, D.; Barman, N.N.; Basu, A. Gut microbiome architecture of wild greater one-horned rhinoceros: A vulnerable species from Kaziranga National Park, India. J. Genet. 2021, 100, 84. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, C.W.; Moley, L.A.; Ivy, J.A.; Metrione, L.C.; LaClaire, S.; Felton, R.G.; Durrant, B.S.; Milnes, M.R. Estrogenicity of captive southern white rhinoceros diets and their association with fertility. Gen. Comp. Endocrinol. 2016, 238, 32–38. [Google Scholar] [CrossRef]
- Kothmann, K.H.; Jons, A.; Wilhelmi, B.; Kasozi, N.; Graham, L.; Gent, R.; Atkin, S.L.; Swart, A.C.; Newell-Fugate, A.E. Non-invasive assessment of fecal glucocorticoid, progesterone, and androgen metabolites and microbiome in free-ranging southern white rhinoceros (Ceratotherium simum simum) in South Africa. Gen. Comp. Endocrinol. 2022, 329, 114099. [Google Scholar] [CrossRef]
- Emslie, R. Diceros bicornis. In The IUCN Red List of Threatened Species; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2020; p. e.T6557A152728945. [Google Scholar] [CrossRef]
- Knight, M.H. African Rhino Specialist Group report. Pachyderm 2019, 60, 14–39. [Google Scholar]
- Edwards, K.L.; McArthur, H.M.; Liddicoat, T.; Walker, S.L. A practical field extraction method for non-invasive monitoring of hormone activity in the black rhinoceros. Conserv. Physiol. 2014, 2, cot037. [Google Scholar] [CrossRef]
- Edwards, K.L.; Shultz, S.; Pilgrim, M.; Walker, S.L. Irregular ovarian activity, body condition and behavioural differences are associated with reproductive success in female eastern black rhinoceros (Diceros bicornis michaeli). Gen. Comp. Endocrinol. 2015, 214, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Antwis, R.E.; Edwards, K.L.; Unwin, B.; Walker, S.L.; Shultz, S. Rare gut microbiota associated with breeding success, hormone metabolites and ovarian cycle phase in the critically endangered eastern black rhino. Microbiome 2019, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.E.; Mylniczenko, N.D.; Nelson, S.E., Jr.; Coffin, B.; Lavin, S.R. Practical management of iron overload disorder (IOD) in black rhinoceros (BR; Diceros bicornis). Animals 2020, 10, 1991. [Google Scholar] [CrossRef]
- Dennis, P.; Ellis, S.; Mellen, J.; Lee, P.; Olea-Popelka, F.; Petric, A.; Ryder, O. IOD in rhinos—Epidemiology Group Report: Report from the Epidemiology Working Group of the International Workshop on Iron Overload Disorder in Browsing Rhinoceros (February 2011). J. Zoo Wildl. Med. 2012, 43, S114–S116. [Google Scholar] [CrossRef]
- Roth, T.; Metrione, L.; Miller, M.; Miller, E.; Roca, A.; Stoops, M. Rhino Research Masterplan; Association of Zoos and Aquariums’ Rhino Advisory Group: Cincinnati, OH, USA, 2019. [Google Scholar]
- Dostal, A.; Baumgartner, J.; Riesen, N.; Chassard, C.; Smuts, C.M.; Zimmermann, M.B.; Lacroix, C. Effects of iron supplementation on dominant bacterial groups in the gut, faecal SCFA and gut inflammation: A randomised, placebo-controlled intervention trial in South African children. Br. J. Nutr. 2014, 112, 547–556. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Gálvez, N.; Martín, J.; Reyes, F.; Pérez-Victoria, I.; Dominguez-Vera, J.M. Identification of the key excreted molecule by Lactobacillus fermentum related to host iron absorption. Food Chem. 2017, 228, 374–380. [Google Scholar] [CrossRef]
- Murphy, S.T.; Subedi, N.; Jnawali, S.R.; Lamichhane, B.R.; Upadhyay, G.P.; Kock, R.; Amin, R. Invasive Mikania in Chitwan National Park, Nepal: The threat to the greater one-horned rhinoceros Rhinoceros unicornis and factors driving the invasion. Oryx 2013, 47, 361–368. [Google Scholar] [CrossRef]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; Janečka, J.E. Pyrosequencing of 16S RRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 8, 231. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Trujillo, S.M.; McKenney, E.A.; Hilderbrand, G.V.; Mangipane, L.S.; Rogers, M.C.; Joly, K.; Gustine, D.D.; Erlenbach, J.A.; Mangipane, B.A.; Lafferty, D.J. Correlating gut microbial membership to brown bear health metrics. Sci. Rep. 2022, 12, 15415. [Google Scholar] [CrossRef]
- Trujillo, S.M.; McKenney, E.A.; Hilderbrand, G.V.; Mangipane, L.S.; Rogers, M.C.; Joly, K.; Gustine, D.D.; Erlenbach, J.A.; Mangipane, B.A.; Lafferty, D.J. Intrinsic and extrinsic factors influence on an omnivore’s gut microbiome. PLoS ONE 2022, 17, e0266698. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, D.J.; Gillman, S.J.; Jeakle, L.K.; Roell, B.J.; McKenney, E.A. Mink (Neovison vison) fecal microbiomes are influenced by sex, temperature, and time post defecation. J. Mammal. 2022, 103, 316–327. [Google Scholar] [CrossRef]
- DeLong, E.F.; Pace, N.R. Environmental diversity of bacteria and archaea. Syst. Biol. 2001, 50, 470–478. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.K.; McKenney, E.A. The inside tract: The appendicular, cecal, and colonic microbiome. Am. J. Phys. Anthropol. 2018, 166, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.K.; McKenney, E.A.; Gasper, W.; Wrampelmeier, C.; Hayer, S.; Ehmke, E.E.; Clayton, J.B. Gut site and gut morphology predict microbiome structure and function in ecologically diverse lemurs. Microb. Ecol. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current sampling methods for gut microbiota: A call for more precise devices. Front. Cell. Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Marneweck, C.; Jürgens, A.; Shrader, A.M. The role of middens in white rhino olfactory communication. Anim. Behav. 2018, 140, 7–18. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef]
- Rausch, P.; Rühlemann, M.; Hermes, B.M.; Doms, S.; Dagan, T.; Dierking, K.; Domin, H.; Fraune, S.; von Frieling, J.; Hentschel, U.; et al. Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome 2019, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, R.; Sylvia, K.E.; Klein, S.L.; Forster, S.C.; Plebanski, M.; Eri, R.; Flanagan, K.L. The microgenderome revealed: Sex differences in bidirectional interactions between the microbiota, hormones, immunity and disease susceptibility. Semin. Immunopathol. 2019, 41, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Tiuria, R.; Primawidyawan, A.; Pangihutan, J.; Warsito, J.; Hariyadi, A.R.S.; Handayani, S.U.; Priosoeryarito, B.P. Identification of endoparasites from faeces of Javan Rhino (Rhinoceros sondaicus) in Ujung Kulon National Park, Indonesia. In Proceedings of the Asian Zoo and Wildlife Medicine Convention, Bangkok, Thailand, 26–29 October 2006; p. 31. [Google Scholar]
- Hariyadi, A.R.S.; Sajuthi, D.; Astuti, D.A.; Maheswari, H.; Alikodra, H.S. Analysis of 3α,11β-Dihydroxy-CM profile for the indicator of stress on male Javan rhinoceros. IOP Conf. Ser. Earth Environ. Sci. 2019, 399, 012066. [Google Scholar] [CrossRef]
- Cardona, S.; Eck, A.; Cassellas, M.; Gallart, M.; Alastrue, C.; Dore, J.; Azpiroz, F.; Roca, J.; Guarner, F.; Manichanh, C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012, 12, 158. [Google Scholar] [CrossRef] [PubMed]
- Tedjo, D.I.; Jonkers, D.M.A.E.; Savelkoul, P.H.; Masclee, A.A.; van Best, N.; Pierik, M.J.; Penders, J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS ONE 2015, 10, e0126685. [Google Scholar] [CrossRef]
| Southern White Rhinoceros (Ceratotherium simum simum) | Black Rhinoceros (Diceros bicornis) | Greater One-Horned Rhinoceros (Rhinoceros unicornis) | Sumatran Rhinoceros (Dicerorhinus sumatrensis) | |
|---|---|---|---|---|
| IUCN status | Near threatened [10] 1 | Critically endangered [72] | Vulnerable [11] | Critically endangered [9] |
| Number in wild | ~18,000 [10] | ~5630 [72] | ~3588 [11] | <80 [9] |
| Social group size [15] | Bulls solitary; cow-calf pairs; female and adolescent groups of <16 individuals | Solitary; cow-calf pairs | Solitary; cow-calf pairs | Solitary; cow-calf pairs |
| Feeding strategy [15] | Grazer | Browser | Primarily grazer, some browsing | Browser |
| Bacteroidetes (%) | 21 [73]–55 [68] | 18 w [62]–49 [69] | 2 w [74]–30 [68] | 39 [16] |
| Firmicutes (%) | 23 [67]–72 [16] | 26 [69]–64 [16] | 20 w [75]–78 [16] | 56 [16] |
| Proteobacteria (%) | <1 [67,73] | <1 [16]–24 w [62] | 1 [16]–63 w [75] | <1 [16] |
| Verrucomicrobia (%) | <1 [69,73] | <1 [16]–3 [69] | <1 [16]–15 w [74] | <1 [16] |
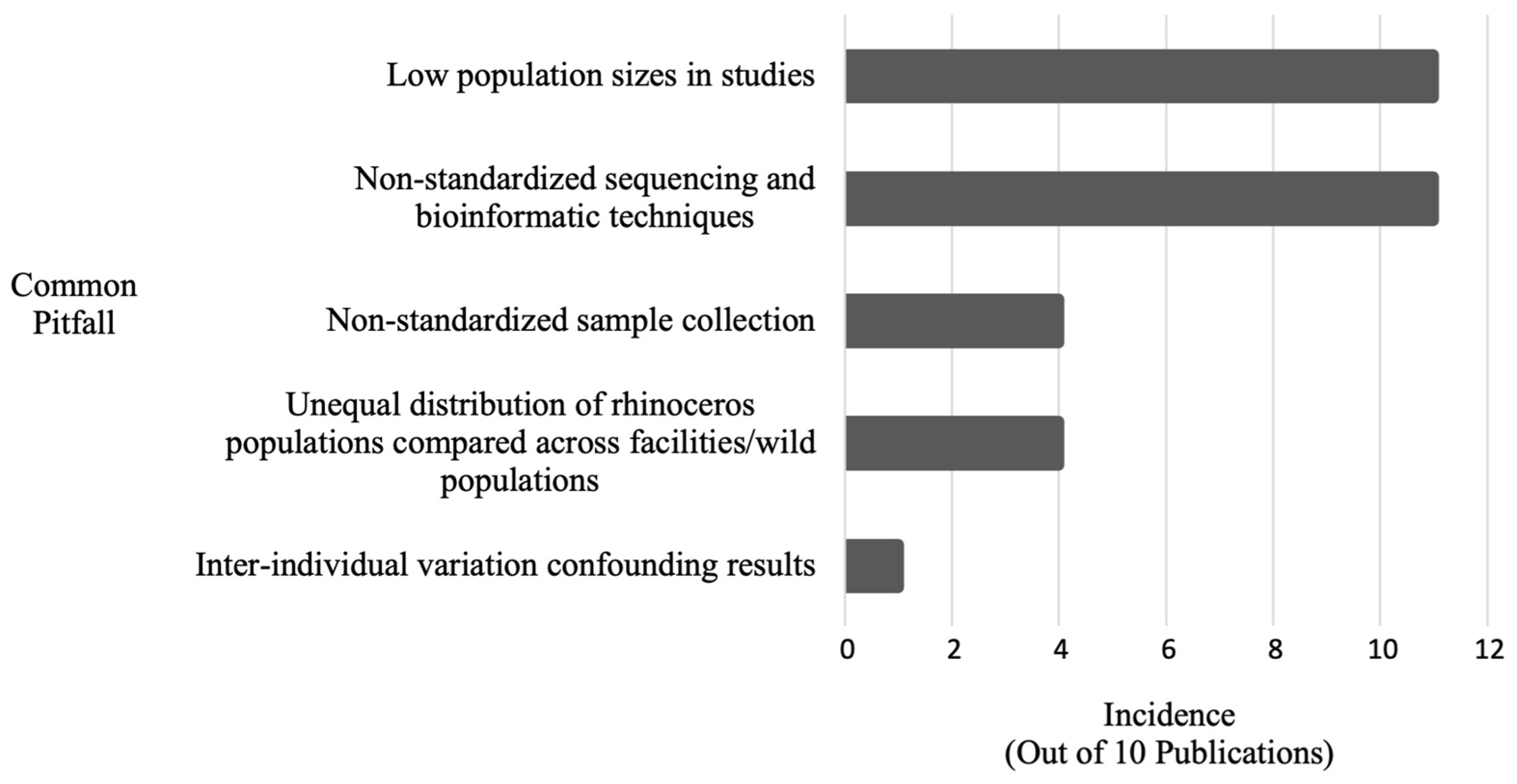
| Common Pitfall | Incidence | Publication(s) |
|---|---|---|
| Low population sizes in studies | 11 | [3,16,62,67,68,69,73,74,75,77,82] |
| Non-standardized sequencing and bioinformatic techniques | 11 | [3,16,62,67,68,69,73,74,75,77,82] |
| Unequal distributions of rhinoceros populations compared across facilities/wild populations | 4 | [3,16,62,82] |
| Non-standardized sample collection (unclear amount of time between defecation and sample collection) | 2 | [74,75] |
| Non-standardized sample collection (time of year/season in which stool specimens were collected not standardized) | 2 | [16,62] |
| Inter-individual variation confounding results | 1 | [82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burnham, C.M.; Ange-van Heugten, K.; McKenney, E.A.; Minter, L.J.; Trivedi, S. Conservation Innovations and Future Directions for the Study of Rhinoceros Gut Microbiome. J. Zool. Bot. Gard. 2023, 4, 396-412. https://doi.org/10.3390/jzbg4020030
Burnham CM, Ange-van Heugten K, McKenney EA, Minter LJ, Trivedi S. Conservation Innovations and Future Directions for the Study of Rhinoceros Gut Microbiome. Journal of Zoological and Botanical Gardens. 2023; 4(2):396-412. https://doi.org/10.3390/jzbg4020030
Chicago/Turabian StyleBurnham, Christina M., Kimberly Ange-van Heugten, Erin A. McKenney, Larry J. Minter, and Shweta Trivedi. 2023. "Conservation Innovations and Future Directions for the Study of Rhinoceros Gut Microbiome" Journal of Zoological and Botanical Gardens 4, no. 2: 396-412. https://doi.org/10.3390/jzbg4020030
APA StyleBurnham, C. M., Ange-van Heugten, K., McKenney, E. A., Minter, L. J., & Trivedi, S. (2023). Conservation Innovations and Future Directions for the Study of Rhinoceros Gut Microbiome. Journal of Zoological and Botanical Gardens, 4(2), 396-412. https://doi.org/10.3390/jzbg4020030







