Investigation of Cross-Reactivity of Anti-Ephrin-B2 Antibody to Other Ephrin-B Members in an Immunohistochemical Study in a Cohort of Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Alignment Analysis
2.2. Transfections, Immunoprecipitation and Western Blotting
2.3. Immunocytochemistry on HEK-293T Cells Transfected with EFNB1, EFNB2 and EFNB3 Expression Constructs
2.4. External Transcriptome Datasets
2.5. FFPE Specimens of Human OSCC
2.6. Immunohistochemistry and Evaluation of Immunoexpression
2.7. Statistical Analysis
3. Results
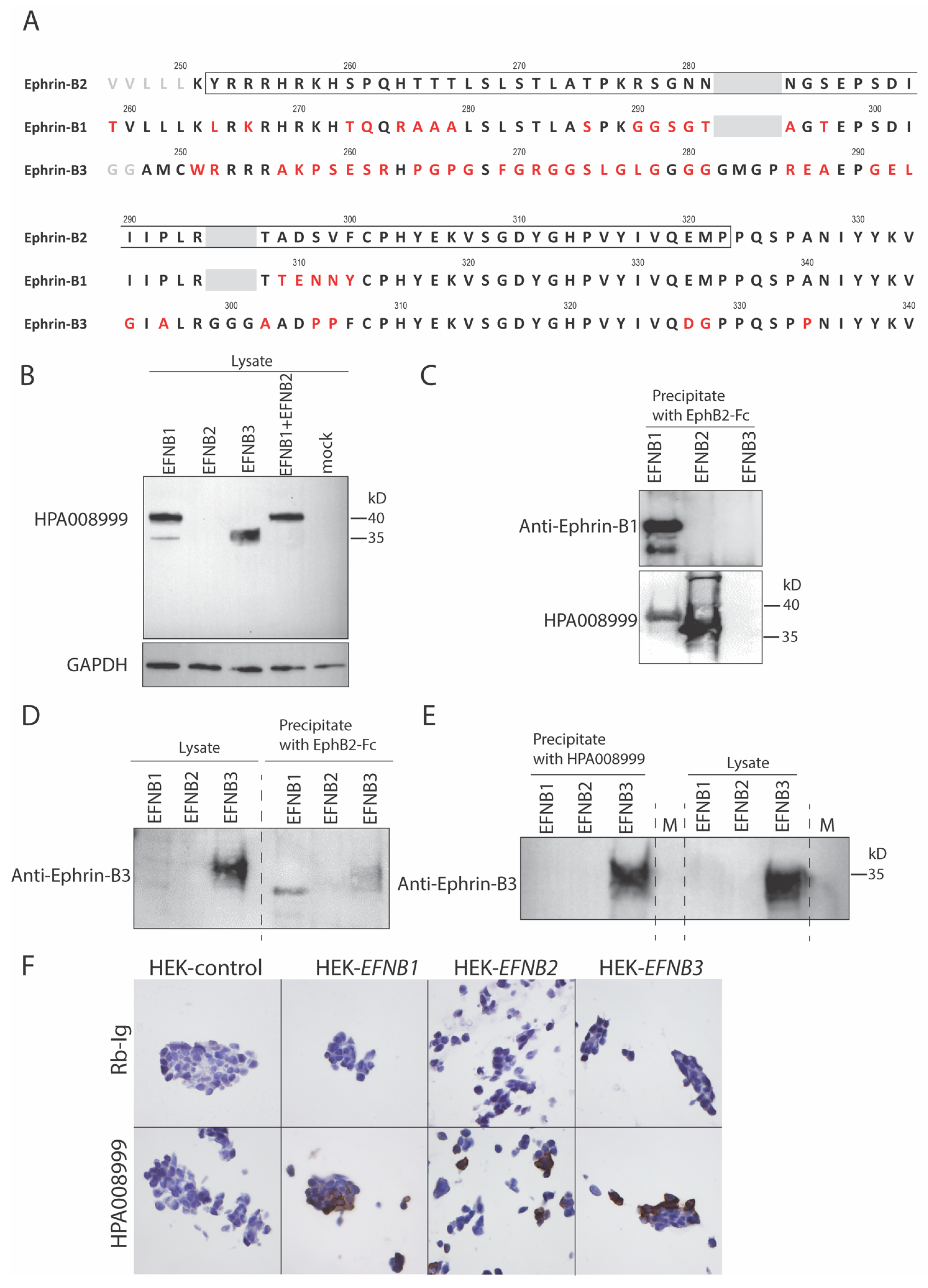
3.1. A High Degree of Sequence Identity Was Observed between Ephrins-B1, -B2 and -B3
3.2. Anti-Ephrin-B2 Antibody HPA008999 Reacted with All Ephrin-B Members
3.3. EFNB1 and EFNB2 mRNA Was Upregulated in OSCC as Compared to the Control Specimens
3.4. HPA008999 Immunoreactivity Scores Were Positively Associated with Clinical Stage, but Negatively with Tumor Differentiation
3.5. Higher Immunoreactivity of HPA008999 at the Tumor Center Was Associated with Reduced 5-Year Overall Survival Probabilities of OSCC Patients
3.6. Higher Immunoreactivity of HPA008999 Was Found to Be an Independent Predictor for Reduced 5-Year Overall Survival of OSCC Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics, 2010. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Eph Nomenclature Committee. Unified nomenclature for Eph family receptors and their ligands, the Ephrins. Cell 1997, 90, 403–404. [Google Scholar] [CrossRef] [Green Version]
- Himanen, J.-P.; Rajashankar, K.R.; Lackmann, M.; Cowan, C.A.; Henkemeyer, M.; Nikolov, D.B. Crystal structure of an Eph receptor–ephrin complex. Nature 2001, 414, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Mosch, B.; Reissenweber, B.; Neuber, C.; Pietzsch, J. Eph receptors and Ephrin ligands: Important players in angiogenesis and tumor angiogenesis. J. Oncol. 2010, 2010, 135285. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph–ephrin promiscuity is now crystal clear. Nat. Neurosci. 2004, 7, 417–418. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef] [Green Version]
- Kaenel, P.; Mosimann, M.; Andres, A.C. The multifaceted roles of Eph/ephrin signaling in breast cancer. Cell Adhes. Migr. 2012, 6, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Mellitzer, G.; Xu, Q.; Wilkinson, D.G. Eph receptors and ephrins restrict cell intermingling and communication. Nature 1999, 400, 77–81. [Google Scholar] [CrossRef]
- Helbling, P.M.; Saulnier, D.M.; Brandli, A.W. The receptor tyrosine kinase EphB4 and ephrin-B ligands restrict angiogenic growth of embryonic veins in Xenopus laevis. Development 2000, 127, 269–278. [Google Scholar] [CrossRef]
- Foo, S.S.; Turner, C.J.; Adams, S.; Compagni, A.; Aubyn, D.; Kogata, N.; Lindblom, P.; Shani, M.; Zicha, D.; Adams, R.H. Ephrin-B2 controls cell motility and adhesion during blood-vessel-wall assembly. Cell 2006, 124, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Adams, R.H.; Wilkinson, G.A.; Weiss, C.; Diella, F.; Gale, N.W.; Deutsch, U.; Risau, W.; Klein, R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: Demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999, 13, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Huynh-Do, U.; Vindis, C.C.; Liu, H.; Cerretti, D.P.; McGrew, J.T.; Enriquez, M.; Chen, J.; Daniel, T.O. Ephrin-B1 transduces signals to activate integrin-mediated migration, attachment and angiogenesis. J. Cell Sci. 2002, 115, 3073–3081. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.G.; Vanderhaeghen, P. The ephrins and Eph receptors in neural development. Annu. Rev. Neurosci. 1998, 21, 309–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimidschstein, J.; Passante, L.; Dufour, A.; Van Den Ameele, J.; Tiberi, L.; Hrechdakian, T.; Adams, R.; Klein, R.; Lie, D.C.; Jossin, Y.; et al. Ephrin-B1 controls the columnar distribution of cortical pyramidal neurons by restricting their tangential migration. Neuron 2013, 79, 1123–1135. [Google Scholar] [CrossRef] [Green Version]
- Hruska, M.; Dalva, M.B. Ephrin regulation of synapse formation, function and plasticity. Mol. Cell. Neurosci. 2012, 50, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhuang, G.; Frieden, L.; Debinski, W. Eph receptors and Ephrins in cancer: Common themes and controversies. Cancer Res. 2008, 68, 10031–10033. [Google Scholar] [CrossRef] [Green Version]
- Nakada, M.; Drake, K.L.; Nakada, S.; Niska, J.A.; Berens, M.E. Ephrin-B3 ligand promotes glioma invasion through activation of Rac1. Cancer Res. 2006, 66, 8492–8500. [Google Scholar] [CrossRef] [Green Version]
- Depner, C.; Zum Buttel, H.; Böğürcü, N.; Cuesta, A.M.; Aburto, M.R.; Seidel, S.; Finkelmeier, F.; Foss, F.; Hofmann, J.; Kaulich, K.; et al. EphrinB2 repression through ZEB2 mediates tumour invasion and anti-angiogenic resistance. Nat. Commun. 2016, 7, 12329. [Google Scholar] [CrossRef]
- Liu, W.; Jung, Y.D.; Ahmad, S.A.; McCarty, M.F.; Stoeltzing, O.; Reinmuth, N.; Fan, F.; Ellis, L.M. Effects of overexpression of ephrin-B2 on tumour growth in human colorectal cancer. Br. J. Cancer 2004, 90, 1620–1626. [Google Scholar] [CrossRef] [Green Version]
- Sasabe, E.; Tomomura, A.; Tomita, R.; Sento, S.; Kitamura, N.; Yamamoto, T. Ephrin-B2 reverse signaling regulates progression and lymph node metastasis of oral squamous cell carcinoma. PLoS ONE 2017, 12, e0188965. [Google Scholar] [CrossRef]
- Beauchamp, A.; Debinski, W. Ephs and ephrins in cancer: Ephrin-A1 signalling. Semin. Cell Dev. Biol. 2012, 23, 109–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barquilla, A.; Pasquale, E.B. Eph receptors and Ephrins: Therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 465–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquale, E.B. Eph-Ephrin Bidirectional Signaling in Physiology and Disease. Cell 2008, 133, 38–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.; Hafner, C.; Guba, M.; Flegel, S.; Geissler, E.K.; Becker, B.; Koehl, G.E.; Orsó, E.; Landthaler, M.; Vogt, T. ephrin-B2 overexpression enhances integrin-mediated ECM-attachment and migration of B16 melanoma cells. Int. J. Oncol. 2005, 27, 1197–1206. [Google Scholar] [CrossRef]
- Bundesen, L.Q.; Scheel, T.A.; Bregman, B.S.; Kromer, L.F. Ephrin-B2 and EphB2 regulation of astrocyte-meningeal fibroblast interactions in response to spinal cord lesions in adult rats. J. Neurosci. 2003, 23, 7789–7800. [Google Scholar] [CrossRef] [Green Version]
- Cramer, K.S.; Karam, S.D.; Bothwell, M.; Cerretti, D.P.; Pasquale, E.B.; Rubel, E.W. Expression of EphB receptors and EphrinB ligands in the developing chick auditory brainstem. J. Comp. Neurol. 2002, 452, 51–64. [Google Scholar] [CrossRef]
- Noberini, R.; Rubio de la Torre, E.; Pasquale, E.B. Profiling Eph receptor expression in cells and tissues: A targeted mass spectrometry approach. Cell Adhes. Migr. 2012, 6, 102–112. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Munthe, E.; Rian, E.; Holien, T.; Rasmussen, A.-M.; Levy, F.O.; Aasheim, H.-C. Ephrin-B2 is a candidate ligand for the Eph receptor, EphB6. FEBS Lett. 2000, 466, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Estilo, C.L.; O-charoenrat, P.; Talbot, S.; Socci, N.D.; Carlson, D.L.; Ghossein, R.; Williams, T.; Yonekawa, Y.; Ramanathan, Y.; Boyle, J.O.; et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer 2009, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Méndez, E.; Houck, J.; Fan, W.; Lohavanichbutr, P.; Doody, D.; Yueh, B.; Futran, N.D.; Upton, M.; Farwell, D.G.; et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2152–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Kenneth Baillie, J.; de Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.; Osman, T.A.; Sharma, S.; Vallenari, E.M.; Shahdadfar, A.; Pun, C.B.; Gautam, D.K.; Uhlin-Hansen, L.; Rikardsen, O.; Johannessen, A.C.; et al. Loss of S100A14 expression at the tumor-invading front correlates with poor differentiation and worse prognosis in oral squamous cell carcinoma. Head Neck 2020, 42, 2088–2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapkota, D.; Bruland, O.; Parajuli, H.; Osman, T.A.; Teh, M.-T.; Johannessen, A.C.; Costea, D.E. S100A16 promotes differentiation and contributes to a less aggressive tumor phenotype in oral squamous cell carcinoma. BMC Cancer 2015, 15, 631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Oweida, A.; Bhatia, S.; Hirsch, K.; Calame, D.; Griego, A.; Keysar, S.; Pitts, T.; Sharma, J.; Eckhardt, G.; Jimeno, A.; et al. Ephrin-B2 overexpression predicts for poor prognosis and response to therapy in solid tumors. Mol. Carcinog. 2017, 56, 1189–1196. [Google Scholar] [CrossRef] [Green Version]
- Yavrouian, E.J.; Sinha, U.K.; Rice, D.H.; Salam, M.T.; Gill, P.S.; Masood, R. The significance of EphB4 and EphrinB2 expression and survival in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2008, 134, 985–991. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, S.; Sharma, J.; Bukkapatnam, S.; Oweida, A.; Lennon, S.; Phan, A.; Milner, D.; Uyanga, N.; Jimeno, A.; Raben, D.; et al. Inhibition of EphB4–Ephrin-B2 signaling enhances response to Cetuximab–radiation therapy in head and neck cancers. Clin. Cancer Res. 2018, 24, 4539–4550. [Google Scholar] [CrossRef] [Green Version]
- Fritschy, J.-M. Is my antibody-staining specific? How to deal with pitfalls of immunohistochemistry. Eur. J. Neurosci. 2008, 28, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, K.J.; Trimmer, J.S. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J. Neurosci. 2006, 26, 8017–8020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.X.; Brodeur, G.M.; Campling, B.G.; Ikegaki, N. Coexpression of transcripts encoding EPHB receptor protein tyrosine kinases and their Ephrin-B ligands in human small cell lung carcinoma. Clin. Cancer Res. 1999, 5, 455–460. [Google Scholar] [PubMed]
- Bryne, M. Is the invasive front of an oral carcinoma the most important area for prognostication? Oral Dis. 1998, 4, 70–77. [Google Scholar] [CrossRef]



| Variables | Low, n (%) | High, n (%) | p |
|---|---|---|---|
| Age a (years) | |||
| ≤68 | 51 (83.6) | 10 (16.4) | 0.187 |
| >68 | 42 (73.7) | 15 (26.3) | |
| Gender | |||
| Female | 40 (80.0) | 20 (20.0) | 0.818 |
| Male | 54 (78.3) | 15 (21.7) | |
| Smoking b | |||
| No | 26 (70.3) | 11 (29.7) | 0.283 |
| Yes | 44 (80.0) | 11 (20.0) | |
| Alcohol c | |||
| No | 33 (76.7) | 10 (23.3) | 0.986 |
| Yes | 20 (76.9) | 6 (23.1) | |
| Anatomic location | |||
| Tongue | 40 (74.1) | 14 (25.9) | 0.654 |
| Gingiva and buccal mucosa | 39 (83.0) | 8 (17.0) | |
| Floor of the mouth | 11 (78.6) | 3 (21.4) | |
| Palate and mucosal lip | 3 (100.0) | 0 (0.0) | |
| Differentiation | |||
| Poor and moderate | 11 (57.9) | 8 (42.1) | 0.014 |
| Well | 83 (83.0) | 17 (17.0) | |
| Lymph node involvement | |||
| Negative (N0) | 66 (82.5) | 14 (17.5) | 0.178 |
| Positive (N1 & N2) | 28 (71.8) | 11 (28.2) | |
| Tumor size d | |||
| T1 & T2 | 62 (82.7) | 13 (17.3) | 0.155 |
| T3 & T4 | 30 (71.4) | 12 (28.6) | |
| Recurrence | |||
| No | 55 (76.4) | 17 (23.6) | 0.388 |
| Yes | 39 (83.0) | 8 (17.0) | |
| Tumor stage | |||
| Early (1 & 2) | 49 (87.5) | 7 (12.5) | 0.032 |
| Late (3 & 4) | 45 (71.4) | 18 (28.6) | |
| Tumor budding e | |||
| <5 | 41(82.0) | 9 (18.0) | 0.430 |
| ≥5 | 37 (75.5) | 12 (24.5) | |
| Depth of invasion f | |||
| <4 cm | 28 (82.4) | 6 (17.6) | 0.491 |
| ≥4 cm | 28 (75.7) | 9 (24.3) |
| Univariate Cox | Multivariate Cox | |||||
|---|---|---|---|---|---|---|
| Variables | Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value |
| HPA008999 | ||||||
| immunoreactivity | ||||||
| Low | 1.98 | 1.19–3.29 | 0.008 | 1.84 | 1.10–3.0 | 0.02 |
| High | ||||||
| Age | ||||||
| ≤68 | 1.16 | 0.74–1.83 | 0.500 | |||
| >68 | ||||||
| Differentiation | ||||||
| Well | 0.695 | 0.38–1.24 | 0.219 | |||
| Moderate & poor | ||||||
| Node status | ||||||
| N0 | 1.372 | 0.853–2.2 | 0.192 | |||
| N1 & N2 | ||||||
| Clinical stage | ||||||
| Early (1 & 2) | 1.75 | 1.1–2.79 | 0.017 | 1.50 | 0.93–2.41 | 0.093 |
| Late (3 & 4) | ||||||
| Recurrence in 5 years | 1.73 | 1.1–2.72 | 0.016 | 1.63 | 1.03–2.58 | 0.034 |
| No | ||||||
| Yes | ||||||
| Depth of invasion | 1.49 | 0.81–2.74 | 0.194 | |||
| <4 cm | ||||||
| ≥4 cm | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapkota, D.; Vallenari, E.M.; Tamatam, D.; Schreurs, O.J.F.; Pandey, S.; Søland, T.M.; Costea, D.-E.; Tokozlu, B.; Åsheim, H.-C. Investigation of Cross-Reactivity of Anti-Ephrin-B2 Antibody to Other Ephrin-B Members in an Immunohistochemical Study in a Cohort of Oral Squamous Cell Carcinoma. Oral 2022, 2, 148-162. https://doi.org/10.3390/oral2020015
Sapkota D, Vallenari EM, Tamatam D, Schreurs OJF, Pandey S, Søland TM, Costea D-E, Tokozlu B, Åsheim H-C. Investigation of Cross-Reactivity of Anti-Ephrin-B2 Antibody to Other Ephrin-B Members in an Immunohistochemical Study in a Cohort of Oral Squamous Cell Carcinoma. Oral. 2022; 2(2):148-162. https://doi.org/10.3390/oral2020015
Chicago/Turabian StyleSapkota, Dipak, Evan M. Vallenari, Dhanalakshmi Tamatam, Olaf Joseph Franciscus Schreurs, Sushma Pandey, Tine Merete Søland, Daniela-Elena Costea, Burcu Tokozlu, and Hans-Christian Åsheim. 2022. "Investigation of Cross-Reactivity of Anti-Ephrin-B2 Antibody to Other Ephrin-B Members in an Immunohistochemical Study in a Cohort of Oral Squamous Cell Carcinoma" Oral 2, no. 2: 148-162. https://doi.org/10.3390/oral2020015
APA StyleSapkota, D., Vallenari, E. M., Tamatam, D., Schreurs, O. J. F., Pandey, S., Søland, T. M., Costea, D.-E., Tokozlu, B., & Åsheim, H.-C. (2022). Investigation of Cross-Reactivity of Anti-Ephrin-B2 Antibody to Other Ephrin-B Members in an Immunohistochemical Study in a Cohort of Oral Squamous Cell Carcinoma. Oral, 2(2), 148-162. https://doi.org/10.3390/oral2020015






