HIV-1 Gag Non-Cleavage Site PI Resistance Mutations Stabilize Protease/Gag Substrate Complexes In Silico via a Substrate-Clamp
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
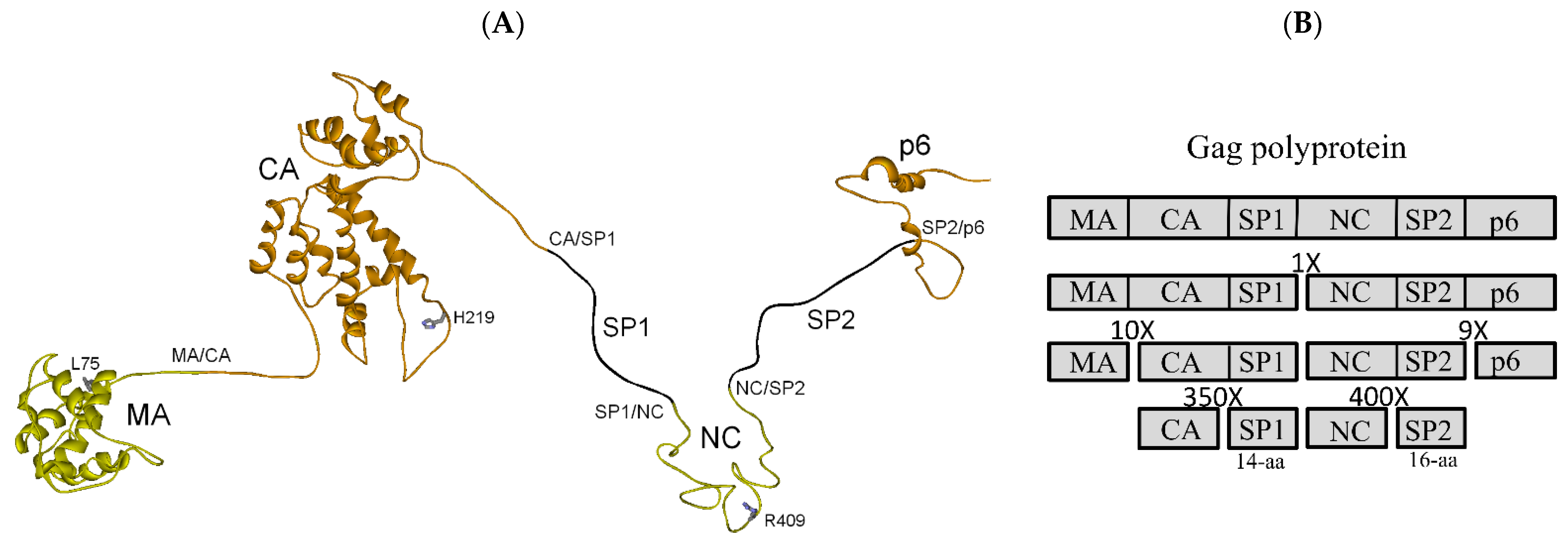
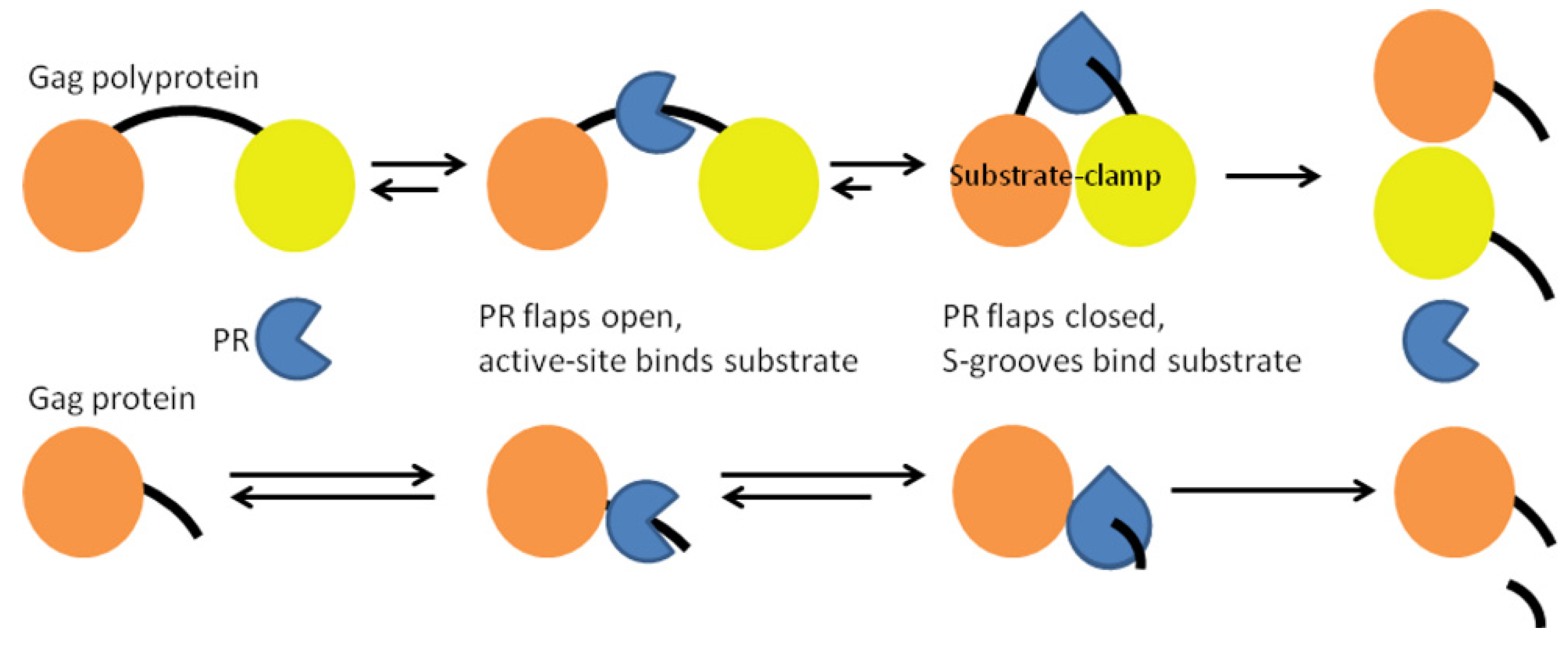
3.1. Full-Length Gag Polyprotein Structure-Based Model, Cleavage Order and Rates
3.2. HIV-1 PR Bound to the Five Gag Substrates
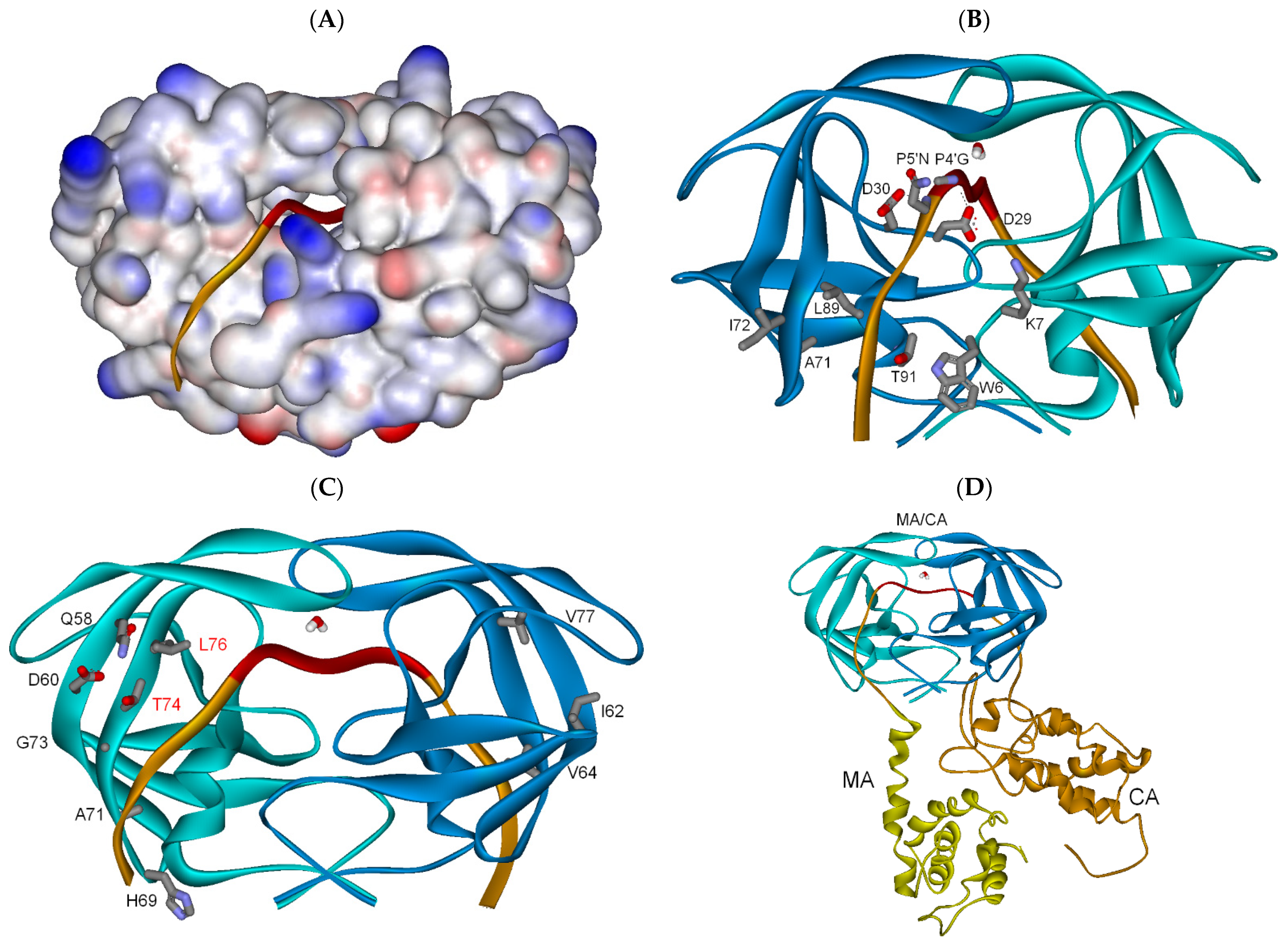
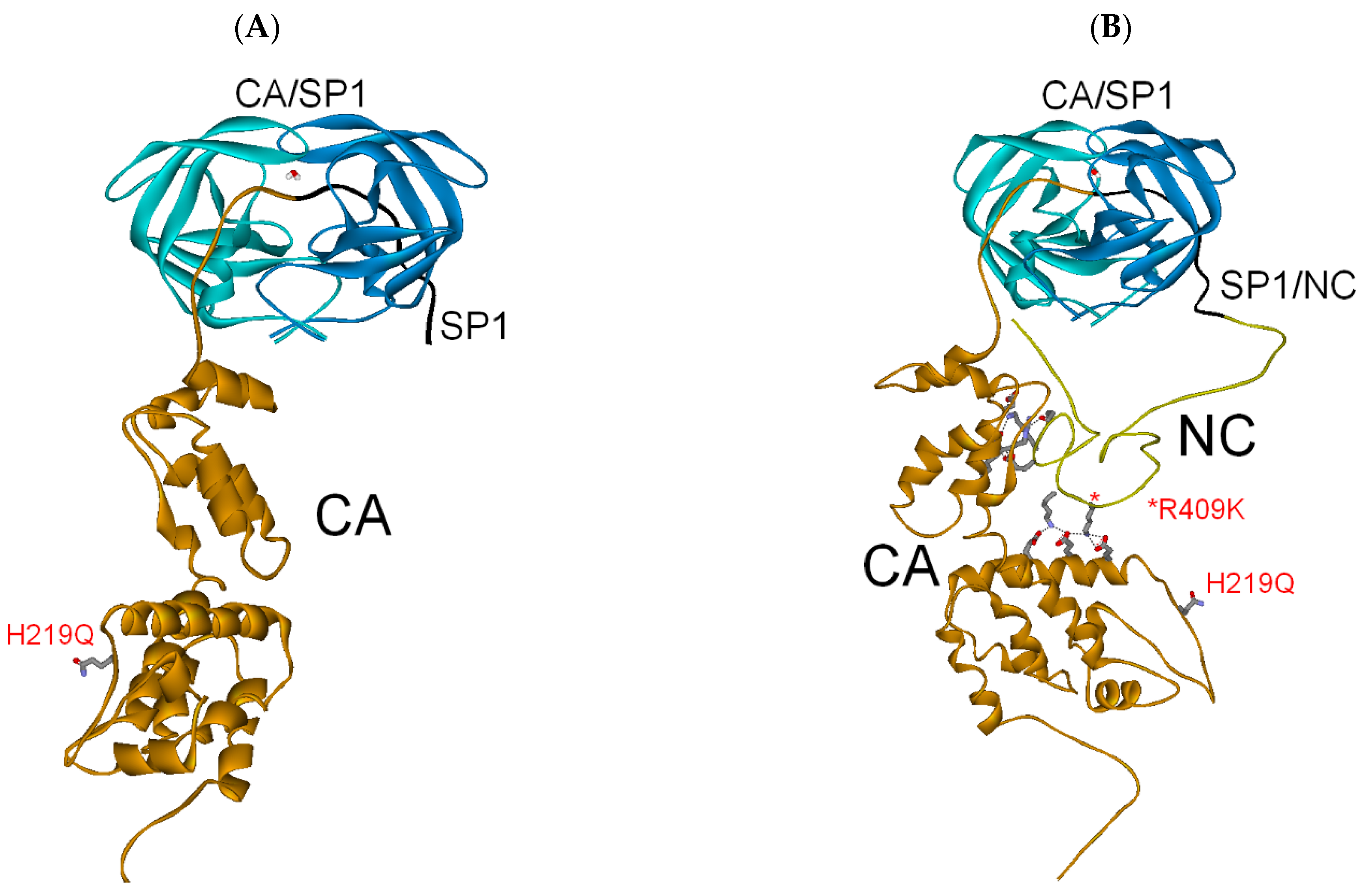
3.2.1. Full-Length Gag Substrate
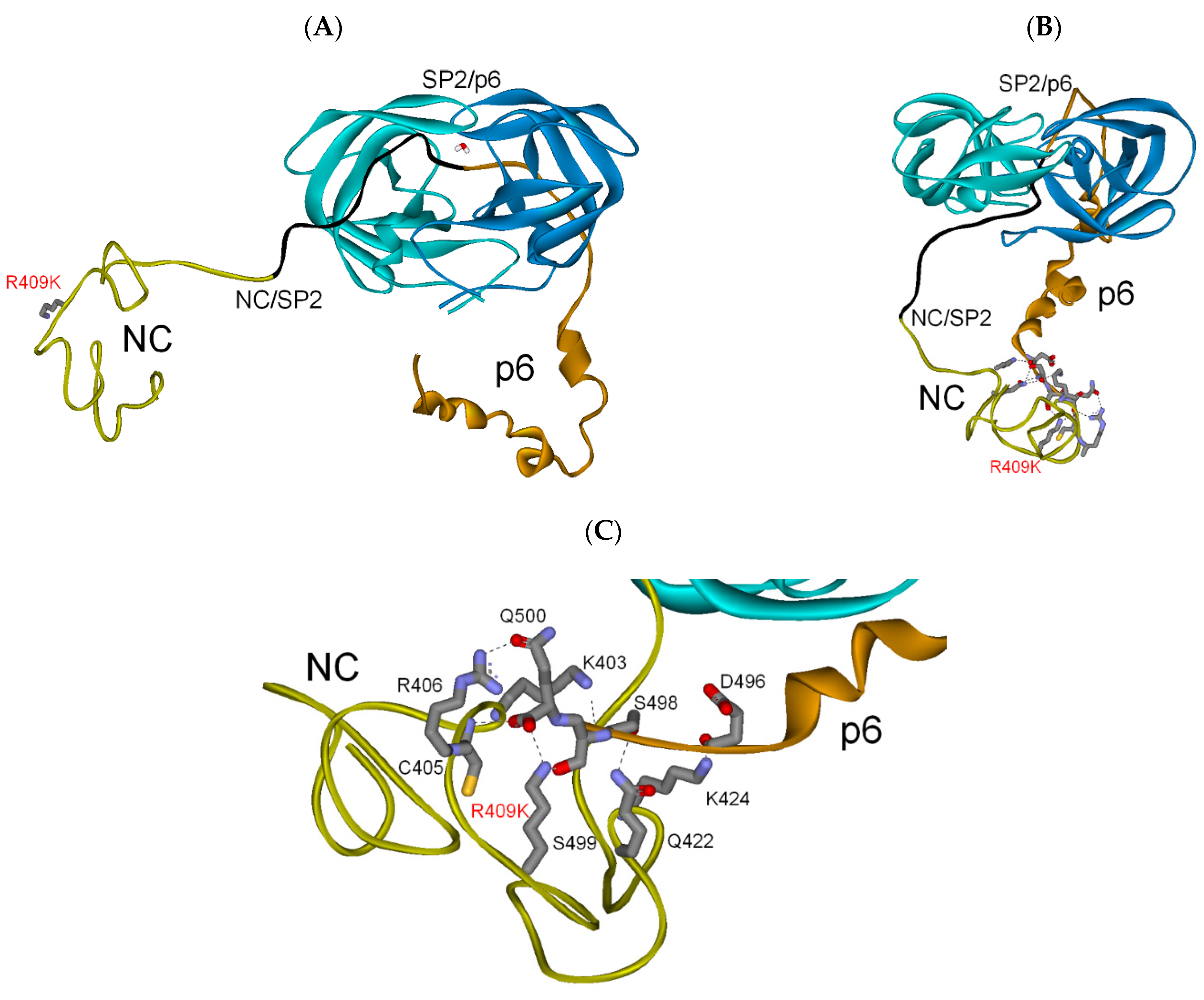
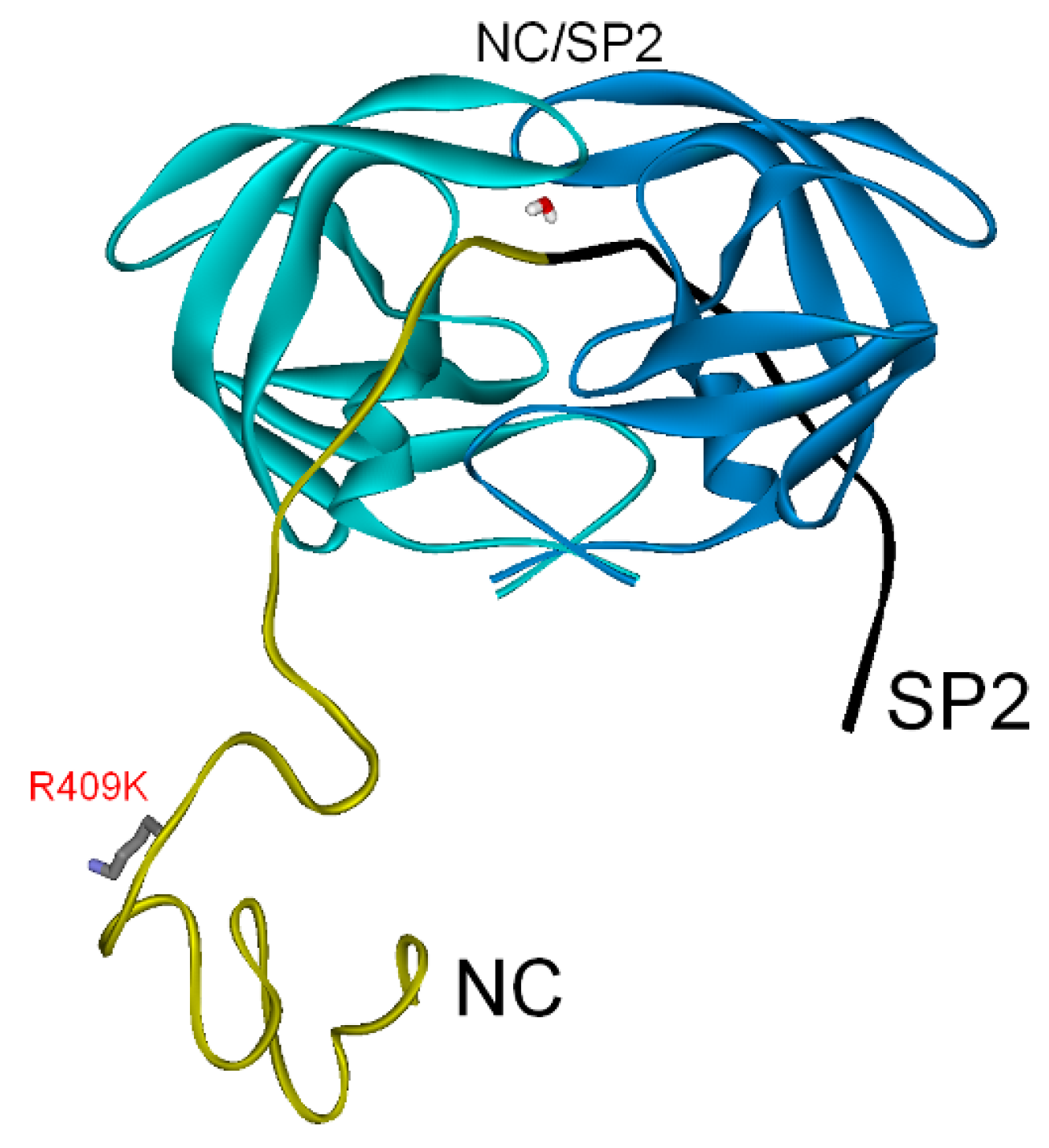
3.2.2. NC-SP2-p6 Substrate
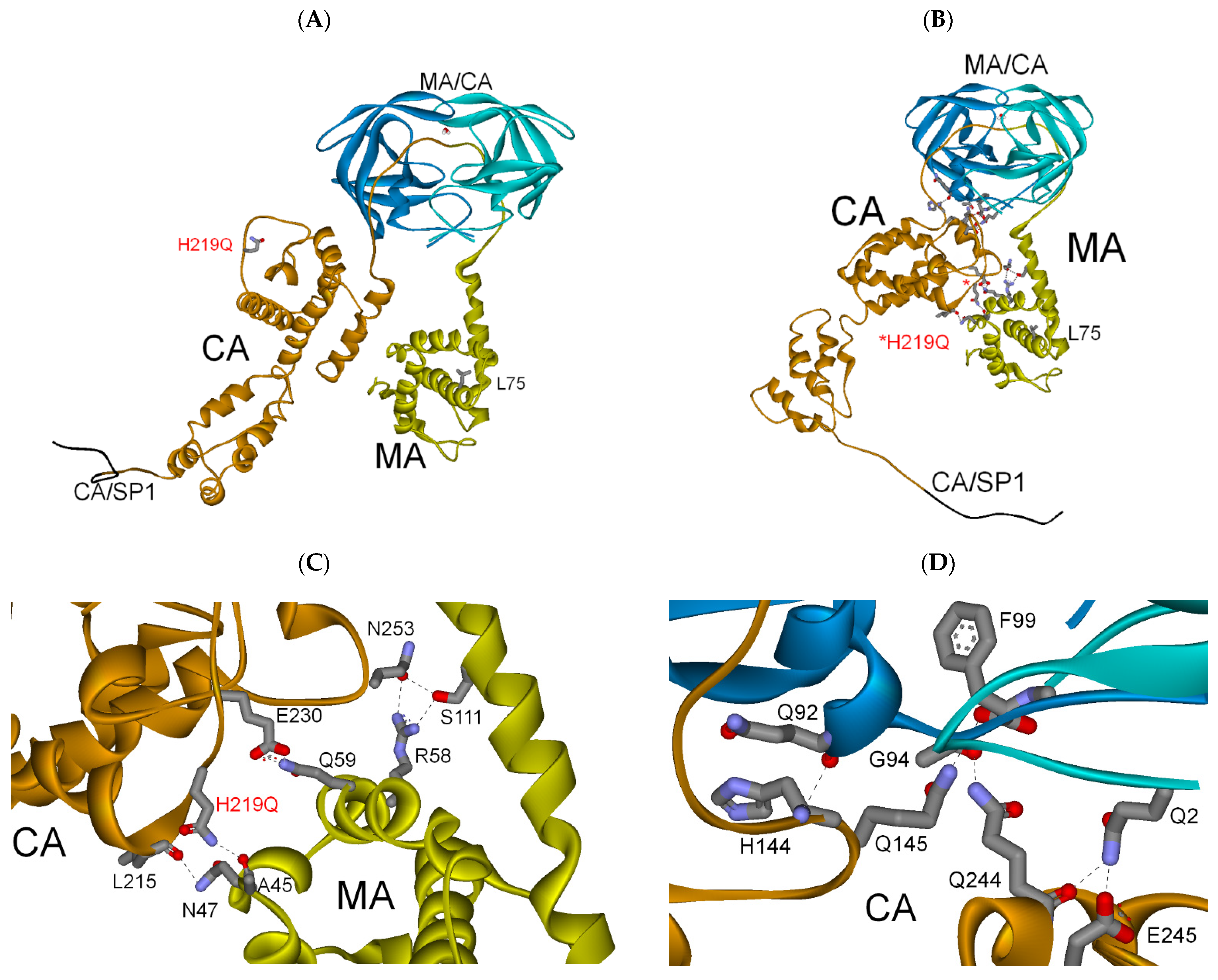
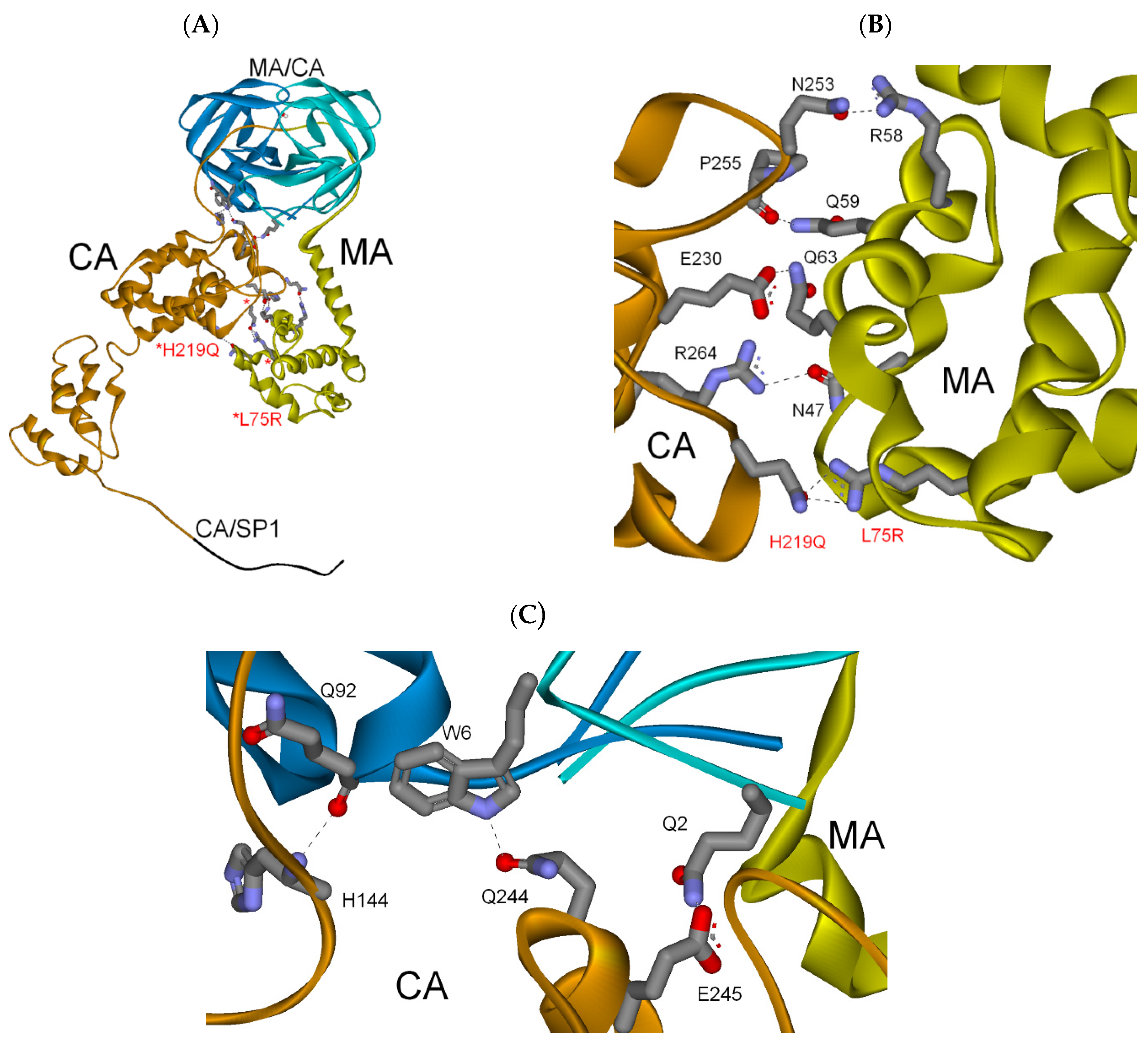
3.2.3. MA-CA-SP1 Substrate (Single and Double Mutation)
3.2.4. CA-SP1 Substrate
3.2.5. NC-SP2 Substrate
3.2.6. Interaction Energy Scores between PR and Substrates, and Substrate Subdomains
3.2.7. Gag Cleavage Order Factors
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef]
- Gallo, R.C.; Sarin, P.S.; Gelmann, E.P.; Robert-Guroff, M.; Richardson, E.; Kalyanaraman, V.S.; Mann, D.; Sidhu, G.D.; Stahl, R.E.; Zolla-Pazner, S.; et al. Isolation of Human T-Cell Leukemia Virus in Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Ratner, L.; Haseltine, W.; Patarca, R.; Livak, K.J.; Starcich, B.; Josephs, S.F.; Doran, E.R.; Rafalski, J.A.; Whitehorn, E.A.; Baumeister, K.; et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nat. Cell Biol. 1985, 313, 277–284. [Google Scholar] [CrossRef]
- Kuritzkes, D.R.; Quinn, J.B.; Benoit, S.L.; Shugarts, D.L.; Griffin, A.; Bakhtiari, M.; Poticha, D.; Eron, J.J.; Fallon, M.A.; Rubin, M. Drug resistance and virologic response in NUCA 3001, a randomized trial of lamivudine (3TC) versus zidovudine (ZDV) versus ZDV plus 3TC in previously untreated patients. AIDS 1996, 10, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Kuritzkes, D.R. Clinical significance of drug resistance in HIV-1 infection. AIDS 1996, 10, S27–S31. [Google Scholar] [CrossRef]
- Shafer, R.W.; Iversen, A.K.N.; Winters, M.A.; Aguiniga, E.; Katzenstein, D.; Merigan, T.C. Drug Resistance and Heterogeneous Long-Term Virologic Responses of Human Immunodeficiency Virus Type 1-Infected Subjects to Zidovudine and Didanosine Combination Therapy. J. Infect. Dis. 1995, 172, 70–78. [Google Scholar] [CrossRef] [PubMed]
- D’Aquila, R.T.; Johnson, V.A.; Welles, S.L.; Japour, A.J.; Kuritzkes, D.R.; DeGruttola, V.; Reichelderfer, P.S.; Coombs, R.W.; Crumpacker, C.S.; Kahn, J.O.; et al. Zidovudine Resistance and HIV-1 Disease Progression during Antiretroviral Therapy. Ann. Intern. Med. 1995, 122, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Lea, A.P.; Faulds, D. Ritonavir. Drugs 1996, 52, 541–546. [Google Scholar] [CrossRef]
- Erickson, J.W.; Burt, S.K. Structural Mechanisms of HIV Drug Resistance. Annu. Rev. Pharmacol. Toxicol. 1996, 36, 545–571. [Google Scholar] [CrossRef] [PubMed]
- Frieden, T.R.; Foti, K.E.; Mermin, J. Applying Public Health Principles to the HIV Epidemic—How Are We Doing? N. Engl. J. Med. 2015, 373, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Kohl, N.E.; Emini, E.A.; Schleif, W.A.; Davis, L.J.; Heimbach, J.C.; Dixon, R.A.; Scolnick, E.M.; Sigal, I.S. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 1988, 85, 4686–4690. [Google Scholar] [CrossRef] [PubMed]
- Bhat, T.N.; Baldwin, E.T.; Liu, B.; Cheng, Y.-S.E.; Erickson, J.W. Crystal structure of a tethered dimer of HIV-1 proteinase complexed with an inhibitor. Nat. Genet. 1994, 1, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Wensing, A.M.; Calvez, V.; Ceccherini-Silberstein, F.; Charpentier, C.; Günthard, H.F.; Paredes, R.; Shafer, R.W.; Richman, D.D. 2019 Update of the Drug Resistance Mutations in HIV-1. Top. Antivir. Med. 2019, 27, 111–121. [Google Scholar] [PubMed]
- Chang, M.W.; Torbett, B.E. Accessory Mutations Maintain Stability in Drug-Resistant HIV-1 Protease. J. Mol. Biol. 2011, 410, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Prabu-Jeyabalan, M.; Nalivaika, E.A.; King, N.M.; Schiffer, C.A. Structural Basis for Coevolution of a Human Immunodeficiency Virus Type 1 Nucleocapsid-p1 Cleavage Site with a V82A Drug-Resistant Mutation in Viral Protease. J. Virol. 2004, 78, 12446–12454. [Google Scholar] [CrossRef] [PubMed]
- Nijhuis, M.; Schuurman, R.; De Jong, D.; Erickson, J.; Gustchina, E.; Albert, J.; Schipper, P.; Gulnik, S.; Boucher, C.A.B. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 1999, 13, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Picado, J.; Savara, A.V.; Sutton, L.; D’Aquila, R.T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 1999, 73, 3744–3752. [Google Scholar] [CrossRef]
- Schock, H.B.; Garsky, V.M.; Kuo, L.C. Mutational Anatomy of an HIV-1 Protease Variant Conferring Cross-resistance to Protease Inhibitors in Clinical Trials. J. Biol. Chem. 1996, 271, 31957–31963. [Google Scholar] [CrossRef] [PubMed]
- Pazhanisamy, S.; Stuver, C.M.; Cullinan, A.B.; Margolin, N.; Rao, B.; Livingston, D.J. Kinetic Characterization of Human Immunodeficiency Virus Type-1 Protease-resistant Variants. J. Biol. Chem. 1996, 271, 17979–17985. [Google Scholar] [CrossRef]
- Gulnik, S.V.; Suvorov, L.I.; Liu, B.; Yu, B.; Anderson, B.; Mitsuya, H.; Erickson, J.W. Kinetic Characterization and Cross-Resistance Patterns Of HIV-1 Protease Mutants Selected under Drug Pressure. Biochemistry 1995, 34, 9282–9287. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.H.; Michael, S.F.; Wehbie, R.S.; Knigge, M.F.; Paul, D.A.; Everitt, L.; Kempf, D.J.; Norbeck, D.W.; Erickson, J.W.; Swanstrom, R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc. Natl. Acad. Sci. USA 1994, 91, 5597–5601. [Google Scholar] [CrossRef]
- Laco, G.S. Retroviral protease: Correlating substrate recognition with both selected and native inhibitor resistance. J. Mol. Biochem. 2017, 6, 45–63. [Google Scholar]
- Laco, G.S. HIV-1 protease substrate-groove: Role in substrate recognition and inhibitor resistance. Biochimie 2015, 118, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, L.; Tugarinov, V.; Louis, J.M.; Clore, G.M. Binding kinetics and substrate selectivity in HIV-1 protease−Gag interactions probed at atomic resolution by chemical exchange NMR. Proc. Natl. Acad. Sci. USA 2017, 114, E9855–E9862. [Google Scholar] [CrossRef]
- Miczi, M.; Diós, Á.; Bozóki, B.; Tőzsér, J.; Mótyán, J. Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model. Viruses 2021, 13, 1183. [Google Scholar] [CrossRef] [PubMed]
- Weikl, T.R.; Hemmateenejad, B. Accessory mutations balance the marginal stability of the HIV-1 protease in drug resistance. Proteins 2020, 88, 476–484. [Google Scholar] [CrossRef]
- Liu, Z.; Tran, T.T.; Pham, L.; Hu, L.; Bentz, K.; Savin, D.A.; Fanucci, G.E. Darunavir-Resistant HIV-1 Protease Constructs Uphold a Conformational Selection Hypothesis for Drug Resistance. Viruses 2020, 12, 1275. [Google Scholar] [CrossRef] [PubMed]
- Bastys, T.; Gapsys, V.; Walter, H.; Heger, E.; Doncheva, N.T.; Kaiser, R.; De Groot, B.L.; Kalinina, O.V. Non-active site mutants of HIV-1 protease influence resistance and sensitisation towards protease inhibitors. Retrovirology 2020, 17, 13. [Google Scholar] [CrossRef]
- Agniswamy, J.; Louis, J.M.; Roche, J.; Harrison, R.; Weber, I.T. Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics. PLoS ONE 2016, 11, e0168616. [Google Scholar] [CrossRef]
- Gatanaga, H.; Suzuki, Y.; Tsang, H.; Yoshimura, K.; Kavlick, M.F.; Nagashima, K.; Gorelick, R.J.; Mardy, S.; Tang, C.; Summers, M.F.; et al. Amino Acid Substitutions in Gag Protein at Non-cleavage Sites Are Indispensable for the Development of a High Multitude of HIV-1 Resistance against Protease Inhibitors. J. Biol. Chem. 2002, 277, 5952–5961. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Lindquist, J.N.; Kaplan, A.H.; Swanstrom, R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology 2005, 2, 66. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Simsic, J.; Loeb, D.D.; Everitt, L.; Hutchison, C.A., 3rd; Swanstorm, R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural re-quirements of the P1 amino acid. J. Biol. Chem. 1991, 266, 14539–14547. [Google Scholar] [CrossRef]
- Datta, S.; Curtis, J.E.; Ratcliff, W.; Clark, P.K.; Crist, R.; Lebowitz, J.; Krueger, S.; Rein, A. Conformation of the HIV-1 Gag Protein in Solution. J. Mol. Biol. 2007, 365, 812–824. [Google Scholar] [CrossRef]
- Tang, C.; Ndassa, Y.; Summers, M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag poly-protein. Nat. Struct. Biol. 2002, 9, 537–543. [Google Scholar] [PubMed]
- Su, C.T.-T.; Koh, D.W.-S.; Gan, S.K.-E. Reviewing HIV-1 Gag Mutations in Protease Inhibitors Resistance: Insights for Possible Novel Gag Inhibitor Designs. Molecules 2019, 24, 3243. [Google Scholar] [CrossRef] [PubMed]
- Fun, A.; Wensing, A.M.; Verheyen, J.; Nijhuis, M. Human Immunodeficiency Virus gag and protease: Partners in resistance. Retrovirology 2012, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Venzon, D.J.; Koh, Y.; Aoki-Ogata, H.; Miyakawa, T.; Yoshimura, K.; Maeda, K.; Mitsuya, H. Non-Cleavage Site Gag Mutations in Amprenavir-Resistant Human Immunodeficiency Virus Type 1 (HIV-1) Predispose HIV-1 to Rapid Acquisition of Amprenavir Resistance but Delay Development of Resistance to Other Protease Inhibitors. J. Virol. 2009, 83, 3059–3068. [Google Scholar] [CrossRef] [PubMed]
- Tritch, R.J.; Cheng, Y.E.; Yin, F.H.; Erickson-Viitanen, S. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J. Virol. 1991, 65, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Erickson-Viitanen, S.; Manfredi, J.; Viitanen, P.; Tribe, D.E.; Tritch, R.; Hutchisoniii, C.A.; Loeb, D.D.; Swanstrom, R. Cleavage of HIV-1gagPolyprotein Synthesized In Vitro: Sequential Cleavage by the Viral Protease. AIDS 1989, 5, 577–591. [Google Scholar] [CrossRef]
- Pettit, S.; Moody, M.D.; Wehbie, R.S.; Kaplan, A.H.; Nantermet, P.V.; Klein, C.A.; Swanstrom, R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994, 68, 8017–8027. [Google Scholar] [CrossRef] [PubMed]
- Laco, G.S. Evaluation of Two Models for Human Topoisomerase I Interaction with dsDNA and Camptothecin Derivatives. PLoS ONE 2011, 6, e24314. [Google Scholar] [CrossRef] [PubMed]
- Krausslich, H.G.; Ingraham, R.H.; Skoog, M.T.; Wimmer, E.; Pallai, P.V.; Carter, C.A. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc. Natl. Acad. Sci. USA 1989, 86, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Veronese, F.D.; Copeland, T.D.; Oroszlan, S.; Gallo, R.C.; Sarngadharan, M.G. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J. Virol. 1988, 62, 795–801. [Google Scholar] [CrossRef]
- Gatanaga, H.; Das, D.; Suzuki, Y.; Yeh, D.D.; Hussain, K.A.; Ghosh, A.K.; Mitsuya, H. Altered HIV-1 Gag Protein Interactions with Cyclophilin A (CypA) on the Acquisition of H219Q and H219P Substitutions in the CypA Binding Loop. J. Biol. Chem. 2006, 281, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.R.; Vajdos, F.; Yoo, S.; Worthylake, D.K.; Houseweart, M.; Sundquist, W.I.; Hill, C.P. Crystal Structure of Human Cyclophilin A Bound to the Amino-Terminal Domain of HIV-1 Capsid. Cell 1996, 87, 1285–1294. [Google Scholar] [CrossRef]
- Deshmukh, L.; Louis, J.M.; Ghirlando, R.; Clore, G.M. Transient HIV-1 Gag–protease interactions revealed by paramagnetic NMR suggest origins of compensatory drug resistance mutations. Proc. Natl. Acad. Sci. USA 2016, 113, 12456–12461. [Google Scholar] [CrossRef] [PubMed]
- Foley, B.T.; Korber, B.T.M.; Leitner, T.K.; Apetrei, C.; Hahn, B.; Mizrachi, I.; Mullins, J.; Rambaut, A.; Wolinsky, S. HIV Sequence Compendium 2018; Los Alamos National Laboratory: Los Alamos, NM, USA, 2018. [Google Scholar]
- Kuiken, C.; Foley, B.; Leitner, T.; Apetrei, C.; Hahn, B.; Mizrachi, I.; Mullins, J.; Rambaut, A.; Wolinsky, S.; Korber, B. HIV Sequence Compendium 2018; Los Alamos National Laboratory: Los Alamos, NM, USA, 2010. [Google Scholar]
- Kuiken, C.; Foley, B.; Freed, E.; Hahn, B.; Marx, P.; McCutchan, F.; Mellors, J.; Wolinsky, S.; Korber, B. HIV Sequence Compendium 2018; Los Alamos National Laboratory: Los Alamos, NM, USA, 2002. [Google Scholar]
- Korber, B.; Foley, B.; Leitner, T. Human retroviruses and AIDS 1997; Los Alamos National Laboratory: Los Alamos, NM, USA, 1997. [Google Scholar]
- Samsudin, F.; Gan, S.K.-E.; Bond, P.J. The impact of Gag non-cleavage site mutations on HIV-1 viral fitness from integrative modelling and simulations. Comput. Struct. Biotechnol. J. 2021, 19, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Su, C.T.-T.; Kwoh, C.-K.; Verma, C.S.; Gan, S.K.-E. Modeling the full length HIV-1 Gag polyprotein reveals the role of its p6 subunit in viral maturation and the effect of non-cleavage site mutations in protease drug resistance. J. Biomol. Struct. Dyn. 2017, 36, 4366–4377. [Google Scholar] [CrossRef]

| Gag Substrate (Cleavage Site) | PR/Gag | Gag Subdomains | Total (PI Res.) |
|---|---|---|---|
| Full-length Gag (SP1/NC) | −253 | 0 | −253 |
| PI res. R409K | −254 | −75 | (−329) |
| NC-SP2-p6 (SP2/p6) | −257 | 0 | −257 |
| PI res. R409K | −264 | −64 | (−328) |
| MA-CA-SP1 (MA/CA) | −280 | −1 | −281 |
| PI res. H219Q | −289 | −48 | (−337) |
| PI res. L75R/H219Q | −282 | −72 | (−354) |
| CA-SP1 * (CA/SP1) | −260 | NA | −260 |
| NC-SP2 * (NC/SP2) | −250 | NA | −250 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laco, G.S. HIV-1 Gag Non-Cleavage Site PI Resistance Mutations Stabilize Protease/Gag Substrate Complexes In Silico via a Substrate-Clamp. BioChem 2021, 1, 190-209. https://doi.org/10.3390/biochem1030015
Laco GS. HIV-1 Gag Non-Cleavage Site PI Resistance Mutations Stabilize Protease/Gag Substrate Complexes In Silico via a Substrate-Clamp. BioChem. 2021; 1(3):190-209. https://doi.org/10.3390/biochem1030015
Chicago/Turabian StyleLaco, Gary S. 2021. "HIV-1 Gag Non-Cleavage Site PI Resistance Mutations Stabilize Protease/Gag Substrate Complexes In Silico via a Substrate-Clamp" BioChem 1, no. 3: 190-209. https://doi.org/10.3390/biochem1030015
APA StyleLaco, G. S. (2021). HIV-1 Gag Non-Cleavage Site PI Resistance Mutations Stabilize Protease/Gag Substrate Complexes In Silico via a Substrate-Clamp. BioChem, 1(3), 190-209. https://doi.org/10.3390/biochem1030015





