Taxonomic Revision of the pulcherrima Clade of Metschnikowia (Fungi): Merger of Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Phenotypic Examination
2.3. Barcode Amplification and Sequencing
2.4. Sequence Analysis
2.5. Hybridisation and Segregation
3. Results
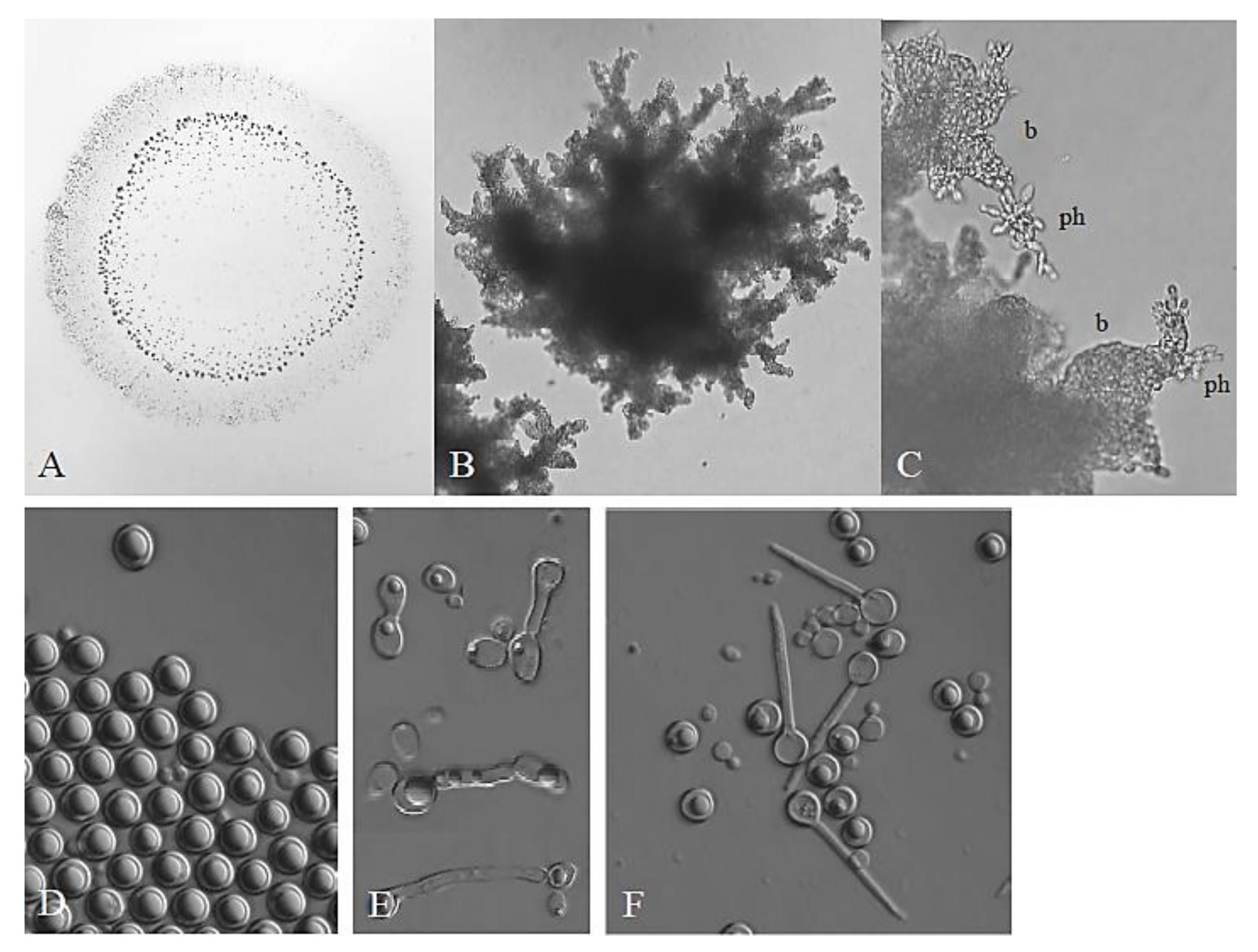
3.1. Phenotypic Examination: Lack of Taxonomically Relevant Differences
3.2. Pulcherrimin Production: A Variable and Unstable Trait
3.3. Analysis of Primary Barcodes
3.3.1. Analysis of D1/D2 Sequences: A Clade-Specific Consensus Sequence Can Be Defined
3.3.2. Analysis of ITS Sequences: No Clade-Specific Consensus Sequence Can Be Defined
3.4. Analysis of Secondary Barcodes
3.4.1. Analysis of ACT1 (Actin) Sequences: Insufficient Variability
3.4.2. Analysis of TEF1 (Translation Elongation Factor 1-α) Sequences: Intragenomic Diversity Obscures Species Boundaries
3.4.3. Analysis of RPB2 (Second Largest Subunit of RNA-Polymerase II) Sequences: Genomes Contain Genes of Different Phylogenetic Relationships
3.5. Hybridisation and Segregation
4. Discussion
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cook, A.H.; Slater, C.A. The structure of pulcherrimin. J. Chem. Soc. 1956, 4133–4135. [Google Scholar] [CrossRef]
- Sipiczki, M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtzman, C.P.; Droby, S. Metschnikowia fructicola, a new ascosporic yeast with potential for biocontrol of postharvest fruit rots. Syst. Appl. Microbiol. 2001, 24, 395–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Türkel, S.; Korukluoglu, M.; Yavuz, M. Biocontrol activity of the local strain of Metschnikowia pulcherrima on different postharvest pathogens. Biotechnol. Res. Int. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.M.; Choi, J.E.; Komakech, R.; Park, J.H.; Kim, D.W.; Cho, K.M.; Kang, S.M.; Choi, S.H.; Song, K.C.; Ryu, C.M.; et al. Characterization of a novel yeast species Metschnikowia persimmonesis KCTC 12991BP (KIOM G15050 type strain) isolated from a medicinal plant, Korean persimmon calyx (Diospyros kaki Thumb). AMB Express 2017, 7, 199. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yao, S.; Deng, L.; Ming, J.; Zeng, K. Metschnikowia citriensis sp. nov., a novel yeast species isolated from leaves with potential for biocontrol of postharvest fruit rot. Biol. Control 2018, 125, 15–19. [Google Scholar] [CrossRef]
- Gore-Lloyd, D.; Sumann, I.; Brachmann, A.O.; Schneeberger, K.; Ortiz-Merino, R.A.; Moreno-Beltrán, M.; Schläfli, M.; Kirner, P.; Santos Kron, A.; Rueda-Mejia, M.P.; et al. Snf2 controls pulcherriminic acid biosynthesis and antifungal activity of the biocontrol yeast Metschnikowia pulcherrima. Mol. Microbiol. 2019, 112, 317–332. [Google Scholar] [CrossRef] [Green Version]
- Saravanakumar, D.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Detection of enzymatic activity and partial sequence of a chitinase gene in Metschnikowia pulcherrima strain MACH1 used as postharvest biocontrol agent. Eur. J. Plant Pathol. 2009, 123, 183–193. [Google Scholar] [CrossRef]
- Oro, L.; Feliziani, E.; Ciani, M.; Romanazzi, G.; Comitini, F. Volatile organic compounds from Wickerhamomyces anomalus, Metschnikowia pulcherrima and Saccharomyces cerevisiae inhibit growth of decay causing fungi and control postharvest diseases of strawberries. Int. J. Food Microbiol. 2018, 265, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Metschnikowia pulcherrima and related pulcherrimin-producing yeasts: Fuzzy species boundaries and complex antimicrobial antagonism. Microorganisms 2020, 8, 1029. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar, E.; Ward, T.J. Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Antonie Van Leeuwenhoek 2018, 111, 2017–2035. [Google Scholar] [CrossRef]
- Pitt, J.I.; Miller, M.W. Sporulation in Candida pulcherrima, Candida reukaufii and Chlamydozyma species; their relationship with Metschnikowia. Mycologia 1968, 60, 663–685. [Google Scholar] [CrossRef]
- Molnar, O.; Prillinger, H. Analysis of yeast isolates related to Metschnikowia pulcherrima using the partialsequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst. Appl. Microbiol. 2005, 28, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.L.; Zhang, L.Q.; Wang, Q.M.; Zhang, J.S.; Bai, F.Y. Metschnikowia sinensis sp. nov., Metschnikowia zizyphicola sp. nov. and Metschnikowia shanxiensis sp. nov., novel yeast species from jujube fruit. Int. J. Syst. Evol. Microbiol. 2006, 56, 2245–2250. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M.; Pfliegler, W.P.; Holb, I.J. Metschnikowia species share a pool of diverse rRNA genes differing in regions that determine hairpin-loop structures and evolve by reticulation. PLoS ONE 2013, 8, e67384. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M.; Horvath, E.; Pfliegler, W.P. Birth-and-death evolution and reticulation of ITS segments of Metschnikowia andauensis and Metschnikowia fructicola rDNA repeats. Front. Microbiol. 2018, 9, 1193. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M. When barcoding fails: Genome chimerisation (admixing) and reticulation obscure phylogenetic and taxonomic relationships. Mol. Ecol. Resour. 2022. [Google Scholar] [CrossRef]
- Wheeler, Q.D.; Platnick, N.I. The Phylogenetic species concept (sensu Wheeler and Platnick). In Species Concepts and Phylogenetic Theory: A Debate; Wheeler, Q.D., Meier, R., Eds.; Columbia University Press: New York, NY, USA, 2000; pp. 55–69. [Google Scholar]
- Zachos, F.E. Species Concepts in Biology: Historical Development, Theoretical Foundations and Practical Relevance; Springer Nature: Cham, Switzerland, 2016. [Google Scholar]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Aime, M.C.; Begerow, D.; Gabaldón, T.; Heitman, J.; Kemler, M.; Khayhan, K.; Lachance, M.-A.; Louis, E.J.; Sun, S.; et al. The evolving species concepts used for yeasts: From phenotypes and genomes to speciation networks. Fungal Divers. 2021, 109, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Yurkov, A.; Alves, A.; Bai, F.Y.; Boundy-Mills, K.; Buzzini, P.; Čadež, N.; Cardinali, G.; Casaregola, S.; Chaturvedi, V.; Collin, V.; et al. Nomenclatural issues concerning cultured yeasts and other fungi: Why it is important to avoid unneeded name changes. IMA Fungus 2021, 12, 18. [Google Scholar] [CrossRef]
- Mayr, E. Speciation phenomena in birds. Am. Nat. 1940, 74, 249–278. [Google Scholar] [CrossRef]
- Naumov, G. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 1996, 17, 295–302. [Google Scholar] [CrossRef]
- Sipiczki, M. Interspecies hybridisation and genome chimerisation in Saccharomyces: Combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 2018, 9, 3071. [Google Scholar] [CrossRef]
- Sipiczki, M. Pichia bruneiensis sp. nov., a biofilm-producing dimorphic yeast species isolated from flowers in Borneo. Int. J. Syst. Evol. Microbiol. 2012, 62, 3099–3104. [Google Scholar] [CrossRef]
- Sipiczki, M. Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int. J. Syst. Evol. Microbiol. 2003, 53, 2079–2083. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K. Fusarium and its near relatives. In The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CAB International: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols. A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Thompson, J.D.; Higgions, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), Version 3.67; Distributed by the Author; Department of Genome Sciences, University of Washington: Seattle, DC, USA, 2007. [Google Scholar]
- Lachance, M.A. Metschnikowia Kamienski (1899). In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 575–620. [Google Scholar]
- Kaur, D.; Singhal, V.K. Meiotic abnormalities affect genetic constitution and pollen viability in dicots from Indian cold deserts. BMC Plant Biol. 2019, 19, 10. [Google Scholar] [CrossRef]
- Mason, A.S.; Wendel, J.F. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 2020, 11, 1014. [Google Scholar] [CrossRef]
- Pitt, J.I.; Miller, M.W. The parasexual cycle in yeasts from the genus Metschnikowia. Mycologia 1970, 62, 462–473. [Google Scholar] [CrossRef]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K.; et al. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Regnum Vegetabile; Koeltz Botanical Books: Glashütten, Germany, 2018; Volume 159. [Google Scholar]
- Mayr, E. Animal Species and Evolution; Harvard University Press: Cambridge, MA, USA, 1963. [Google Scholar]
- Mayr, E. Populations, Species, and Evolution; Harvard University Press: Cambridge, MA, USA, 1970. [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sipiczki, M. Taxonomic Revision of the pulcherrima Clade of Metschnikowia (Fungi): Merger of Species. Taxonomy 2022, 2, 107-123. https://doi.org/10.3390/taxonomy2010009
Sipiczki M. Taxonomic Revision of the pulcherrima Clade of Metschnikowia (Fungi): Merger of Species. Taxonomy. 2022; 2(1):107-123. https://doi.org/10.3390/taxonomy2010009
Chicago/Turabian StyleSipiczki, Matthias. 2022. "Taxonomic Revision of the pulcherrima Clade of Metschnikowia (Fungi): Merger of Species" Taxonomy 2, no. 1: 107-123. https://doi.org/10.3390/taxonomy2010009
APA StyleSipiczki, M. (2022). Taxonomic Revision of the pulcherrima Clade of Metschnikowia (Fungi): Merger of Species. Taxonomy, 2(1), 107-123. https://doi.org/10.3390/taxonomy2010009





