Genomic Insights into the Taxonomy and Metabolism of the Cyanobacterium Pannus brasiliensis CCIBt3594
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture and DNA Extraction
2.2. Genome Sequencing and Assembly
2.3. Functional Annotation
2.4. Potential for Nitrogen Fixation
2.5. 16S rRNA Gene Phylogeny
2.6. Phylogenomic Analysis
3. Results and Discussion
3.1. Genome Assembly and Annotation
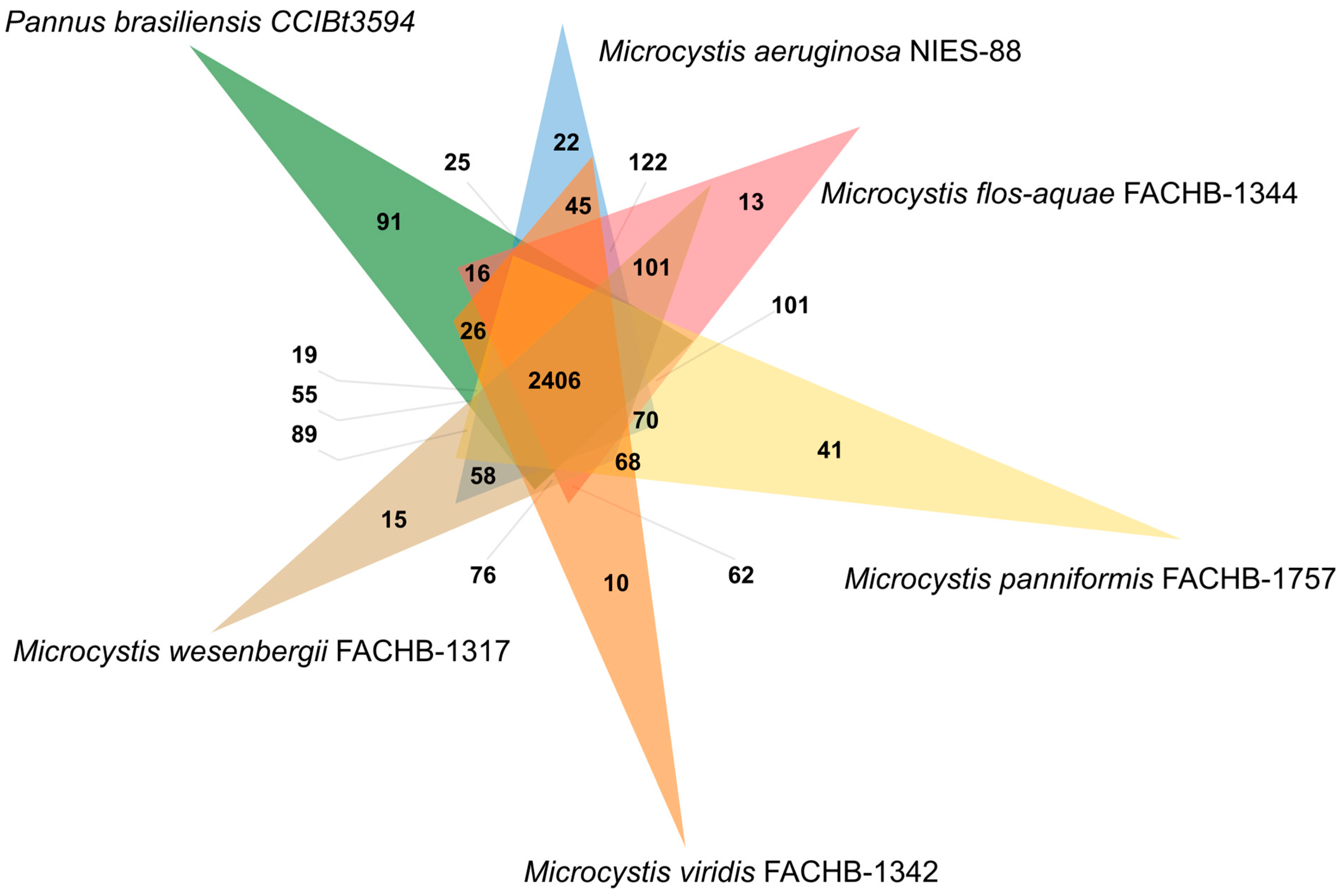
3.2. Identification of Genes Involved in Secondary Metabolites Products
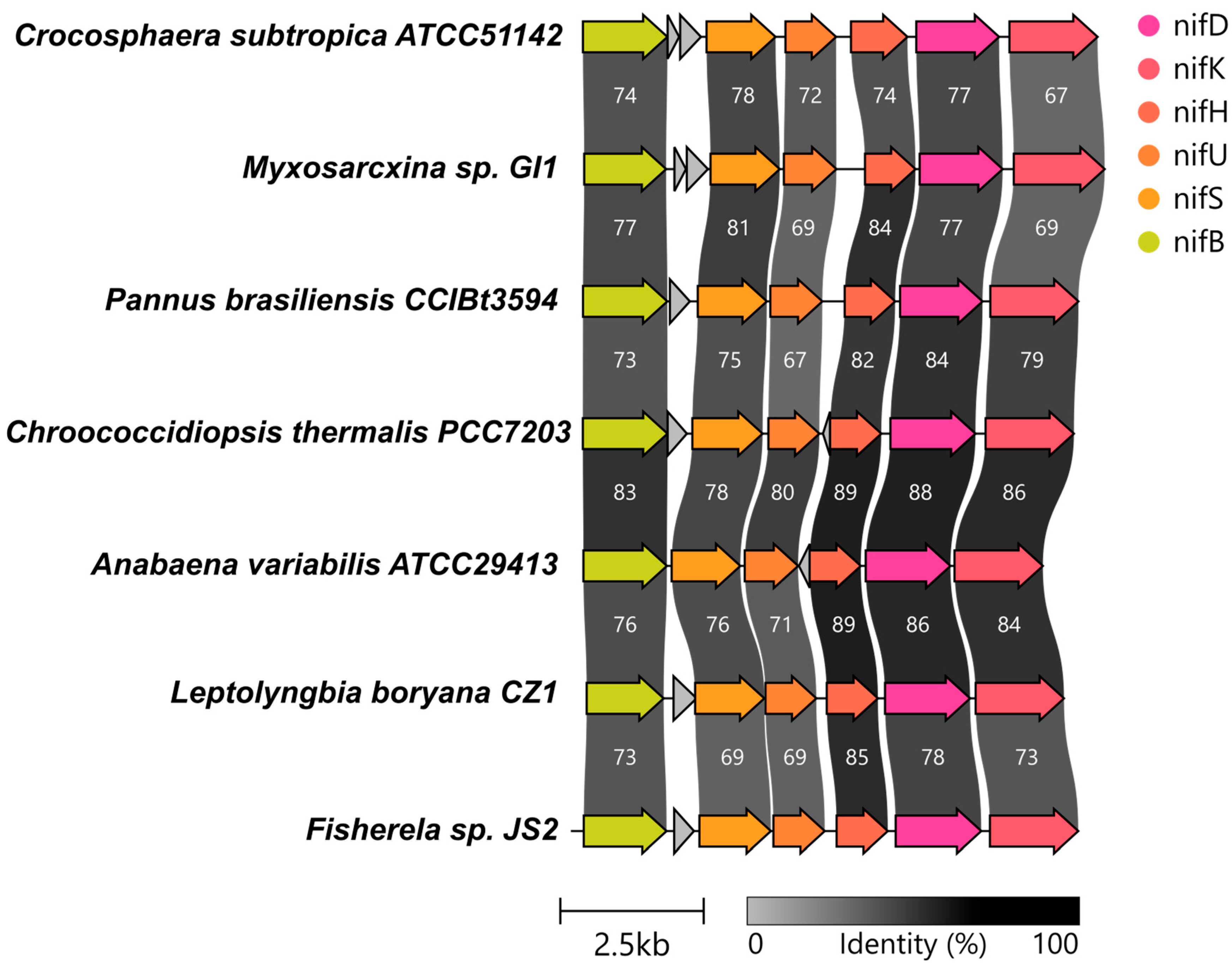
3.3. Genes Involved in Nitrogen Fixation
3.4. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Archibald, J.M. The Puzzle of Plastid Evolution. Curr. Biol. 2009, 19, R81–R88. [Google Scholar] [CrossRef]
- Schirrmeister, B.E.; Gugger, M.; Donoghue, P.C.J. Cyanobacteria and the Great Oxidation Event: Evidence from genes and fossils. Palaeontology 2015, 58, 769–785. [Google Scholar] [CrossRef]
- Maya, Y.; López-Cortés, A.; Soeldner, A. Cyanobacterial Microbiotic Crusts in Eroded Soils of a Tropical Dry Forest in the Baja California Peninsula, Mexico. Geomicrobiol. J. 2002, 19, 505–518. [Google Scholar] [CrossRef]
- Kobos, J.; Błaszczyk, A.; Hohlfeld, N.; Toruńska-Sitarz, A.; Krakowiak, A.; Hebel, A.; Sutryk, K.; Grabowska, M.; Toporowska, M.; Kokociński, M.; et al. Cyanobacteria and Cyanotoxins in Polish Freshwater Bodies. Oceanol. Hydrobiol. Stud. 2013, 42, 358–378. [Google Scholar] [CrossRef]
- Costa, M.; Garcia, M.; Costa-Rodrigues, J.; Costa, M.S.; Ribeiro, M.J.; Fernandes, M.H.; Barros, P.; Barreiro, A.; Vasconcelos, V.; Martins, R. Exploring Bioactive Properties of Marine Cyanobacteria Isolated from the Portuguese Coast: High Potential as a Source of Anticancer Compounds. Mar. Drugs 2014, 12, 98–114. [Google Scholar] [CrossRef]
- Mataloni, G.; Komárek, J. Gloeocapsopsis aurea, a New Subaerophytic Cyanobacterium from Maritime Antarctica. Polar Biol. 2004, 27, 623–628. [Google Scholar] [CrossRef]
- Bravakos, P.; Kotoulas, G.; Skaraki, K.; Pantazidou, A.; Economou-Amilli, A. A Polyphasic Taxonomic Approach in Isolated Strains of Cyanobacteria from Thermal Springs of Greece. Mol. Phylogenet. Evol. 2016, 98, 147–160. [Google Scholar] [CrossRef]
- Bergman, B.; Johansson, C.; Soderback, E. Tansley Review No. 42: The Nostoc—Gunnera Symbiosis. Rev. Lit. Arts Am. 1992, 42, 379–400. [Google Scholar] [CrossRef]
- Baker, J.A.; Entsch, B.; McKay, D.B. The Cyanobiont in an Azolla Fern is Neither Anabaena nor Nostoc. FEMS Microbiol. Lett. 2003, 229, 43–47. [Google Scholar] [CrossRef]
- Gutierrez-Garcıa, K.; Bustos-Dıaz, E.D.; Corona-Gomez, J.A.; Ramos-Aboites, H.E.; Selem-Mojica, N.; Cruz-Morales, P.; Perez-Farrera, M.A.; Barona-Gomez, F.; Cibrian-Jaramillo, A. Cycad Coralloid Roots Contain Bacterial Communities Including Cyanobacteria and Caulobacter spp. That Encode Niche-Specific Biosynthetic Gene Clusters. Genome Biol. Evol. 2019, 11, 319–334. [Google Scholar] [CrossRef]
- Hilton, J.A.; Foster, R.A.; James Tripp, H.; Carter, B.J.; Zehr, J.P.; Villareal, T.A. Genomic Deletions Disrupt Nitrogen Metabolism Pathways of a Cyanobacterial Diatom Symbiont. Nat. Commun. 2013, 4, 1767. [Google Scholar] [CrossRef]
- Rikkinen, J. Algal and Cyanobacteria Symbioses. In Symbiotic Cyanobacteria in Lichens; World Scientific: London, UK, 2017; Chapter 5; pp. 147–167. [Google Scholar]
- Konstantinou, D.; Gerovasileiou, V.; Voultsiadou, E.; Gkelis, S. Sponges-Cyanobacteria Associations: Global Diversity Overview and New Data from the Eastern Mediterranean. PLoS ONE 2018, 13, e0195001. [Google Scholar] [CrossRef] [PubMed]
- Oswald, F.; Schmitt, F.; Leutenegger, A.; Ivanchenko, S.; D’Angelo, C.; Salih, A.; Maslakova, S.; Bulina, M.; Schirmbeck, R.; Nienhaus, G.U.; et al. Contributions of Host and Symbiont Pigments to the Coloration of Reef Corals. FEBS J. 2007, 274, 1102–1122. [Google Scholar] [CrossRef] [PubMed]
- Berman-Frank, I.; Lundgren, P.; Falkowski, P. Nitrogen Fixation and Photosynthetic Oxygen Evolution in Cyanobacteria. Res. Microbiol. 2003, 154, 157–164. [Google Scholar] [CrossRef]
- Hartmann, M.; Gomez-Pereira, P.; Grob, C.; Ostrowski, M.; Scanlan, D.J.; Zubkov, M.V. Efficient CO2 Fixation by Surface Prochlorococcus in the Atlantic Ocean. ISME J. 2014, 8, 2280–2289. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Menakha, M. Pharmaceutical Applications of Cyanobacteria—A Review. J. Acute Med. 2015, 5, 15–23. [Google Scholar] [CrossRef]
- Zahra, Z.; Choo, D.H.; Lee, H.; Parveen, A. Cyanobacteria: Review of Current Potentials and Applications. Environments 2020, 7, 13. [Google Scholar] [CrossRef]
- Hickel, B. Two New Chroococcal Cyanophytes from a Brackish Environment (Schlei-Fjord) Germany. Algol. Stud./Arch. Hydrobiol. 1991, 64, 97–104. [Google Scholar]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic Classification of Cyanoprokaryotes (Cyanobacterial genera) 2014, Using a Polyphasic Approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Da Silva Malone, C.F.; Alvarenga, D.O.; Fiore, M.F.; Sant’Anna, C.L. Towards a Phylogenetic Position for the Morphologically-Defined Genus Pannus (Cyanobacteria). Nova Hedwig. 2014, 99, 511–524. [Google Scholar] [CrossRef]
- Werner, V.R.; Martins, M.D.; da Silva, F.O. Cyanobacteria from a Brazilian Subtropical Freshwater Water Body. Braz. J. Bot. 2018, 41, 901–921. [Google Scholar] [CrossRef]
- Willis, A.; Woodhouse, J.N. Defining Cyanobacterial Species: Diversity and Description Through Genomics. Crit. Rev. Plant Sci. 2020, 39, 101–124. [Google Scholar] [CrossRef]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; de Marsac, N.T.; Rippka, R.; et al. Improving the Coverage of the Cyanobacterial Phylum Using Diversity-Driven Genome Sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Abreu, V.A.; Popin, R.V.; Alvarenga, D.O.; Schaker, P.D.; Hoff-Risseti, C.; Varani, A.M.; Fiore, M.F. Genomic and Genotypic Characterization of Cylindrospermopsis raciborskii: Toward an Intraspecific Phylogenetic Evaluation by Comparative Genomics. Front. Microbiol. 2018, 9, 306. [Google Scholar] [CrossRef]
- Pérez-Carrascal, O.M.; Terrat, Y.; Giani, A.; Fortin, N.; Greer, C.W.; Tromas, N.; Shapiro, B.J. Coherence of Microcystis Species Revealed Through Population Genomics. ISME J. 2019, 13, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.B. Genome Sequences as the Type Material for Taxonomic Descriptions of Prokaryotes. Syst. Appl. Microbiol. 2015, 38, 217–222. [Google Scholar] [CrossRef]
- Ramasamy, D.; Mishra, A.K.; Lagier, J.C.; Padhmanabhan, R.; Rossi, M.; Sentausa, E.; Raoult, D.; Fournier, P.E. A Polyphasic Strategy Incorporating Genomic Data for the Taxonomic Description of Novel Bacterial Species. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 384–391. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Alvarenga, D.O.; Fiore, M.F.; Varani, A.M. A Metagenomic Approach to Cyanobacterial Genomics. Front. Microbiol. 2017, 8, 809. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome Analysis Using the Kraken Software Suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Gilchrist, C.L.; Chooi, Y.H. Clinker & Clustermap.js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic Use of DNA G+C Content and DNA–DNA Hybridization in the Genomic Age. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 352–356. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An Integrated Platform for Exploring and Visualizing Orthologous Data Across Genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- De Las Rivas, J.; Lozano, J.J.; Ortiz, A.R. Comparative Analysis of Chloroplast Genomes: Functional Annotation, Genome-Based Phylogeny, and Deduced Evolutionary Patterns. Genome Res. 2002, 12, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Cornet, L.; Meunier, L.; Van Vlierberghe, M.; Léonard, R.R.; Durieu, B.; Lara, Y.; Misztak, A.; Sirjacobs, D.; Javaux, E.J.; Philippe, H.; et al. Consensus Assessment of the Contamination Level of Publicly Available Cyanobacterial Genomes. PLoS ONE 2018, 13, e0200323. [Google Scholar] [CrossRef] [PubMed]
- Frangeul, L.; Quillardet, P.; Castets, A.M.; Humbert, J.F.; Matthijs, H.C.; Cortez, D.; Tolonen, A.; Zhang, C.-C.; Gribaldo, S.; Kehr, J.-C.; et al. Highly Plastic Genome of Microcystis aeruginosa PCC 7806, a Ubiquitous Toxic Freshwater Cyanobacterium. BMC Genom. 2008, 9, 274. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Guan, R.; Zhang, H.-J.; Li, H.; Xiao, P.; Yu, G.-L.; Du, L.; Cao, D.-M.; Zhu, B.-C.; Li, R.-H.; et al. Complete Genome Sequence and Genomic Characterization of Microcystis panniformis FACHB 1757 by Third-Generation Sequencing. Stand. Genom. Sci. 2016, 11, 11. [Google Scholar] [CrossRef]
- Lefler, F.W.; Barbosa, M.; Berthold, D.E.; Laughinghouse IV, H.D. Genome Sequences of Two Microcystis aeruginosa (Chroococcales, Cyanobacteria) Strains from Florida (United States) with Disparate Toxigenic Potentials. Microbiol. Resour. Announc. 2020, 9, 10–1128. [Google Scholar] [CrossRef]
- Shestakov, S.V.; Karbysheva, E.A. The Role of Viruses in the Evolution of Cyanobacteria. Biol. Bull. Rev. 2015, 5, 527–537. [Google Scholar] [CrossRef]
- Melnikov, O.; Zaritsky, A.; Zarka, A.; Boussiba, S.; Malchin, N.; Yagil, E.; Kolot, M. Site-specific Recombination in the Cyanobacterium Anabaena sp. Strain PCC 7120 Catalyzed by the Integrase of Coliphage HK022. J. Bacteriol. 2009, 191, 4458–4464. [Google Scholar] [CrossRef]
- Zhong, K.X.; Suttle, C.A.; Baudoux, A.C.; Derelle, E.; Colombet, J.; Cho, A.; Caleta, J.; Six, C.; Jacquet, S. A New Freshwater Cyanosiphovirus Harboring Integrase. Front. Microbiol. 2018, 9, 2204. [Google Scholar] [CrossRef]
- Kultschar, B.; Llewellyn, C. Secondary metabolites in cyanobacteria. In Secondary Metabolites—Sources and Applications; Vijayakumar, R., Ed.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Grant Burgess, J.; Wright, P.C. Marine Cyanobacteria—A Prolific Source of Natural Products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural Products from Cyanobacteria: Focus on Beneficial Activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef]
- Yadav, P.; Singh, R.P.; Rana, S.; Joshi, D.; Kumar, D.; Bhardwaj, N.; Gupta, R.K.; Kumar, A. Mechanisms of Stress Tolerance in Cyanobacteria under Extreme Conditions. Stresses 2022, 2, 531–549. [Google Scholar] [CrossRef]
- Micallef, M.L.; D’Agostino, P.M.; Al-Sinawi, B.; Neilan, B.A.; Moffitt, M.C. Exploring Cyanobacterial Genomes for Natural Product Biosynthesis Pathways. Mar. Genom. 2015, 21, 1–12. [Google Scholar] [CrossRef]
- Coates, R.C.; Podell, S.; Korobeynikov, A.; Lapidus, A.; Pevzner, P.; Sherman, D.H.; Allen, E.E.; Gerwick, W.H. Characterization of Cyanobacterial Hydrocarbon Composition and Distribution of Biosynthetic Pathways. PLoS ONE 2014, 9, e85140. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.; Gu, L.; Zhu, H.; Gibbons, W.; Zhou, R. Identification of Two Genes Required for Heptadecane Production in a N2-Fixing Cyanobacterium Anabaena sp. Strain PCC 7120. AMB Express 2018, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Nitnaware, K.M.; Raskar, K.B.; Agarwal, G.; Montes, R.A.C.; Chopra, R.; López-Arredondo, D.L.; Nikam, T.D.; Patil, G.B. Whole-Genome Characterization and Comparative Genomics of a Novel Freshwater Cyanobacteria Species: Pseudanabaena punensis. Mol. Phylogenet. Evol. 2021, 164, 107272. [Google Scholar] [CrossRef] [PubMed]
- Kudo, H.; Hayashi, Y.; Arai, M. Improving Hydrocarbon Production by Engineering Cyanobacterial Acyl-(Acyl Carrier Protein) Reductase. Biotechnol. Biofuels 2019, 12, 291. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Dong, S.H.; Sarkar, S.; Nair, S.K.; Schmidt, E.W. The Biochemistry and Structural Biology of Cyanobactin Pathways: Enabling Combinatorial Biosynthesis. Methods Enzymol. 2018, 604, 113–163. [Google Scholar] [CrossRef] [PubMed]
- Pearson, A.; Rusch, D.B. Distribution of Microbial Terpenoid Lipid Cyclases in the Global Ocean Metagenome. ISME J. 2009, 3, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Poulter, C.D. Cloning, Solubilization, and Characterization of Squalene Synthase from Thermosynechococcus elongatus BP-1. J. Bacteriol. 2008, 190, 3808–3816. [Google Scholar] [CrossRef]
- Englund, E.; Pattanaik, B.; Ubhayasekera, S.J.K.; Stensjö, K.; Bergquist, J.; Lindberg, P. Production of Squalene in Synechocystis sp. PCC 6803. PLoS ONE 2014, 9, e90270. [Google Scholar] [CrossRef]
- Fagundes, M.B.; Alvarez-Rivera, G.; Vendruscolo, R.G.; Voss, M.; da Silva, P.A.; Barin, J.S.; Jacob-Lopes, E.; Zepka, L.Q.; Wagner, R. Green microsaponification-based method for gas chromatography determination of sterol and squalene in cyanobacterial biomass. Talanta 2021, 224, 121793. [Google Scholar] [CrossRef]
- Pattanaik, B.; Englund, E.; Nolte, N.; Lindberg, P. Introduction of a Green Algal Squalene Synthase Enhances Squalene Accumulation in a Strain of Synechocystis sp. PCC 6803. Metab. Eng. Commun. 2020, 10, e00125. [Google Scholar] [CrossRef]
- Fagundes, M.B.; Falk, R.B.; Facchi, M.M.X.; Vendruscolo, R.G.; Maroneze, M.M.; Zepka, L.Q.; Jacob-Lopes, E.; Wagner, R. Insights in Cyanobacteria Lipidomics: A Sterols Characterization from Phormidium autumnale Biomass in Heterotrophic Cultivation. Food Res. Int. 2019, 119, 777–784. [Google Scholar] [CrossRef]
- Jiang, Y.; Fan, K.W.; Wong, R.T.Y.; Chen, F. Fatty Acid Composition and Squalene Content of the Marine Microalga Schizochytrium mangrovei. J. Agric. Food Chem. 2004, 52, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.C.; Fernandes, P. Production of Metabolites as Bacterial Responses to the Marine Environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar] [CrossRef] [PubMed]
- Belin, B.J.; Busset, N.; Giraud, E.; Molinaro, A.; Silipo, A.; Newman, D.K. Hopanoid Lipids: From Membranes to Plant–Bacteria Interactions. Nat. Rev. Microbiol. 2018, 16, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Takaichi, S.; Mochimaru, M. Carotenoids and Carotenogenesis in Cyanobacteria: Unique Ketocarotenoids and Carotenoid Glycosides. Cell. Mol. Life Sci. 2007, 64, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Gan, F.; Bryant, D.A. Adaptive and Acclimative Responses of Cyanobacteria to Far-Red Light. Environ. Microbiol. 2015, 17, 3450–3465. [Google Scholar] [CrossRef]
- Latifi, A.; Ruiz, M.; Zhang, C.C. Oxidative Stress in Cyanobacteria. FEMS Microbiol. Rev. 2009, 33, 258–278. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox Homeostasis and Antioxidant Signaling: A Metabolic Interface between Stress Perception and Physiological Responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Averina, S.; Velichko, N.; Senatskaya, E.; Pinevich, A. Far-red light photoadaptations in aquatic cyanobacteria. Hydrobiologia 2018, 813, 1–17. [Google Scholar] [CrossRef]
- Gallon, J.R. The oxygen sensitivity of nitrogenase: A problem for biochemists and micro-organisms. Trends Biochem. Sci. 1981, 6, 19–23. [Google Scholar] [CrossRef]
- Stal, L.J. Nitrogen fixation in cyanobacteria. eLS 2015, 1–9. [Google Scholar] [CrossRef]
- Colón-López, M.S.; Sherman, D.M.; Sherman, L.A. Transcriptional and translational regulation of nitrogenase in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 1997, 179, 4319–4327. [Google Scholar] [CrossRef]
- Lin, S.; Henze, S.; Lundgren, P.; Bergman, B.; Carpenter, E.J. Whole-cell immunolocalization of nitrogenase in marine diazotrophic cyanobacteria, Trichodesmium spp. Appl. Environ. Microbiol. 1998, 64, 3052–3058. [Google Scholar] [CrossRef]
- Fujita, Y.; Uesaka, K. Nitrogen fixation in cyanobacteria. In Cyanobacterial Physiology: From Fundamentals to Biotechnology; Kageyama, H., Waditee-Sirisattha, R., Eds.; Academic Press: London, UK, 2022; pp. 29–45. [Google Scholar] [CrossRef]
- Zehr, J.P.; Waterbury, J.B.; Turner, P.J.; Montoya, J.P.; Omoregie, E.; Steward, G.F.; Hansen, A.; Karl, D.M. Unicellular cyanobacteria fix N2 in the subtropical north Pacific Ocean. Nature 2001, 412, 635–638. [Google Scholar] [CrossRef]
- Zehr, J.P.; Capone, D.G. Unsolved mysteries in marine nitrogen fixation. Trends Microbiol. 2023, 31, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, J.T.; Silvey, J.K.G. Nitrogen fixation by Gloeocapsa. Science 1969, 165, 908–909. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, A.; Cao, S.; Takahashi, A.; Arai, T. Growth synchrony and cellular parameters of the unicellular nitrogen-fixing marine cyanobacterium, Synechococcus sp. strain Miami BG043511, under continuous illumination. Physiol. Plant 1987, 69, 1–8. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Cavalcanti, J.H.F.; Vaz, M.G.M.V.; Alvarenga, L.V.; Nunes-Nesi, A.; Araújo, W.L. Cyanobacterial Nitrogenases: Phylogenetic Diversity, Regulation and Functional Predictions. Genet. Mol. Biol. 2017, 40, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Horiike, T. The evolution of molybdenum-dependent nitrogenase in cyanobacteria. Biology 2021, 10, 329. [Google Scholar] [CrossRef] [PubMed]
- Nowruzi, B.; Hutarova, L. Structural and Functional Genes, and Highly Repetitive Sequences Commonly Used in the Phylogeny and Species Concept of the Phylum Cyanobacteria. Cryptogam. Algol. 2023, 44, 59–84. [Google Scholar] [CrossRef]
- Thiel, T.; Pratte, B.S. Regulation of Three Nitrogenase Gene Clusters in the Cyanobacterium Anabaena variabilis ATCC 29413. Life 2014, 4, 944–967. [Google Scholar] [CrossRef]
- Schrautemeier, B.; Cassing, A.; Böhme, H. Characterization of the Genome Region Encoding an fdxH-Type Ferredoxin and a New 2 [4Fe-4S] Ferredoxin from the Nonheterocystous, Nitrogen-Fixing Cyanobacterium Plectonema boryanum PCC 73110. J. Bacteriol. 1994, 176, 1037–1046. [Google Scholar] [CrossRef]
- Dos Santos, P.C.; Johnson, D.C.; Ragle, B.E.; Unciuleac, M.C.; Dean, D.R. Controlled Expression of nif and isc Iron-Sulfur Protein Maturation Components Reveals Target Specificity and Limited Functional Replacement Between the Two Systems. J. Bacteriol. 2007, 189, 2854–2862. [Google Scholar] [CrossRef] [PubMed]
- Böhme, H. Regulation of Nitrogen Fixation in Heterocyst-Forming Cyanobacteria. Trends Plant Sci. 1998, 3, 346–351. [Google Scholar] [CrossRef]
- Latysheva, N.; Junker, V.L.; Palmer, W.J.; Codd, G.A.; Barker, D. The Evolution of Nitrogen Fixation in Cyanobacteria. Bioinformatics 2012, 28, 603–606. [Google Scholar] [CrossRef]
- Zehr, J.P.; Bench, S.R.; Carter, B.J.; Hewson, I.; Niazi, F.; Shi, T.; Tripp, H.J.; Affourtit, J.P. Globally Distributed Uncultivated Oceanic N2-Fixing Cyanobacteria Lack Oxygenic Photosystem II. Science 2008, 322, 1110–1112. [Google Scholar] [CrossRef]
- Dick, G.J.; Duhaime, M.B.; Evans, J.T.; Errera, R.M.; Godwin, C.M.; Kharbush, J.J.; Nitschky, H.S.; Powers, M.A.; Vanderploeg, H.A.; Schmidt, K.C.; et al. The Genetic and Ecophysiological Diversity of Microcystis. Environ. Microbiol. 2021, 23, 7278–7313. [Google Scholar] [CrossRef]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An Updated Classification of Cyanobacterial Orders and Families Based on Phylogenomic and Polyphasic Analysis. J. Phycol. 2023, 59, 12–51. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota. 1. Teil Chroococcales. In Süsswasserflora von Mitteleuropa; Gärtner, E.H., Heying, H., Möllenhauer, D., Eds.; G. Fischer: Jena, Germany, 1998. [Google Scholar]
- Hoffmann, L.; Komárek, J.; Kaštovský, J. System of Cyanoprokaryotes (Cyanobacteria) State in 2004. Algol. Stud./Arch. Hydrobiol. 2005, 117, 95–115. [Google Scholar] [CrossRef]
- Dextro, R.B.; Delbaje, E.; Cotta, S.R.; Zehr, J.P.; Fiore, M.F. Trends in Free-access Genomic Data Accelerate Advances in Cyanobacteria Taxonomy. J. Phycol. 2021, 57, 1392–1402. [Google Scholar] [CrossRef]
- Zhang, X.; Xiao, L.; Liu, J.; Tian, Q.; Xie, J. Trade-off in Genome Turnover Events Leading to Adaptive Evolution of Microcystis aeruginosa Species Complex. BMC Genom. 2023, 24, 462. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.W.; Foster, R.A.; Krupke, A.; Carter, B.J.; Musat, N.; Vaulot, D.; Kuypers, M.M.M.; Zehr, J.P. Unicellular Cyanobacterium Symbiotic with a Single-Celled Eukaryotic Alga. Science 2012, 337, 1546–1550. [Google Scholar] [CrossRef] [PubMed]
- Temraleeva, A.D.; Dronova, S.A.; Moskalenko, S.V.; Didovich, S.V. Modern Methods for Isolation, Purification, and Cultivation of Soil Cyanobacteria. Microbiology 2016, 85, 389–399. [Google Scholar] [CrossRef]
- Velichko, N.; Smirnova, S.; Averina, S.; Pinevich, A. A Survey of Antarctic Cyanobacteria. Hydrobiologia 2021, 848, 2627–2652. [Google Scholar] [CrossRef]
- Ullah, I.; Sjöstrand, J.; Andersson, P.; Sennblad, B.; Lagergren, J. Integrating Sequence Evolution into Probabilistic Orthology Analysis. Syst. Biol. 2015, 64, 969–982. [Google Scholar] [CrossRef]
- Bobay, L.M.; Ochman, H. Factors Driving Effective Population Size and Pan-Genome Evolution in Bacteria. BMC Evol. Biol. 2018, 18, 153. [Google Scholar] [CrossRef]
- Goyal, A. Metabolic Adaptations Underlying Genome Flexibility in Prokaryotes. PLoS Genet. 2018, 14, e1007763. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, D.; Zhang, T.; Ren, Q.; Xiang, L.; Ning, C.; Zhang, Y.; Gao, R. Comprehensive and Functional Analyses Reveal the Genomic Diversity and Potential Toxicity of Microcystis. Harmful Algae 2022, 113, 102186. [Google Scholar] [CrossRef] [PubMed]

| Attributes | Value |
|---|---|
| Genome size (bp) | 5,152,646 |
| G + C content (%) | 52.04 |
| Completeness (%) | 99.12 |
| Contamination (%) | 0.29 |
| Contigs | 117 |
| Largest contig (bp) | 248,692 |
| N50 length (bp) | 83,624 |
| Protein encoding gene | 4988 |
| Total tRNAs | 44 |
| 16S rRNA copies | 2 |
| Contig | Location (nt) | Product Type | Most Similar Known Cluster | Similarity |
|---|---|---|---|---|
| 8 | 80,421–126,963 | T1PKS; NRPS | 1-heptadecene | 100% |
| 22 | 5103–76,035 | Terpene | Phytoene synthase | 31% |
| 51 | 1–19,543 | Terpene | Terpene cyclase | 11% |
| 57 | 12,658–28,163 | RRE-Containing | RiPP | 12% |
| 76 | 1–13,978 | Cyanobactin | RiPP | 13% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, M.J.; Botero, N.B.; Andreote, A.P.D.; Feitosa, A.M.T.; Popin, R.V.; Sivonen, K.; Fiore, M.F. Genomic Insights into the Taxonomy and Metabolism of the Cyanobacterium Pannus brasiliensis CCIBt3594. Taxonomy 2024, 4, 184-198. https://doi.org/10.3390/taxonomy4010010
Machado MJ, Botero NB, Andreote APD, Feitosa AMT, Popin RV, Sivonen K, Fiore MF. Genomic Insights into the Taxonomy and Metabolism of the Cyanobacterium Pannus brasiliensis CCIBt3594. Taxonomy. 2024; 4(1):184-198. https://doi.org/10.3390/taxonomy4010010
Chicago/Turabian StyleMachado, Mauricio Junior, Natalia Betancurt Botero, Ana Paula Dini Andreote, Anderson Miguel Teixeira Feitosa, Rafael Vicentini Popin, Kaarina Sivonen, and Marli F. Fiore. 2024. "Genomic Insights into the Taxonomy and Metabolism of the Cyanobacterium Pannus brasiliensis CCIBt3594" Taxonomy 4, no. 1: 184-198. https://doi.org/10.3390/taxonomy4010010
APA StyleMachado, M. J., Botero, N. B., Andreote, A. P. D., Feitosa, A. M. T., Popin, R. V., Sivonen, K., & Fiore, M. F. (2024). Genomic Insights into the Taxonomy and Metabolism of the Cyanobacterium Pannus brasiliensis CCIBt3594. Taxonomy, 4(1), 184-198. https://doi.org/10.3390/taxonomy4010010






