Minimally Monophyletic Genera Present within Meso- and Macrogenera
Abstract
:1. Introduction
2. Materials and Methods
3. Results
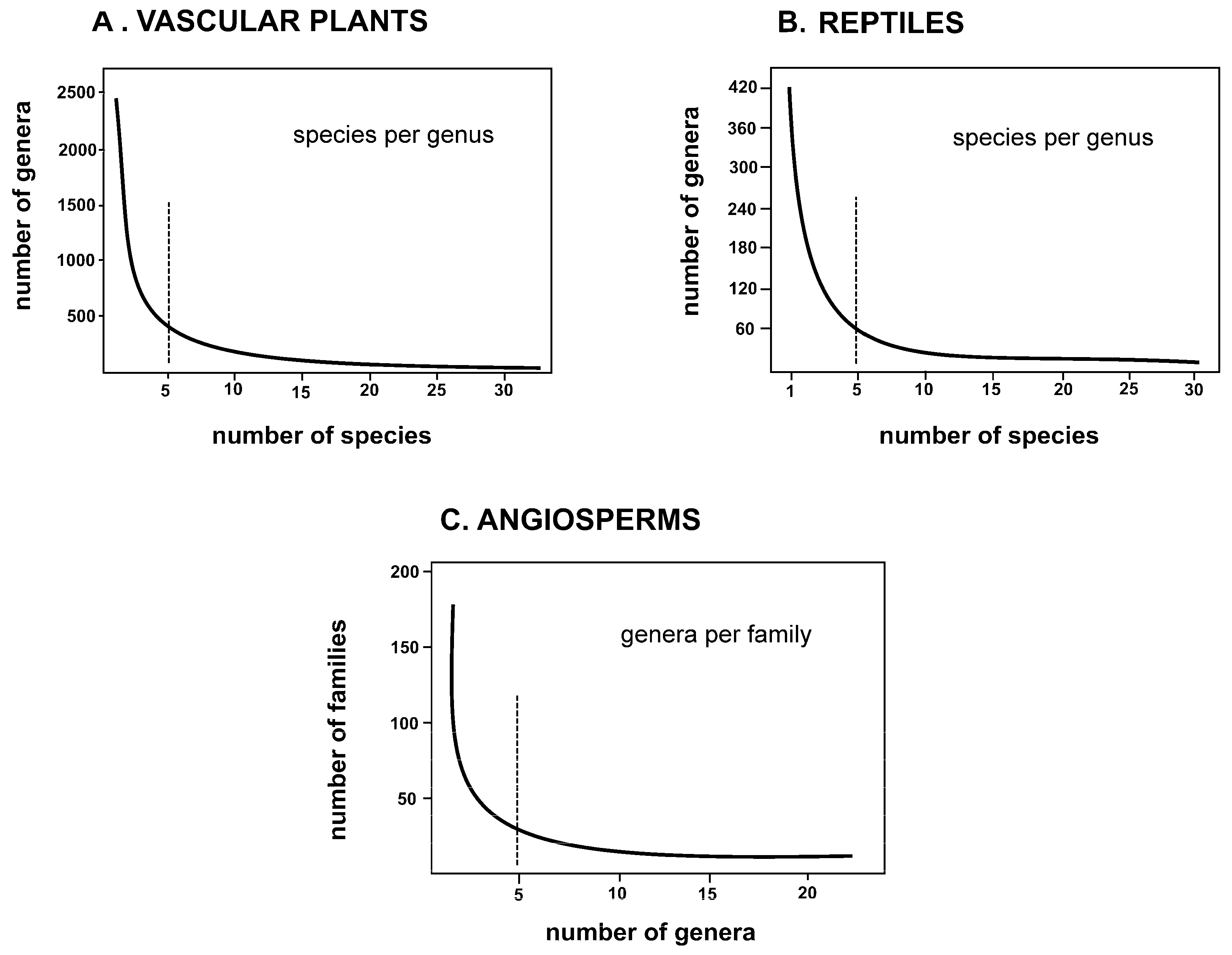
3.1. Evolutionary Models of the Mesogenera of Classical Systematics
3.2. Evaluation of Macrogenera
3.2.1. Morphologically Based Cladistics
3.2.2. Granularity in Molecular Cladograms
Syntrichia Molecular Cladogram
Chionoloma Molecular Cladogram
4. New Taxonomic Combination
5. Discussion and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zander, R.H. Minimally monophyletic genera are the cast-iron building blocks of evolution. Ukr. Bot. J. 2024, 81, 87–99. [Google Scholar] [CrossRef]
- Zander, R.H. Lineages of fractal genera comprise the 88-million-year steel evolutionary spine of the ecosphere. Plants 2024, 13, 1559. [Google Scholar] [CrossRef]
- Kauffman, S.A. The Origins of Order, Self-Organization and Selection in Evolution; Oxford University Press: New York, NY, USA, 1993; 709p. [Google Scholar]
- Kauffman, S.A. Investigations; Oxford University Press: Oxford, UK, 2000; 300p. [Google Scholar]
- Zander, R.H. Infraspecific molecular trees are associated with serial macroevolution in Pottiaceae (Bryophyta). Ukr. Bot. J. 2019, 76, 390–405. [Google Scholar] [CrossRef]
- Zander, R.H. Fractal Evolution, Complexity and Systematics; Zetetic Publications: St. Louis, MO, USA, 2023. [Google Scholar]
- Van Wijnen, A.; Lewallen, E. Natural selection and evolution: Evolving concepts. Acad. Biol. 2024, 2, 1–4. [Google Scholar] [CrossRef]
- Stanley, S.M. A Theory of Evolution above the Species Level. Proc. Natl. Acad. Sci. USA 1975, 72, 646–650. [Google Scholar] [CrossRef]
- Rodriquez, C.R.L. A graphic representation of Bessy’s taxonomic sytem. Madroño 1950, 10, 214–218. [Google Scholar]
- Zander, R.H. Evolutionary analysis of five bryophyte families using virtual fossils. Anales Jard. Bot. Madrid 2009, 77, 263–277. [Google Scholar] [CrossRef]
- Nicolis, G.; Prigogine, I. Exploring Complexity: An Introduction; W.H. Freeman and Company: New York, NY, USA, 1989; 313p. [Google Scholar]
- Gazzarrini, E.; Cersonsky, R.K.; Bercx, M.; Adorf, C.S.; Marzari, N. The rule of four: Anomalous distributions in the stoichiometries of inorganic compounds. NPJ Comput. Mater. 2024, 10, 73. [Google Scholar] [CrossRef]
- Lloyd, A.C. Genus, Species and Ordered Series in Aristotle. Phronesis 1962, 7, 67–90. [Google Scholar] [CrossRef]
- Sokal, R.R.; Sneath, P.H.A. Principles of Numerical Taxonomy; W.H. Freeman: San Francisco, CA, USA, 1963; 359p. [Google Scholar]
- Wiley, E.O. Phylogenetics: The Theory and Practice of Phylogenetic Systematics; John Wiley: New York, NY, USA, 1981. [Google Scholar]
- Abbott, L.A.; Bisby, F.A.; Rogers, D.J. Taxonomic Analysis in Biology; Columbia University Press: New York, NY, USA, 1985; 336p. [Google Scholar]
- Sneath, P.H.; Sokal, R.R. The Principles and Practice of Numerical Classification; W.H. Freeman: San Francisco, CA, USA, 1973; 557p. [Google Scholar]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K.; et al. (Eds.) International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Regnum Vegetabile 159; Koeltz Botanical Books: Glashütten, Germany, 2018. [Google Scholar] [CrossRef]
- Humphreys, A.M.; Linder, H.P. Concept versus data in delimitation of plant genera. Taxon 2009, 58, 1054–1074. [Google Scholar] [CrossRef]
- Stebbins, G.L. Taxonomy and the evolution of genera, with special reference to the family Gramineae. Evolution 1956, 10, 235–245. [Google Scholar] [CrossRef]
- Clayton, W.D. The genus concept in practice. Kew Bull. 1983, 38, 149–153. [Google Scholar] [CrossRef]
- Bessey, C.E. The phylogenetic taxonomy of flowering plants. Ann. Missouri Bot. Gard. 1915, 2, 109–164. [Google Scholar] [CrossRef]
- Cuerrier, A.; Kiger, R.W.; Stevens, P.F. Charles Bessey, evolution, classification and the New Botany. Huntia 1996, 9, 179–213. [Google Scholar]
- Stevens, P.F. Metaphors and typology in the development of botanical systematics 1690–1960, or the art of putting new wine in old bottles. Taxon 1984, 33, 169–211. [Google Scholar] [CrossRef]
- Stevens, P.F. Evolutionary classification in botany. J. Arnold Arbor. 1986, 67, 313–339. [Google Scholar] [CrossRef]
- Stevens, P.F. The genus concept in practice: But for what practice? Kew Bull. 1985, 40, 457–465. [Google Scholar] [CrossRef]
- Stevens, P.F. Why do we name organisms? Some reminders from the past. Taxon 2002, 51, 11–26. [Google Scholar] [CrossRef]
- Stevens, P.F. How to interpret botanical classifications: Suggestions from history. BioScience 1997, 47, 243–250. [Google Scholar] [CrossRef]
- Schroeder, M. Fractals, Chaos, Power Laws, Minutes from an Infinite Paradise; W.H. Freeman and Company: New York, NY, USA, 1991; 429p. [Google Scholar]
- François, O. A multi-epoch model for the number of species within genera. Theor. Pop. Biol. 2020, 133, 97–103. [Google Scholar] [CrossRef]
- Clayton, W.D. The logarithmic distribution of angiosperm families. Kew Bull. 1974, 9, 271–279. [Google Scholar] [CrossRef]
- Binning, G. The fractal structure of evolution. Phys. D Nonlinear Phenom. 1989, 8, 32–36. [Google Scholar] [CrossRef]
- West, G.B.; Brown, J.H.; Enquist, B.J. The fourth dimension of life: Fractal geometry and the allometric scaling of organisms. Science 1999, 284, 1677–1679. [Google Scholar] [CrossRef] [PubMed]
- Carine, M.A.; Christenhusz, M.J.M. About this volume: The monograph of Hypericum by Norman Robson. Phytotaxa 2013, 4, 1–4. [Google Scholar] [CrossRef]
- Welch, S.L. Astragalus. In Flora of North America North of Mexico; Flora of North America Editorial Committee, Ed.; Oxford University Press: New York, NY, USA, 2024; Volume 13, pp. 584–900. [Google Scholar]
- Humphreys, A.M.; Barraclough, T.G. The evolutionary reality of higher taxa in mammals. Proc. R. Soc. B 2014, 281, 20132750. [Google Scholar] [CrossRef]
- de Queiroz, K.; Cantino, P. International Code of Phylogenetic Nomenclature (PhyloCode); CRC Press: Boca Raton, FL, USA, 2020; 190p. [Google Scholar]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; Chapman and Hall: New York, NY, USA, 1993. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Van Valen, L. A new evolutionary law. Evol. Theory 1973, 1, 1–30. [Google Scholar]
- Simpson, G.G. The Major Features of Evolution; Columbia University Press: New York, NY, USA, 1944; 434p. [Google Scholar]
- Vos, J.M.; Joppa, L.N.; Gittleman, J.L.; Stephens, P.R.; Pimm, S.L. Estimating the normal background rate of species extinction. Conserv Biol. 2015, 29, 452–462. [Google Scholar] [CrossRef]
- Alonso, M.; Jiménez, J.A.; Nylinder, S.; Hedenäs, L.; Cano, M.J. Disentangling generic limits in Chionoloma, Oxystegus, Pachyneuropsis and Pseudosymblepharis (Bryophyta: Pottiaceae): An inquiry into their phylogenetic relationships. Taxon 2016, 65, 3–18. [Google Scholar] [CrossRef]
- Alonso, M.; Jiménez, J.A.; Cano, M.J. Taxonomic revision of Chionoloma (Pottiaceae, Bryophyta). Ann. Missouri Bot. Gard. 2019, 104, 563–632. [Google Scholar] [CrossRef]
- Barido-Sottani, J.; Schwery, O.; Warnock, R.C.M.; Zhang, C.; Wright, A.M. Practical guidelines for Bayesian phylogenetic inference using Markov chain Monte Carlo (MCMC). Open. Res. Europe 2023, 3, 204. [Google Scholar] [CrossRef] [PubMed]
- Jauregui-Lazo, J.; Brinda, J.C.; Consortium, G.; Mishler, B.D. The Phylogeny of Syntrichia: An Ecologically Diverse Clade of Mosses with an Origin in South America. Am. J. Bot. 2023, 110, e16103. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.K. The strong law of small numbers. Amer. Math. Mon. 1988, 95, 697–712. [Google Scholar] [CrossRef]
- Wheeler, Q. Species, Science and Society; Routledge: London, UK; New York, NY, USA, 2024; 246p. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zander, R.H. Minimally Monophyletic Genera Present within Meso- and Macrogenera. Taxonomy 2024, 4, 649-660. https://doi.org/10.3390/taxonomy4030033
Zander RH. Minimally Monophyletic Genera Present within Meso- and Macrogenera. Taxonomy. 2024; 4(3):649-660. https://doi.org/10.3390/taxonomy4030033
Chicago/Turabian StyleZander, Richard H. 2024. "Minimally Monophyletic Genera Present within Meso- and Macrogenera" Taxonomy 4, no. 3: 649-660. https://doi.org/10.3390/taxonomy4030033
APA StyleZander, R. H. (2024). Minimally Monophyletic Genera Present within Meso- and Macrogenera. Taxonomy, 4(3), 649-660. https://doi.org/10.3390/taxonomy4030033





