Abstract
In scenarios when the morphological identification of samples is not feasible, the utilization of molecular analyses becomes an imperative. However, that can often be challenging in conditions where standard molecular laboratories cannot be established. In this study, we present a simplified and field-friendly protocol of the previously published Triplex PCR Assay for the identification of mammalian and plant sample origin using a fully portable molecular laboratory from Bento Lab (Bento Bioworks Ltd., London, UK), effectively eliminating the need for a traditional laboratory setup. The protocol in combination with correctly selected kits enables DNA extraction, result evaluation based on electrophoresis, and direct use of the PCR products for downstream analyses such as Sanger sequencing and third-generation sequencing; therefore, enabling the use of molecular analysis directly in situ or for educational purposes in a classroom.
1. Introduction
Over the past few decades, molecular analyses has become a part of standard procedure in research and applied sciences across different fields, from species determination (barcoding) to the identification of unknown samples [1,2,3,4,5]. The previously published Triplex Assay enables us to determine the clear origin of a sample in single-tube PCR, distinguishing between plant and mammalian sample origin [6] in cases when morphological analyses cannot be implemented (e.g., powdered samples, plant/animal extracts dissolved in water). The Triplex Assay is designed for standard laboratory arrangement, which can be often challenging to setup, as the equipment is expensive and needs regular calibration/maintenance, making it unfeasible in settings like remote field workstations. The fully mobile molecular laboratory Bento Lab (Bento Bioworks Ltd., London, UK) offers an affordable and fully mobile alternative, which has been already applied for several in situ research works such as the barcoding of Nematodes [7], delimitation of Rhodocybe species [8], on-site detection of Campylobacter in broilers [9], SARS-CoV-2 detection in low-income countries [10], or in situ rapid big-scale barcoding conducted on a marine vessel [11]. Bento Lab Pro contains a thermocycler cycler with a temperature range of 12–105 °C, a ramp rate of up to 2.5 °C/s, supports touchdown range, a centrifuge speed that ranges between 3000 and 13,500 rpm, and has a capacity for 6 × 2 mL tubes, electrophoresis, and transilluminator (further specifications https://bento.bio/product/bento-lab/; accessed on 1 June 2024), which is sufficient machine equipment for preliminary molecular analyses and visualization when combined with suitable DNA extraction kits and PCR master mixes for a simplified workflow.
This paper aims to simplify the protocol for the Triplex Assay so that it can be performed using a mobile molecular laboratory, primarily allowing for in situ execution. Additionally, we aim to demonstrate that the results of this single-tube reaction can be used as a preliminary step in downstream analyses, such as Sanger sequencing and third-generation sequencing. Furthermore, the simplified protocol can be used by educators at the high school level to practically demonstrate the principles of PCR in classrooms without the need for a designated molecular laboratory.
2. Materials and Methods
To simulate field conditions, the entire process of extraction and downstream analyses preceding sequencing was conducted outside of a standard laboratory environment and was performed using the Bento Lab mobile molecular laboratory, pipettes, 1.5 mL tubes (Eppendorf, Hamburg, Germany), PCR strips (Eppendorf, 0.2 mL tubes), and grinding pestles. The surface of the desk was treated with Microzid AF wipes (Schülke & Mayr, Norderstedt, Germany) and was divided into “pre-PCR” and “post-PCR” sections to prevent contamination and simulate a proper molecular laboratory setup. General protection, such as gloves, laboratory coats, and masks, was used. DNA was eluted in to 100 µL of Elution Buffer (Zymo Research, Irvine, CA, USA).
DNA was extracted from freshly collected Hibiskus sp. leaves using a Quick-DNA Plant/Seed Kit (Zymo Research, Irvine, CA, USA), with slight modification. As the mobile laboratory does not contain a grinding mill, the leaves where transferred in to a 1.5 mL tube and ground using plastic grinding pestles and then moved into a ZR BashingBead™ Lysis Tube (2.0 mm) (Zymo Research). DNA from mammalian tissue was extracted from the muscle tissue of Bos taurus using a Quick-DNA Miniprep Plus Kit (Zymo Research); the incubation was conducted in the 0.2 mL tubes (Eppendorf) in a total volume of 200 µL (≤25 mg of muscle tissue, 95 µL ddH2O, 95 µL Solid Tissue Buffer (Blue) and 10 µL of Proteinase K) using a hot plate from the thermocycler. Both extractions were performed in duplicate. Genomic DNA obtained from all six extractions was quantified using a Qubit 4 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) to ensure that the extraction was successful. This step was included only to ensure that the following steps would not be hindered by unsuccessful extraction and that samples were not adjusted for an equal concentration for initial PCR.
To detect either plant or mammalian DNA in an unknown sample, a PCR was performed based on a previously published Triplex Assay using the Bento Lab cycler under unchanged cycling conditions (95 °C 120 s, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 45 s, ending with a final extension step at 72 °C for 10 min) [6]. The PCR reaction was conducted in 30.5 µL PPP Master Mix (Top Bio, s.r.o., Vestec, Czech Republic) (12.5 µL) containing DNA/RNA-free water (9.5 µL), 5 µL of extracted DNA of the sample used for the analysis/positive control (PC)/negative control (NC) and 1 µL of internal positive control (IPC) (1 pg/µL), and Triplex Assay mix (2.5 µL) targeting the plant rbcL gene (RuBisCO), the mammalian COI gene (cytochrome oxidase I) (Table 1), and the artificial internal positive control in single reaction. The post-PCR samples were treated with DNA Clean & Concentrator-5 (Zymo Research). The mixture of plant DNA and mammalian DNA in a ratio 1:4 was used as an unknown sample (samples 052023_Mix).

Table 1.
Primers for individual products.
The samples were consequently visualized using electrophoresis (1% gel, 30 µL 1×TBE buffer, 0.3 g agaroses, 3 µL of dsGreen Nucleic Acid Gel Staining Solution (Lumiprobe Life science solutions, Hannover, Germany)). A total of 5 µL of the post-PCR sample mixed with 1 µL of Orange DNA Loading Dye (6×) (Thermo Fisher Scientific) was loaded into each well. The gel electrophoresis was run for 45 min at 65 V.
For Sanger sequencing, the sequencing PCR was conducted under the following conditions: 96 °C for 1 min, 96 °C for 10 s, 50 °C for 5 s, 60 °C for 4 min in a total volume of 10 µL (1.75 µL BigDye™ Terminator v1.1 and v3.1 5× Sequencing Buffer (ThermoFisher Scientific), 0.5 µL Big Dye® Terminator v3.1 Cycle Sequencing RR-100 (ThermoFisher Scientific), 1 µL primer (3.2 µM), 6.25 µL molecular-grade water, and 0.5 µL of purified PCR product); the primers used for the sequencing were identical to the ones used for the initial amplification, depending on the result of electrophoresis. After PCR sequencing, the samples were treated with the ZR DNA Sequencing Clean-Up kit (Zymo Research) and sequenced using SeqStudio™ 3200 Genetic Analyzer Systems (Applied Biosystems, Waltham, MA, USA). The obtained sequences were visually checked for quality in Chromas v2.6.6. (Technelysiun Pty Ltd., Brisbane, Australia) and trimmed. The sequences were consequently compared to an NCBI reference database using BLAST (Basic Local Alignment Search Tool) (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 22 February 2024) for species identification.
The purified PCR products were quantified using a Qubit 4 Fluorometer (Table 2). Samples with a concentration > 25 ng/µL (total of >200 ng) were sent to SEQme s.r.o. (Dobříš, Czech Republic) for library preparation and sequencing. The libraries were prepared using a Ligation Sequencing Kit V14 (Oxford Nanopore, Oxford, UK) and sequenced using the Flongle Flow Cell (Oxford Nanopore). Obtained sequences were consequently compared to an NCBI reference database using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 22 February 2024) for species identification.

Table 2.
Concentrations of individual samples determined by Qubit 4 Fluorometer.
Sequences obtained by Sanger sequencing and MinION were consequently aligned and manually compared for mismatches using BioEdit v5.0.9. [15].
3. Results
The quantification of all samples using a Qubit 4 Fluorometer (ThermoFisher Scientific) was successful (Table 2). The visualized samples (Figure 1) showed the successful detection of plant, mammalian, and mixed DNA in the individual samples. The sequences obtained by Sanger sequencing targeting plant and mammalian genes were of sufficient quality and scored the expected species, Bos taurus and Hibiscus sp., in BLAST (Table 3). The sequences obtained by MinION sequencing were of sufficient quality and matched the expected species, Bos taurus and Hibiscus sp., in BLAST (Table 4).
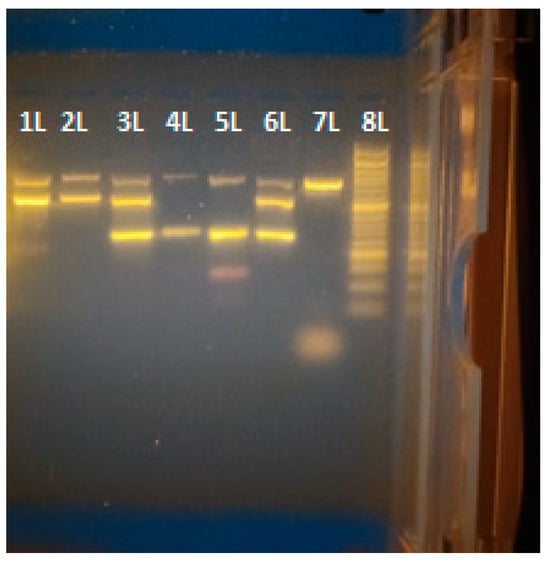
Figure 1.
Gel electrophoresis of Triplex Assay samples are ordered in lanes from left to right 1–8L as follows—012023_Bos_taurus, 022023_Bos_taurus, 052023_Mix, 032023_Hibiskus_sp, 042023_Hibiskus_sp., PC, NC, Ladder 100 bp.

Table 3.
Sample identification using BLAST (Sanger sequencing) (https://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 22 February 2024).

Table 4.
Sample identification using BLAST (MinION) (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 22 February 2024).
The sequences obtained by Sanger sequencing and by MinION were compared for discrepancies; there were no mismatches across the selected sequence between the samples and/or between the sequencing methods (COI 410 bp and Rubisco 230 bp).
4. Discussion
The presented paper demonstrates the practical use of a simplified Triplex Assay protocol that can be easily employed in field work stations or institutions that are not primarily laboratories, as well as for educational purposes such as in classrooms or extracurricular activities. The rapid determination of sample origin can be crucial in fields such as forensic sciences, where identifying the origin of a sample is step one for any downstream analyses. The simplicity of the Triplex Assay lies in it straight away differentiating between plant/mammalian/mixed sample origin using electrophoresis in a sample of unknown origin, and the fact that single-tube PCR can be used for downstream sequencing analyses [6]. All the steps before sequencing can be conducted with the mobile Bento Lab Pro. For Sanger sequencing, primers identical to those for the initial PCR can be used for the PCR sequencing, regardless of the result (mixed sample, single-origin sample). Furthermore, we show that the extracts obtained solely using machines implemented in Bento Lab Pro for the preparation of Sanger sequencing are suitable for third-generation sequencing. Replacing the original master mix protocol [6] with a manufactured master mix (e. g. PPP Master Mix, Top Bio, s.r.o., 5× HOT FIREPol®Blend Master Mix Ready to Load) with a slightly adjusted protocol further simplifies and speeds up the process not only in the out of regular laboratory setup, but also in a standard molecular laboratory. We observed that in plant samples with an initial concentration of template DNA higher than 5 ng/uL, the visibility of the IPC on gel can be affected. That can be caused by the large difference between fragments (Rubisco 280 bp, IPC 702 bp) (Figure S1). However, if the IPC band is showing in the NC, amplifying the targeted Rubisco fragment in a sample used for the analysis of a focal sample subjected to the identification is still feasible for sequencing. If the IPC band (702 bp) is required, dilution of the initial extract used for PCR amplification is recommended.
The use of a simplified Triplex Assay protocol for Bento Lab combined with MinION has the potential to serve in field work stations and outside of standard laboratory environments, not only for scientific research and applied environmental protection but also for educational purposes. The protocol easily can be applied to find the origin of unknown samples, in cases such as the interception of protected species of animals and plants during illegal trade transport. Other protocols can also be adjusted for the Bento Lab Pro and MinION, and they should be encouraged for facilities that do not possess standard molecular laboratories. The implementation of these simplified protocols for initial data analyses can rapidly speed up the process of sample identification, as the sample does not need to be sent out to additional facilities; therefore, it potentially prevents the loss of samples in the posting process and sample contamination. Furthermore, the all-in-one Bento Lab is at least 20-times cheaper than the combination of standalone instruments, and 10-times less heavy, thus allowing for easier manipulation and faster instrument setup. The presented paper provides more evidence of the demand for mobile genetic laboratories. The financial resources for the Bento Lab mobile laboratory were originally obtained through crowd funding (Kickstarter, Inc., Brooklyn, NY, USA), showing that the interest in developing such an instrument exceeds standard laboratory users. Bento Lab is not the only currently available portable molecular laboratory. For example, during the COVID-19 pandemic, other lightweight devices were tested, and the near-patient portable device BioBox was developed to ensure rapid results for patients, again eliminating the time needed for transportation [16]; additionally, the Biomeme Franklin™ three9 Real-Time PCR device was proven reliable for the detection of COVID-19 and other vector-borne diseases [17,18].
When conducting protocol optimalisation, we came across several limitations of the Bento Lab Pro; while these can be overcome, they should be addressed before in situ field work is started. Bento Lab Pro does not possess a 2 mL tube heating block, vortex, or a cell disruptor. To overcome the lack of a heating block for 2 mL tubes, an extraction kit or another alternative method that does not require lengthy above-room-temperature incubation in a 2 mL tube should be selected. We selected kits from Zymo Research that were easily compatible with Bento Lab; however, other extraction protocols such as HotShot [19], the Dipstick DNA Extraction Kit [20], or phenol–chloroform extraction [21] could be used. A cell disrupter can be substituted with plastic pestles, which can be used for homogenizing the sample, as we demonstrated through the slight adjustment of the Quick-DNA Plant/Seed Kit (Zymo Research). A vortex can be substituted with flicking the tube in combination with mixing inside the pipette tip. The limitations of Bento Lab have been previously addressed [22]; however, the Pro version offers a suitable and affordable alternative to the standard molecular laboratory setup and has the potential to serve in remote field stations and educational environments.
5. Conclusions
As sequencing has become a routine procedure in the field work of many specialisms over the past decades, having a mobile laboratory is a crucial advantage for in situ research. The results obtained by the simple Triplex Assay can rapidly uncover the type of sample that is being dealt with and consequently speeds up the downstream process for sequencing. Implementing mobile laboratories such as Bento Lab can rapidly accelerate the process of data analysis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/forensicsci4040038/s1, Figure S1: Troubleshooting.
Author Contributions
Conceptualization, D.V.; methodology, L.V., D.V. and K.M.; data curation K.M.; formal analysis, K.M., Writing—original draft, K.M.; Writing—review and editing, D.V., M.T., L.V. and K.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Czech Ministry of the Interior from the Program Strategic support for the development of security research 2019–2025 (IMPAKT 1) [grant number VJ01010026].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Waugh, J. DNA Barcoding in Animal Species: Progress, Potential and Pitfalls. BioEssays 2007, 29, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Gathier, G.; van der Niet, T.; Peelen, T.; van Vugt, R.R.; Eurlings, M.C.M.; Gravendeel, B. Forensic Identification of CITES Protected Slimming Cactus (Hoodia) Using DNA Barcoding. J. Forensic Sci. 2013, 58, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Jafar, S.; Ashraf Raja, N.; Mahar, J. Use of DNA Barcoding to Control the Illegal Wildlife Trade: A CITES Case Report from Pakistan. J. Bioresour. Manag. 2015, 2, 3. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding Animal Life: Cytochrome Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. B Biol. Sci. 2003, 270 (Suppl. S1), S96–S99. [Google Scholar] [CrossRef]
- Saskova, L.; Votrubova, J.; Vanek, D. Rapid Classification of Unknown Biological Material Using a Novel Triplex Assay. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e132–e134. [Google Scholar] [CrossRef]
- Knot, I.E.; Zouganelis, G.D.; Weedall, G.D.; Wich, S.A.; Rae, R. DNA Barcoding of Nematodes Using the MinION. Front. Ecol. Evol. 2020, 8, 100. [Google Scholar] [CrossRef]
- Aplin, N.; Cullington, P.; Douglas, B.; Janke, E. DNA Barcoding Reveals Three Species New to Britain. Field Mycol. 2023, 23, 124–132. [Google Scholar]
- Marin, C.; Marco-Jiménez, F.; Martínez-Priego, L.; De Marco-Romero, G.; Soriano-Chirona, V.; Lorenzo-Rebenaque, L.; D’Auria, G. Rapid Oxford Nanopore Technologies MinION Sequencing Workflow for Campylobacter Jejuni Identification in Broilers on Site—A Proof-of-Concept Study. Animals 2022, 12, 2065. [Google Scholar] [CrossRef] [PubMed]
- Alcántara, R.; Peñaranda, K.; Mendoza-Rojas, G.; Nakamoto, J.A.; Martins-Luna, J.; del Valle-Mendoza, J.; Adaui, V.; Milón, P. Unlocking SARS-CoV-2 Detection in Low-and Middle-Income Countries. Cell Rep. Methods 2021, 1, 100093. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.J.M.; Ip, Y.C.A.; Ng, C.S.L.; Huang, D. Takeaways from Mobile DNA Barcoding with BentoLab and MinION. Genes 2020, 11, 1121. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding RbcL Gene Complements the Non-Coding TrnH-PsbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-Z.; Fan, X.; Sha, L.-N.; Zeng, J.; Wang, Y.; Chen, Q.; Kang, H.-Y.; Zhang, H.-Q.; Zhou, Y.-H. Phylogeny and Molecular Evolution of the RbcL Gene of St Genome in Elymus Sensu Lato (Poaceae: Triticeae). Biochem. Syst. Ecol. 2013, 50, 322–330. [Google Scholar] [CrossRef]
- Kitpipit, T.; Chotigeat, W.; Linacre, A.; Thanakiatkrai, P. Forensic Animal DNA Analysis Using Economical Two-Step Direct PCR. Forensic Sci. Med. Pathol. 2014, 10, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Toppings, N.B.; Mohon, A.N.; Lee, Y.; Kumar, H.; Lee, D.; Kapoor, R.; Singh, G.; Oberding, L.; Abdullah, O.; Kim, K.; et al. A Rapid Near-Patient Detection System for SARS-CoV-2 Using Saliva. Sci. Rep. 2021, 11, 13378. [Google Scholar] [CrossRef] [PubMed]
- Valtier, S. Development of Assay Panel for Vector-Borne Disease; Research Cooperative Agreement D17AC00026; 59th Medical Wing, Office of the Chief Scientist: Lackland, TX, USA, 2020. [Google Scholar]
- Voelker, C.R.; Ochoa, A.R.; Armstrong-Spenrath, L.; Lott, L.; McDaniel, J.S.; Blackburn, A.N.; Cornell, L.E.; Mahoney, R.; Asin, S.N. Evaluating Sensitivity and Specificity of the Biomeme FranklinTM Three9 Real-Time PCR Device and SARS-CoV-2 Go-Strips Assay Using Clinical Samples. J. Clin. Virol. 2022, 146, 105046. [Google Scholar] [CrossRef] [PubMed]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-Quality Mouse Genomic DNA with Hot Sodium Hydroxide and Tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Mason, M.G.; Wang, Y.; Wee, E.; Turni, C.; Blackall, P.J.; Trau, M.; Botella, J.R. Nucleic Acid Purification from Plants, Animals and Microbes in under 30 Seconds. PLoS Biol. 2017, 15, e2003916. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Kambouris, M.E.; Siamoglou, S.; Kordou, Z.; Milioni, A.; Vassilakis, S.; Goudoudaki, S.; Kritikou, S.; Manoussopoulos, Y.; Velegraki, A.; Patrinos, G.P. Point-of-Need Molecular Processing of Biosamples Using Portable Instrumentation to Reduce Turnaround Time. Biosaf. Health 2020, 2, 177–182. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).