Abstract
The Eugénio de Almeida Foundation’s Casa de Fresco is a historical monument of valuable historic–artistic significance, which currently reveals an assortment of biofilms due to the proliferation of microorganisms in the stone and rocaille elements. The biodeterioration in this area was studied as part of the Conservation and Restoration Project. We effectively characterized the local microbial community using modern high-throughput DNA analysis. Our results suggested the existence of a variety of lichens or lichenized fungi, including genera such as Variospora, Verrucaria, Circinaria, and Caloplaca. Furthermore, we detected several prokaryote microorganisms related to the identification of these lichens. To properly deal with this microbiological issue and avoid fungal recolonization, we evaluated available commercial antimicrobial treatments.
1. Introduction
Biodeterioration of cultural heritage is the consequence of interactions between living organisms, material support, and environmental conditions [1]. The process is related to surface bioreceptivity, that is, “the totality of properties or characteristics of a material that controls its probability of being colonized by one or more groups of living microorganisms” [2,3,4].
Stones’ weathering characteristics, particularly bioreceptivity, are impacted by their chemical composition, physical structure, and geological origin. The microbial colonization of stones is influenced by external factors such as water availability, pH, humidity, climatic exposure, nutrition sources, and petrologic features such as mineral composition, stone type, porosity, and permeability. The inherent porosity of materials renders their surfaces susceptible to the deposition of microbial spores and vegetative cells carried by airborne particles, thus facilitating their growth [1,5]. Conditions are suitable for the generation of microbial spores when they are mild and appropriately warm and combined with a nutrient-rich substrate. Sunlight offers the energy required for photosynthetic organisms to convert inorganic CO2 into biomass, which is subsequently deposited and enriched on the stone’s surface. As soon as the initial carbon immobilization happens on the fresh stone surface by the carbon cycle of phototrophs, the microbial community modifies the surface qualities and, more crucially, its composition by developing into a more complex microbial community [6].
Fungi sensitive to humidity and heat emit hyphae that penetrate the substrate, with mechanically destructive effects [7]. Due to their enormous enzymatic activity, metabolic versatility (greater than all other biodeteriogens), and ability to grow in low water activity (aw) values, fungi are able to inhabit and deteriorate a wide variety of substrates. Also, their ability to withstand extreme environmental conditions, establish mutualistic association with other microorganisms, and adopt various structural and morphological strategies further enhances their adaptability. As a result, they are currently understood to be the main agents contributing to the degradation of structures and heritage sites [6,7]. Lichens, in particular, are recognized for producing organic acids to release minerals and micronutrients from stone, allowing their development to be sustained and maintained through a symbiotic relationship between a fungi and algae, which receives energy from sunlight for photosynthesis [6].
In stone materials, deterioration by this type of microorganism is mainly the result of mechanical, chemical, and physical processes, consisting essentially of penetration of hyphae, production of fruiting bodies, production and release of destructive extracellular organic acids, enzymes, and metabolites, as well as the development of biofilms and chemical reactions with inorganic compounds [6,8].
Due to the well-known problems of biodeterioration resulting from microbial proliferation, the precise identification of species and a consequent deteriorative profile of the isolates are crucial steps for the development and establishment of adequate protection and conservation–restoration measures for the various cultural heritage assets. It is crucial to have effective methods for identifying potentially dangerous microorganisms as well as methods for preserving and removing their contamination. The development of innovative culture-independent methodologies, such as metagenomic techniques, has become increasingly important for the identification of biodeteriogens, analysis of their diversity, characterization of their metabolic functions, and their deteriorating by-products [9]. In addition, using biocides is a critical step in preventing and controlling microbial growth/recolonization. Color changes, variations in water absorption capacity, permeability, and surface tension must all be considered while performing this approach. To evaluate the effectiveness of biocides against microorganisms, treatments must be carried out on small areas (in vitro test), particularly on the deteriorated monument [10,11].
The Fundação Eugénio de Almeida was in the process of preserving the Casa de Fresco in the Condes de Basto Palace, a heritage site of inestimable historic–artistic value. In the latter half of the 16th century, D. Diogo de Castro introduced a remarkable array of visually captivating elements to this garden, with a particular focus on the Casa de Fresco. This structure was adorned with rocaille decoration, featuring an intricate assemblage of embellishments such as shells, glass fragments, porcelain, mosaics, tiles, crystals, fossils, and more, used to ornament its walls. Originally, this small space served as a place for conversation and leisure. During the hottest periods of the year, it’s cool and refreshing atmosphere provides comfort for moments of contemplation and reflection in direct communication with nature [12].
This stone monument, being exposed to sunlight through the entrance via an arch on the central frontage, and having a water source nearby, creates an environment with temperature and humidity conducive to microbial growth. Also, being located in an open garden, it is a place that is also not free from deteriorating factors such as erosion by wind or rain. Therefore, external environmental factors are inevitably linked to microbial growth in this location. Unfortunately, this space has undergone numerous aesthetic, physical, and chemical changes due to the growth and action of fungi, bacteria, and lichens.
In this work, the biodeterioration of Casa de Fresco located in the Condes de Basto Palace in Fundação Eugénio de Almeida (Évora, Portugal) was investigated within the scope of a Conservation and Restoration Project contributing to the “Study of Characterization and Diagnosis of the State of Conservation”. The study was carried out with the main objectives of: (i) detection of microbial colonization; (ii) evaluation and characterization of the microbial community through phylogenetic metagenomic techniques; and (iii) investigation of the effectiveness of commercial antimicrobials, in order to promote the safeguarding of this cultural heritage asset. Thus, this study aimed to ensure the conservation of Casa de Fresco, and took place between April and September 2022, as an architectural, decorative, and functional element essential to the harmonization of the entire set formed by the garden. This work was an opportunity to deepen knowledge about the historical, symbolic, and material dimensions of this heritage.
2. Materials and Methods
2.1. Sampling Process
The sampling process (Figure 1) was carried out in representative areas of the Casa de Fresco with significant signs of contamination and alteration, under the coordination of conservators–restorers, complying with the requirements of conservation and minimization of the structural and aesthetic impact on the work of art, collecting the minimum amount necessary for the assays. Non-invasive methods (which include the swab samples Z1 to Z10) and semi-invasive methods were used for sample collection, under semi-aseptic conditions (sampling carried out with sterilized material in outside conditions). The collection of stone microfragments, taken with sterilized scalpels (lichens identified as B2, B3, B4, B6, and B8), was performed near to losses or cracks to prevent additional damage [1,13].

Figure 1.
Casa de Fresco located in Condes de Basto Palace (Évora, Portugal) with some sampling process details. The red circles represent the sampling sites.
2.2. Detection of Microbiological Contamination
The microfragments were collected from sites with evidence of biodeterioration, observed under an optical microscope (Motic BA410E), and images acquired with a MoticamPro 282B camera; evidence of contamination was also visualized using the LEICA M205C magnifier.
2.3. Characterization of Microbiota through Next Generation Sequencing (NGS)
To use the NGS approach, metagenomic DNA was extracted from original microfragments (taken with scalpels) and swabs, for eukaryotes and prokaryotes, using the Soil DNA Purification Kit (OMNI International, Bethesda, Md, USA), according to the manufacturer’s instructions. The DNA was quantified using a fluorometric technique with the Quantus™ Fluorometer equipment (E6150, Promega, USA) using the QuantiFluor® dsDNA System Kit (E487, Promega, USA).
Bacterial communities were characterized by Illumina Sequencing for the 16S rRNA V3–V4 region. DNA was amplified for the hypervariable regions with specific primers and further reamplified in a limited-cycle PCR (Polymerase Chain Reaction) to add sequencing adaptor and dual indexes. For bacteria, the following primers were used: forward primer Bakt_341F 5′-CCTACGGGNGGCWGCAG-3′ and reverse primer Bakt_805R 5′-GACTACHVGGGTATCTAATCC-3′. For eukaryotic communities, a pool of forward primers was used: ITS3NGS1_F 5′-CATCGATGAAGAACGCAG-3′, ITS3NGS2_F5′-CAACGATGAAGAACGCAG-3′, ITS3NGS3_F5′-CACCGATGAAGAACGCAG-3′, ITS3NGS4_F 5′-CATCGATGAAGA ACGTAG-3′, ITS3NGS5_F 5′-CATCGATGAAGAACGTGG-3′, and ITS3NGS10_F 5′-CATCGATGAAGAACGCTG-3′ with the reverse primer ITS3NGS001_R 5′-TCCTSCGCTTATTGATATGC-3′ [14].
The DNA amplification, purification, quantification of the amplicons, and the analysis data for the V3–V4 and ITS2 samples were performed according to Rosado et al. (2020) [11].
2.4. Evaluation of Antimicrobial Activity
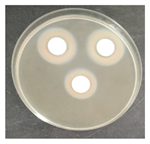
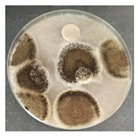
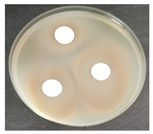
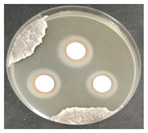
Laboratory tests were carried out to determine the effect of two commercial biocides to be used by the restoration team in inhibiting microbial growth, namely, Preventol Ri 50 (Lanxess, Köln, Germany) and Biotin-T (C.T.S., Madrid, Spain). Mixed cultures, made from swabs collected from places with visible contamination (Z1, Z3, Z4, and Z7) were carried out, incubating each corresponding swab separately in malt extract medium, with agitation at 120 rpm, at 28 °C, for 7 days, for the growth of microorganisms.
To test the effectiveness of each antimicrobial, the agar disk diffusion method was used. Bioactivity tests were performed by incorporating 1 × 108 cells of each of the liquid mixed culture in corresponding malt extract agar (MEA, Himedia Laboratories, Modautal, Germany—malt extract 30 g/L, peptone mycologic 5 g/L and agar 15 g/L) petri dishes (9 cm), and the biocides to be tested were placed in 13 mm sterile paper discs (Macherey-Nagel 827 ATD), impregnated with 20 µL of the antimicrobial substance. The petri dishes were incubated at 28 °C, and the growth was monitored daily. The microbial growth inhibition halos around the discs were measured.
3. Results and Discussion
3.1. Detection of Microbial Contamination
The goal of the microbiological analysis was to know the microbial community present in this monument and find biodeteriogenous agents. The materials exhibited visible evidence of alterations, with the appearance of lichens on the surface being highly noticeable and seemingly connected to the observed aesthetic changes.
The collected lichens’ microfragments were observed and photographed with a LEICA M205C binocular microscope, as shown in Table 1.

Table 1.
Detection of microbial contamination by LEICA M205C binocular microscope.
3.2. Phylogenetic Metagenomic Approach
3.2.1. Eukaryote Communities
A phylogenetic metagenomic approach is used as a culture-independent method to detect the communities of microorganisms responsible for the biocolonization of the monument. Figure 2, Figure 3 and Figure 4 show the predominant eukaryote communities present in the stone microfragments.
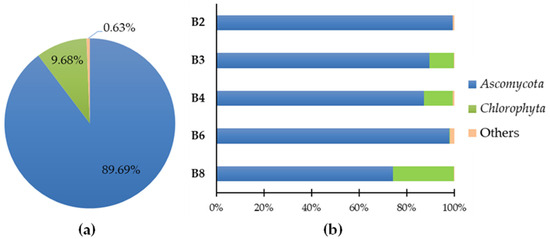
Figure 2.
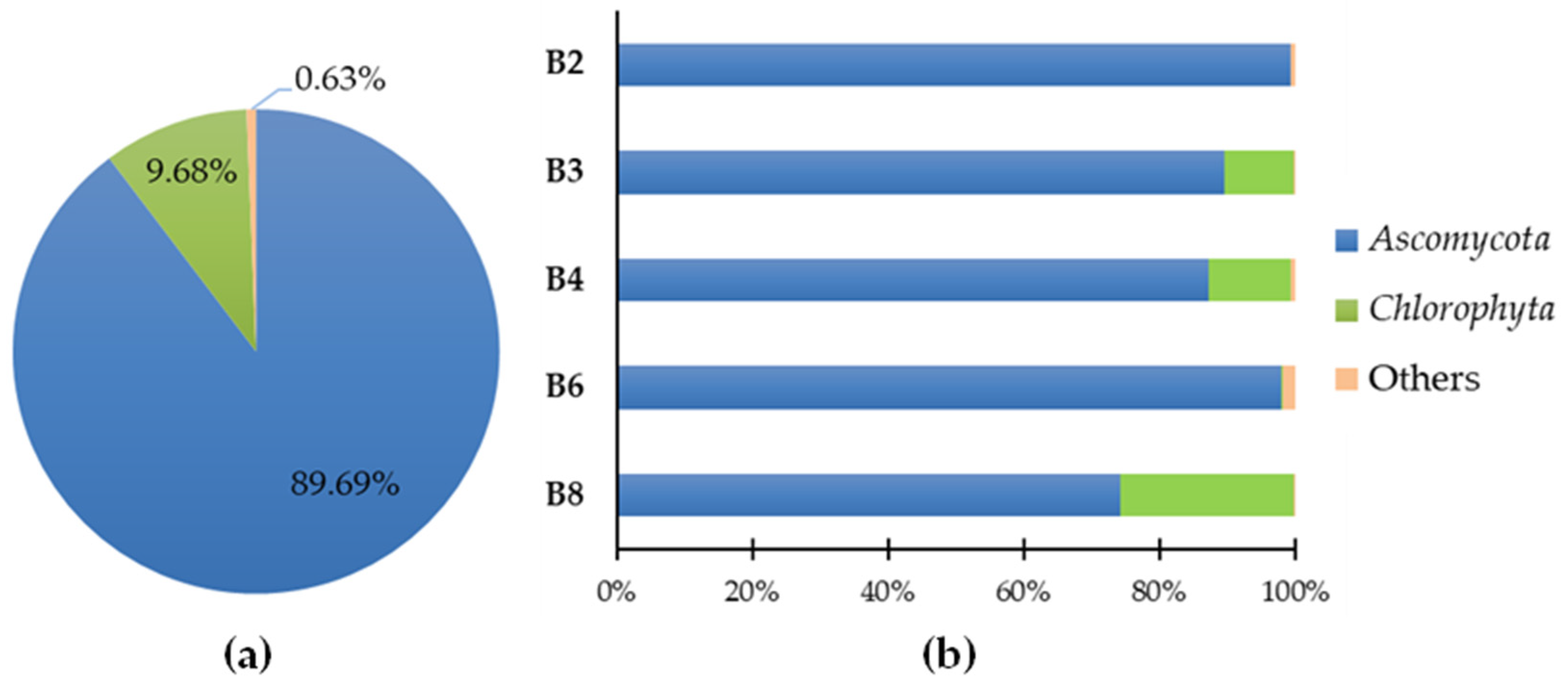
Predominant general phyla (a) and sample-specific phyla (b) of eukaryote populations present in the microbial samples.
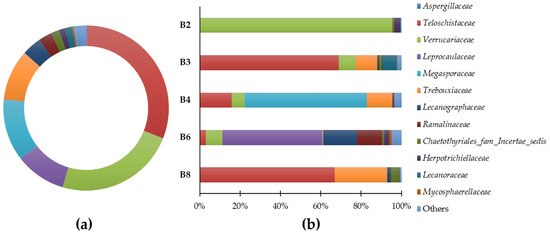
Figure 3.
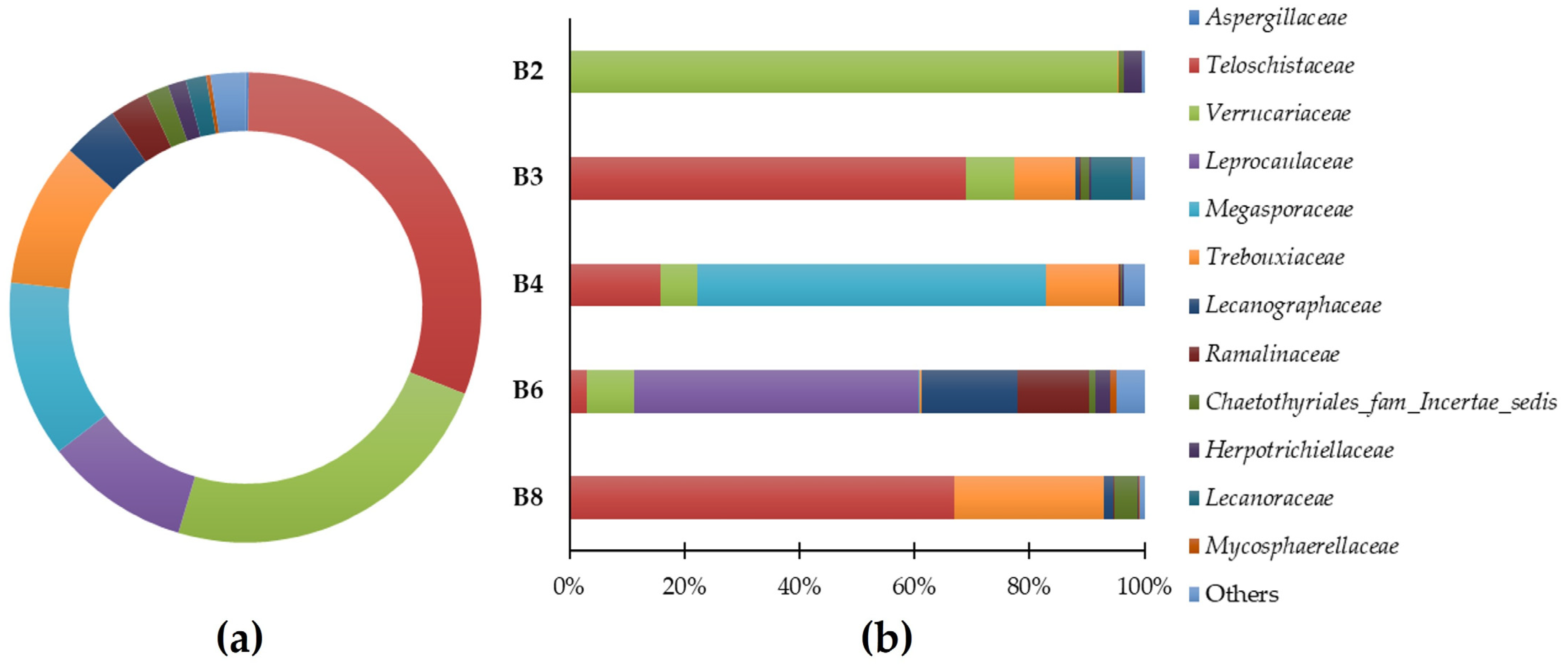
Predominant general families (a) and sample-specific phyla (b) of eukaryote populations present in the microbial samples.
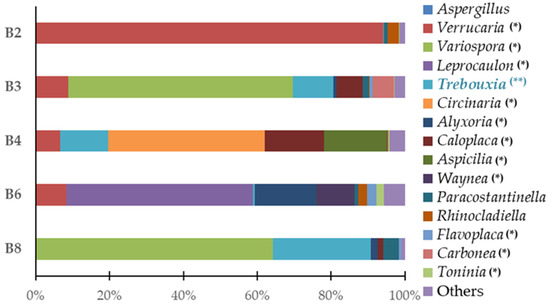
Figure 4.
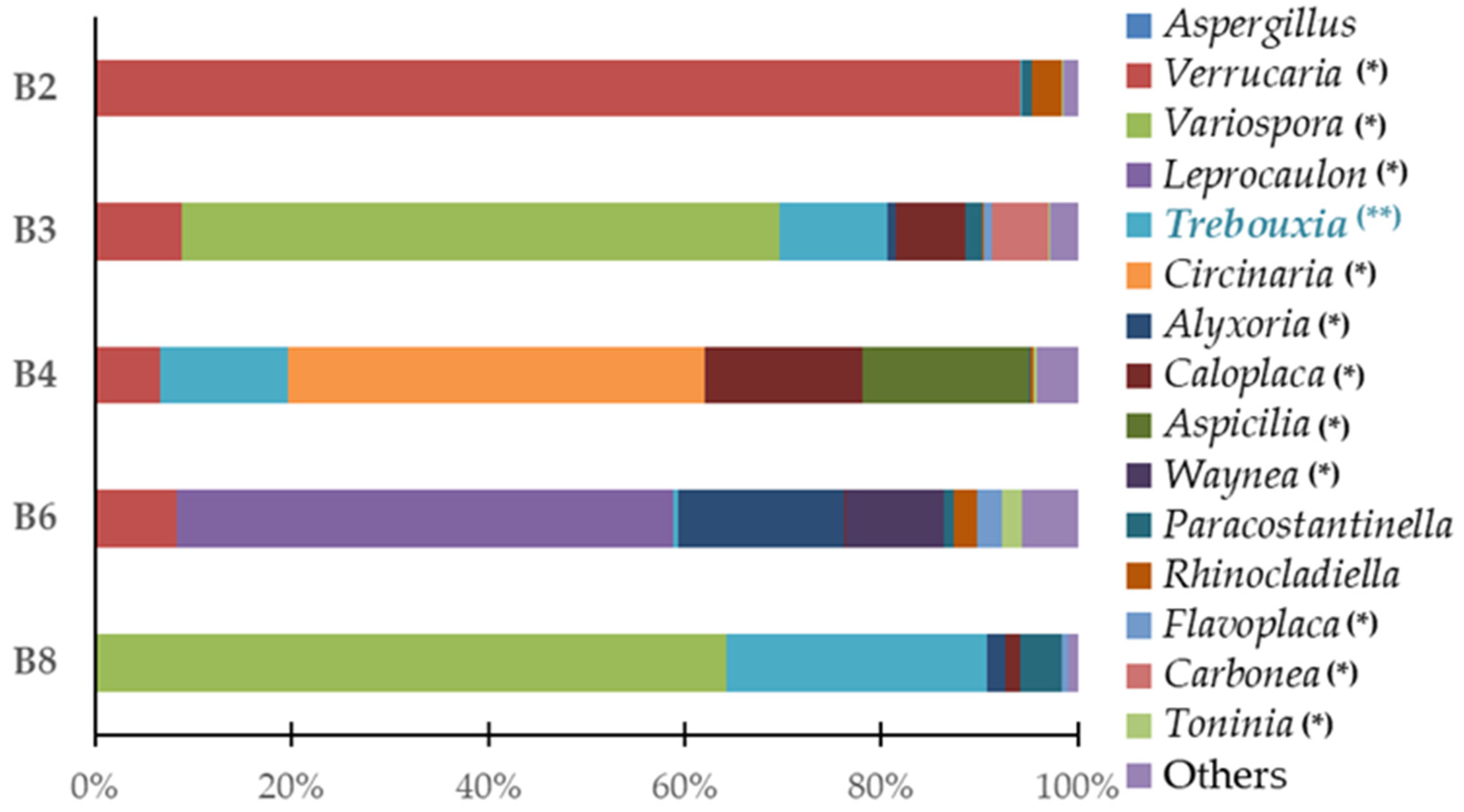
Predominant genus of eukaryote populations present in microfragments samples. (*) represents the genera of lichenized fungi; (**) highlights the photobiont of the lichens identified in the study.
Considering the set of all microfragment samples, the dominant phylum (Figure 2a) is Ascomycota (89.69%), followed by Chlorophyta (9.68%), and 0.63% for “others” (generic designation corresponding to microorganisms with lower representation). In Figure 2b, the predominant identified phyla in each of the samples are highlighted.
Ascomycota is one of the most diverse and ubiquitous phylum of eukaryotes, present in numerous ecological habitats [15]. More than 40% of all Ascomycota are lichenized and, according to the literature, as a result of their fungal metabolic activity and ability to hydrolyze short-chain fatty acids, in addition to amylolytic, and occasionally proteolytic and glycolytic activities, most of the fungal species that are involved in the deterioration of cultural heritage belong to this phylum [16,17]. Chlorophyta is a taxon of eukaryotic green algae that often assembles symbiotic relationships with fungi to form lichens. As the primary producer, this algal partner feeds the fungi with photosynthesis-derived carbohydrates to maintain the lichen relationship [18].
The results suggest that the eukaryotic population of Casa de Fresco belongs to several different families: (Figure 3a): Teloschistaceae (30.77%), Verrucariaceae (23.61%), Megasporaceae (12.16%), Trebouxiaceae (9.95%), Leprocaulaceae (9.93%), Lecanographaceae (3.83%), Ramalinaceae (2.65%), Chaetothyriales_fam_Incertae_sedis (1.54%), Lecanoraceae (1.40%), Herpotrichiellaceae (1.26%), Mycosphaerellaceae (0.30%), and Aspergillaceae (0.22%). “Others” comprised 2.38% of the families. In Figure 3b, the predominant identified families in each of the samples are highlighted.
Regarding the genus level (Figure 4), considering the set of all samples, Variospora (24.95%) was the most abundant followed by Verrucaria (23.42%), Trebouxia (10.22%), Leprocaulon (10.14%), Circinaria (8.48%), Caloplaca (4.99%), Alyxoria (3.92%), Aspicilia (3.38%), Waynea (1.99%), Paracostantinella (1.58%), Rhinocladiella (1.19%), Carbonea (1.14%), Flavoplaca (0.82%), Toninia (0.51%), and Aspergillus (0.18%); 3.09% of the genera were classified as “others”.
Lichens are formed by the combination of a fungus, often an ascomycete (mycobiont), with one or more photosynthetic members, most commonly green algae, or cyanobacteria. The fungus forms a thallus or lichenized stroma that is able to penetrate deeply into the stone substrate due to the proliferation of fungal hyphae [19].
It should be mentioned that the Teloschistaceae family of fungi, which has the highest relative abundance in this study’s samples, is one of the biggest families of lichenized fungi. Many species in this family are among the most beautiful and diverse lichens with the potential to be researched due to their vivid orange color and distinctive tallus [20]. Namely, the Variospora genus are remarkably tolerant of severe aridity, direct sunlight, high eutrophication, and air pollution [21]. This genus of lichen represents 60.8% of the yellow/orange lichen (B3, Table 1). We were able to identify the species Variospora thallincola, which is characterized by a crusty, golden-yellow shiny thallus, that seems to be the reason why the lichen B3 has the same color. Also, the other main eukaryotic genus associated with this lichen were identified as Verrucaria, Caloplaca, Paracostantinella and Carbonea.
Verrucaria is a genus of epilithic crusted lichen from the Verrucariaceae family, also notable in this study for its significant presence. It is a highly diversified genus characterized by crustose, heteromerous, areolate, or placodioid thalli that is typically seen as a black crust coating the rocks. This genus comprises 94.0% of the brown lichen (B2, Table 1), which corresponds to the aesthetic evidence of biodeterioration in this sample. Verrucaria nigrescens was also identified in this study, as a species of the previously described genus, which is a brownish-black lichen, often found in stone materials (like limestone or siliceous rocks), in several buildings and heritage monuments. V. nigrescens is described as a highly aggressive biodeterioration agent for causing aesthetic damage, mainly darkening of the substrate, and disintegration/increased susceptibility to the breakdown of the stone [22,23,24]. In addition to the predominant genus Verrucaria, the main genera Rhinocladiella, Paracostantinella, and Aspergillus were also identified.
The white spots of lichen B4 (Table 1) were associated with the notable presence of the genera Circinaria and Caloplaca with the former comprising 42.4% and the latter 16.0% of the lichen. Regarding these genera lichens’, we identified Circinaria Calcarea and Caloplaca teicholyta in our study which, according to the literature, are usually discovered in the stone components of historical sites across the world, such as monasteries, sculptures, cathedrals, and temples [21,25,26,27,28,29,30]. In addition to the aesthetic observations resulting from lichen colonization, the crusted epilithic thallus of the genus Caloplaca appears to become fully integrated with the substrate, which means that, when changes in the volume of the thallus occur due to expansion and contraction, it contributes to the processes of biodeterioration through the changes generated by its internal penetration and mechanical alteration capacity [31]. Other significant genera, associated with these lichenized genera, Aspicilia and Verrucaria were also identified in this sample.
The greenish and fruity appearance of the green lichen (B6, Table 1) might be explained by the majority composition of the lichenized fungus Leprocaulon (50.6%) in this sample. Verrucaria, Alyxoria, Waynea, Rhinocladiella, Flavoplaca, and Toninia are genera that were also identified as relevant in this sample.
Finally, the majority lichen genus in sample B8 was Variospora (64.0%), and the most significant species was identified as Variospora thallincola. Other major genera associated with this sample were identified as Alyxoria, Caloplaca, and Paracostantinella.
Lichens damage the aesthetics of stone monuments and modify the physicochemical characteristics of the monument’s structure. In addition to developing colored structures on the stone’s surface, lichens negatively impact stone materials by penetration with hyphae and secreting acidic metabolites including carbon dioxide, lichenic acids, and oxalic acid. Finally, these acids dissolve calcareous rocks, which results in the production of calcified encrustation and irreparable injury to outdoor stone monuments [32].
On the other hand, Trebouxia is a genus of unicellular green algae also relevant in this study as it is the photobiont associated with more than 15.000 species of existing lichens, which makes it the most common and widely distributed algae in the world [33,34]. In this study, it is present in all the lichens’ samples; it is known for essentially colonizing stone monuments in areas protected by shade and characterized by low substrate humidity or even prone to desiccation [26].
3.2.2. Prokaryote Communities
The DNA extracted from 10 different swabs were recovered and used for NGS study (16S and ITS). Regarding the bacterial characterization, the main taxonomic levels are discussed below.
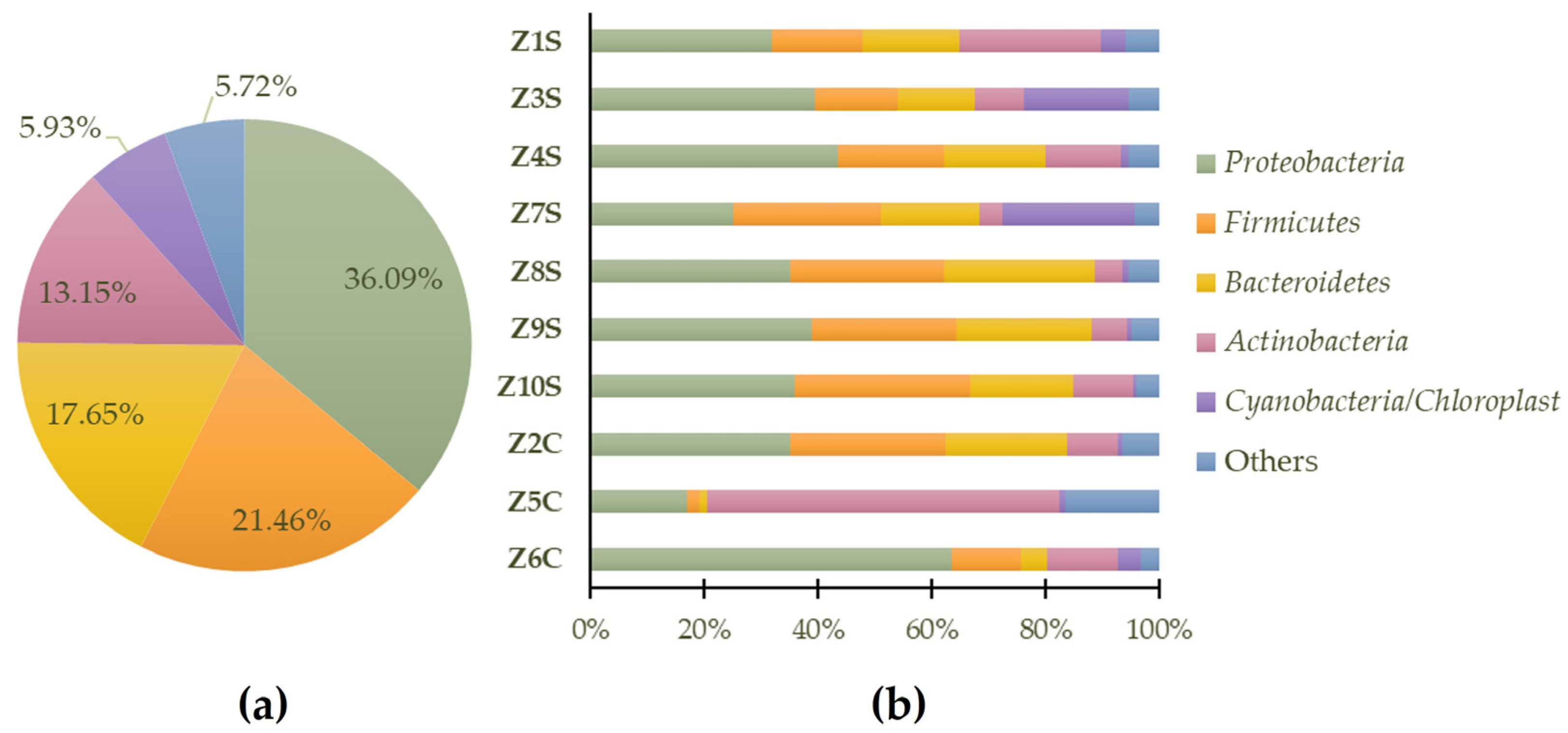
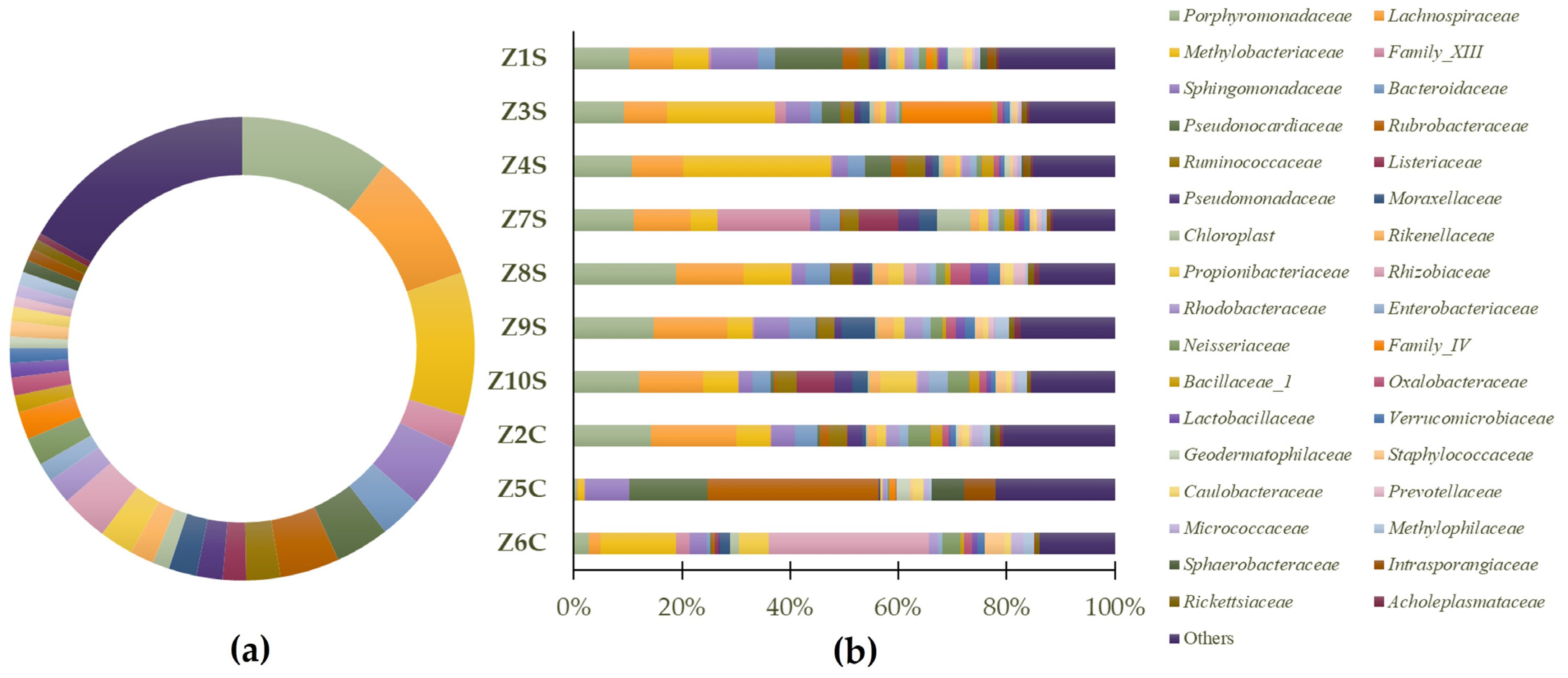
Considering the set of all samples, the dominant phylum (Figure 5a) is Proteobacteria (36.09%), followed by Firmicutes (21.46%), Bacteroidetes (17.65%), Actinobacteria (13.15%), Cyanobacteria/Chloroplast (5.93%), and 5.72% for “others” (generic designation corresponding to phyla with lower representation). In Figure 5b, the predominant identified phyla in each of the samples are highlighted.
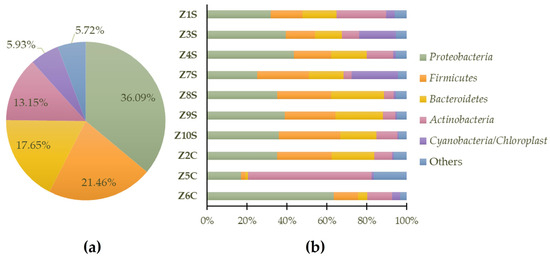
Figure 5.
Predominant general phyla (a) and sample-specific phyla (b) of prokaryote populations present in swab samples.
Proteobacteria are currently the largest phylum within the domain of bacteria and represent the vast majority of known Gram-negative bacteria. This group of microorganisms is ubiquitous, being able to colonize almost all types of materials; it has the ability to form biofilms or aggregates of microorganisms that adhere to each other to facilitate community survival on stone surfaces to retain water and nutrients [35]. In recent studies, through molecular assays, it was revealed that a high diversity of bacteria of the phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes are also related to lichen symbiotic relationships which play several roles such as providing nutrients, mobilizing iron and phosphate, and fixing nitrogen for symbiotic lichen growth; they could provide resistance to both abiotic and biotic factors, detoxification of metabolites, a source of vitamin B12 for photosynthesis, the generation of hormones that promote fungal and algal development, and the breakdown of older sections of the lichen thallus [36,37].
The results suggest that the prokaryote population of Casa de Fresco belongs to several different families: (Figure 6a): Porphyromonadaceae (10.47%), Methylobacteriaceae (10.04%), Lachnospiraceae (9.20%), Sphingomonadaceae (4.50%), Rubrobacteraceae (4.13%), Pseudonocardiaceae (3.76%), Rhizobiaceae (3.30%), Bacteroidaceae (2.95%), Ruminococcaceae (2.37%), Propionibacteriaceae (2.37%), Family_XIII (2.31%), Moraxellaceae (1.95%), Neisseriaceae (1.95%), Family_IV (1.92%), Pseudomonadaceae (1.82%), Rhodobacteraceae (1.80%), Rikenellaceae (1.64%), Listeriaceae (1.62%), Enterobacteriaceae (1.30%), Oxalobacteraceae (1.26%), Bacillaceae_1 (1.18%), Chloroplast (1.17%), Staphylococcaceae (1.09%), Verrucomicrobiaceae (1.04%), Lactobacillaceae (1.02%), Caulobacteraceae (1.01%), Methylophilaceae (0.93%), Sphaerobacteraceae (0.86%), Micrococcaceae (0.83%), Intrasporangiaceae (0.81%), Geodermatophilaceae (0.78%), Rickettsiaceae (0.74%), Prevotellaceae (0.70%), Acholeplasmataceae (0.49%), and 16.71% were considered as “others”. In Figure 6b, the predominant identified phyla in each of the samples are highlighted.
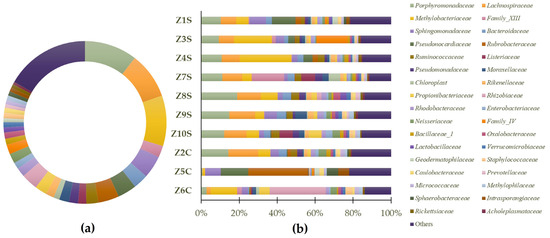
Figure 6.
Predominant general families (a) and sample-specific families (b) of prokaryote populations present in swab samples.
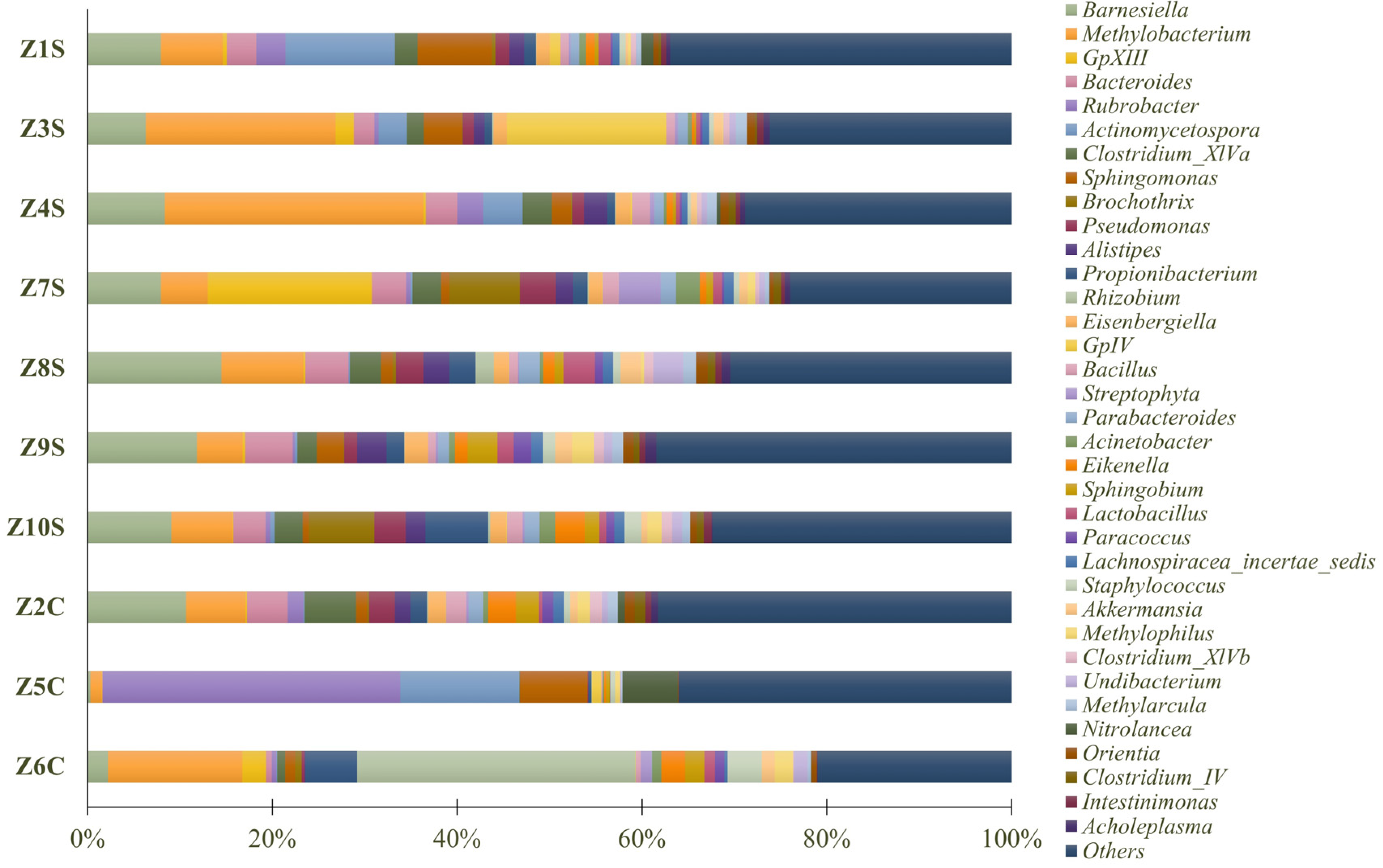
Regarding the genus level, Methylobacterium (10.30%) was the most abundant (Figure 7), followed by Barnesiella (7.90%), Rubrobacter (4.22%), Actinomycetospora (3.33%), Rhizobium (3.29%), Bacteroides (3.09%), Sphingomonas (2.96%), Clostridium_XlVa (2.55%), GpXIII (2.38%), Propionibacterium (2.38%), GpIV (1.99%), Pseudomonas (1.89%), Alistipes (1.71%), Brochothrix (1.66%), Eikenella (1.41%), Eisenbergiella (1.37%), Sphingobium (1.25%), Bacillus (1.20%), Parabacteroides (1.14%), Staphylococcus (1.11%), Lactobacillus (1.05%), Akkermansia (0.96%), Undibacterium (0.96%), Methylophilus (0.94%), Nitrolancea (0.86%), Acinetobacter (0.84%), Methylarcula (0.82%), Lachnospiracea_incertae_sedis (0.81%), Orientia (0.76%), Streptophyta (0.75%), Paracoccus (0.68%), Clostridium_XlVb (0.66%), Clostridium_IV (0.54%), Acholeplasma (0.52%), and Intestinimonas (0.51%). “Others” comprised 31.24% of the identified genera.
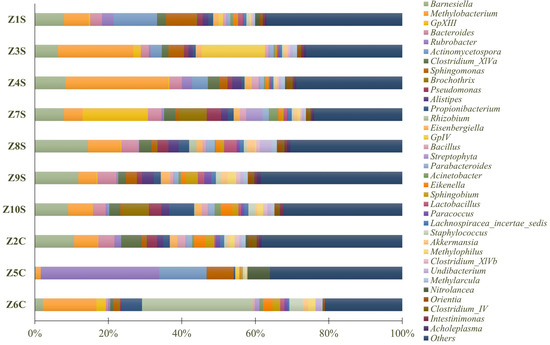
Figure 7.
Predominant genus of prokaryote populations present in swab samples.
Previous studies’ by Grube et al. (2015) revealed a wide range of bacteria that are associated with lichens [38]. This evidence has been provided by a series of studies based on morphological evidence using cultivable isolates of bacteria in lichens [19]. Bates et al. (2011) corroborate the idea that bacterial communities associated with lichens are highly organized, possibly as a result of the functional role they play in lichen symbiosis, and they also reveal that some bacterial taxa are prevalent across various lichen species [37]. On the lichen thalli, bacteria frequently form biofilm-like communities and are typically closely linked to fungal structures, especially with the hydrophilic surfaces of the lichens. Furthermore, the bacteria are occasionally seen inside the hyphae of the fungal hosts and may penetrate the intercellular matrix of the upper cortex at different depths [39].
Methylobacterium was the bacterial genus with the greatest representation in the samples under study. Members of the Methylobacterium genus are ubiquitous and provide crucial functions in environments that are natural as well as artificial. Most are pink-pigmented and facultative methylotrophs, able to grow on methanol and other one-carbon compounds as sources of energy and carbon. Bacteria of this genus have strong biofilm-producing ability, are pigment-producing microorganisms, and frequent colonizers stone substrates. Other studies have shown that this genus has a high tolerance to various stress factors (resistance to high temperatures, drying, and disinfecting agents), particularly under conditions of extreme nutrient limitation, making it difficult to remove the pink-pigmented biofilms already established by these microorganisms [40,41,42].
Among the identified bacteria, we can also highlight the presence of Rubrobacter, previously identified as being part of the microbial communities frequently associated with the presence of lichens [43] and extremely resistant to gamma and UV radiation [44], which has been identified in the literature as a significant contributor to biodeterioration, primarily because (i) it produces pink carotenoids (bacterioruberin and monoanhydrobacterioruberin) that result in the formation of colored spots on a variety of substrates [45]; (ii) it actively participates in efflorescence phenomena and mineral precipitation [46]; and (iii) it has the benefit of growing on stone surfaces even in direct sunlight due to its halotolerance and UV radiation resistance [47].
3.3. Evaluation of Antimicrobial Activity
With a view to choosing the biocides Preventol Ri 50 (Lanxess, Köln, Germany) and Biotin-T (C.T.S., Madrid, Spain) to be used in the conservation and restoration campaign of this monument, bioactivity tests were performed with mixed cultures carried out in the laboratory. These cultures were inoculated with swabs collected from four zones with notable evidence of biodeterioration inside the Casa de Fresco.
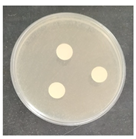
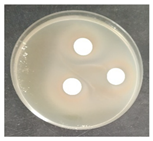
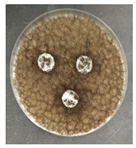
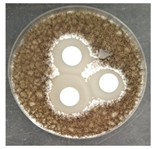
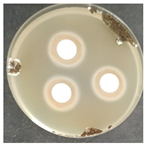
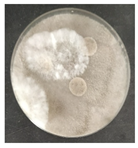
The antifungal activity was tested against a high concentration of microbial cells in mixed culture (Table 2) by swab collection from four sampling zones (namely Z1, Z3, Z4, and Z7). The formation of inhibition halos around the disks indicated the antifungal activity of the tested compounds and their diameter was considered as a measure of sensitivity (Table 3).

Table 2.
Antifungal activity assay.

Table 3.
Effect of commercial compounds against microbial communities from Casa de Fresco.
The microorganisms in sample Z1 displayed the highest susceptibility to the antimicrobial agent, as evidenced by complete inhibition. Following this, sample Z3 showed the second highest susceptibility, characterized by the formation of a 47.0 ± 3.0 mm inhibition halo. In contrast, when considering the microbial inhibition caused by commercial compounds, those found in sampling zone Z7 exhibited the greatest resistance to the action of Preventol, resulting in a very small inhibition halo of only 4.0 ± 2.0 mm. As expected, the control assay did not reveal any inhibition zones around the discs.
In summary, the effectiveness of inhibiting microbial growth varies across different sampling zones. Among the two commercial compounds tested, Biotin-T demonstrated the highest efficiency in inhibiting microbial growth in all sampling zones. This antimicrobial, based on quaternary ammonium salts, has a curative and preventive effect, resulting in the elimination of biological contamination and protecting materials from new contamination.
4. Conclusions
Through the process of biodeterioration—unwanted alterations to a valuable material caused by the action of living organisms—fungal species’ development and biological activity in cultural heritage materials are known to result in notable damage. Fungi are very adaptable, widespread, chemoheterotrophic microorganisms that can grow in a wide range of substrates and play a role in a number of biodeterioration processes.
We were able to identify lichenized fungi from the predominant phylum Ascomycota, with potential biodeteriogenic roles, such as Variospora, Verrucaria, Circinaria, Caloplaca, and Leprocaulon, which might be the explanation for the change in the color of the stone. In addition, some bacteria associated with lichens that might have important roles were identified, mainly from the phylum Proteobacteria, in which the genus Methylobacterium was dominant. Bacteria from the phyla Bacteroidetes, Actinobacteria, and Firmicutes were also identified.
In an attempt to mitigate or reduce fungal contamination, commercial antimicrobials were tested in four sampling zones, in which Biotin-T was the most efficient biocide in inhibiting microbial growth, producing an inhibition halo of 47, 35, and 27 mm for the sampling zone Z3, Z4, and Z7, respectively, and caused total inhibition in sample Z1. Thus, with biological colonization being the most extensive pathology present in the decorative elements, the organic biocide Biotin-T was the product chosen and the treatment methodology effectively applied to the different sampling areas.
We believe that this study was an asset for the implementation of an effective and precise plan for the conservation and restoration of the Casa de Fresco in the garden of the Paço de São Miguel, which took place between April and September 2022, preventing destruction and promoting its preservation.
Author Contributions
Conceptualization, I.S., A.C. and A.T.C.; methodology, C.S. and S.A.; formal analysis, I.S., C.S. and S.A.; investigation, I.S, C.S., A.Z.M., A.C., S.A. and A.T.C.; writing—original draft preparation, I.S.; writing—review and editing, A.C., A.T.C., C.S. and A.Z.M.; supervision, A.T.C.; funding acquisition, A.T.C., A.C., C.S. and A.Z.M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge financial support for the FCT—Foundation for Science and Technology, I.P.—within the scope of the projects UIDB/04449/2020 (HERCULES Lab), 2022.07303.PTDC/FCT, 10,54499/PTDC/CTA-AMB/0608/2020, 2022.10305.PTDC/FCT, and gratefully acknowledges the City University of Macau endowment to the Sustainable Heritage Chair and Sino-Portugal Joint Laboratory of Cultural Heritage Conservation Science supported by the Belt and Road Initiative. The authors also wish to acknowledge Fundação Eugénio de Almeida for allowing them to perform this study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequence data presented in this study has been uploaded to the NCBI Sequence Read Archive (SRA) under Bioproject PRJNA1076809 (SRR27975197-SRR27975201 and SRR27975458-SRR27975467). This data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA1076809 (accessed on 1 February 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Rosado, T.; Silva, M.; Dias, L.; Candeias, A.; Gil, M.; Mirão, J.; Pestana, J.; Caldeira, A.T. Microorganisms and the Integrated Conservation-Intervention Process of the Renaissance Mural Paintings from Casas Pintadas in Évora—Know to Act, Act to Preserve Tânia. J. King Saud Univ.—Sci. 2017, 29, 478–486. [Google Scholar] [CrossRef]
- Joseph, E. (Ed.) Microorganisms in the Deterioration and Preservation of Cultural Heritage; Springer: Berlin/Heidelberg, Germany, 2021; ISBN 9783030694104. [Google Scholar]
- Dakal, T.C.; Cameotra, S.S. Microbially Induced Deterioration of Architectural Heritages: Routes and Mechanisms Involved. Environ. Sci. Eur. 2012, 24, 1–13. [Google Scholar] [CrossRef]
- Gaylarde, C. Influence of Environment on Microbial Colonization of Historic Stone Buildings with Emphasis on Cyanobacteria. Heritage 2020, 3, 1469–1482. [Google Scholar] [CrossRef]
- Warscheid, T.; Braams, J. Biodeterioration of Stone: A Review. Int. Biodeterior. Biodegrad. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Zhang, G.; Gong, C.; Gu, J.; Katayama, Y.; Someya, T.; Gu, J.D. Biochemical Reactions and Mechanisms Involved in the Biodeterioration of Stone World Cultural Heritage under the Tropical Climate Conditions. Int. Biodeterior. Biodegrad. 2019, 143, 104723. [Google Scholar] [CrossRef]
- Milanesi, C.; Baldi, F.; Vignani, R.; Ciampolini, F.; Faleri, C.; Cresti, M. Fungal Deterioration of Medieval Wall Fresco Determined by Analysing Small Fragments Containing Copper. Int. Biodeterior. Biodegrad. 2006, 57, 7–13. [Google Scholar] [CrossRef]
- Tyagi, P.; Verma, R.K.; Jain, N. Fungal Degradation of Cultural Heritage Monuments and Management Options. Curr. Sci. 2021, 121, 1553–1560. [Google Scholar] [CrossRef]
- Zucconi, L.; Canini, F.; Isola, D.; Caneva, G. Fungi Affecting Wall Paintings of Historical Value: A Worldwide Meta-Analysis of Their Detected Diversity. Appl. Sci. 2022, 12, 2988. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A. Current Knowledge on the Fungal Degradation Abilities Profiled through Biodeteriorative Plate Essays. Appl. Sci. 2021, 11, 4196. [Google Scholar] [CrossRef]
- Rosado, T.; Dias, L.; Lança, M.; Nogueira, C.; Santos, R.; Martins, M.R.; Candeias, A.; Mirão, J.; Caldeira, A.T. Assessment of Microbiota Present on a Portuguese Historical Stone Convent Using High-Throughput Sequencing Approaches. MicrobiologyOpen 2020, 9, 1067–1084. [Google Scholar] [CrossRef]
- Lourenço e Silva, A. Conservação e Restauro de Embrechados. In Proceedings of the Conservação e Restauro de Artes Decorativas de Aplicação Arquitectónica, Porto, Portugal, 29 September 2012; p. 167. [Google Scholar]
- Da Silva, M.T.C. Novel Biocides for Cultural Heritage; Universidade de Évora: Évora, Portugal, 2017. [Google Scholar]
- Salvador, C.; Silva, M.; Rosado, T.; Freire, R.V.; Bordalo, R.; Candeias, A.; Caldeira, A.T. Biodeterioração de Pinturas de Cavalete: Desenvolvimento de Novas Estratégias de Mitigação. Conserv. Património 2016, 23, 119–124. [Google Scholar] [CrossRef]
- Schoch, C.L.; Sung, G.H.; López-Giráldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Matheny, P.B.; Kauff, F.; Wang, Z.; et al. The Ascomycota Tree of Life: A Phylum-Wide Phylogeny Clarifies the Origin and Evolution of Fundamental Reproductive and Ecological Traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef]
- Văcar, C.L.; Mircea, C.; Pârvu, M.; Podar, D. Diversity and Metabolic Activity of Fungi Causing Biodeterioration of Canvas Paintings. J. Fungi 2022, 8, 589. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and Diversity of Bacteria and Fungi Colonization in Stone Monuments Analyzed by High-Throughput Sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef] [PubMed]
- Sanders, W.B.; Masumoto, H. Lichen Algae: The Photosynthetic Partners in Lichen Symbioses. Lichenologist 2021, 53, 347–393. [Google Scholar] [CrossRef]
- Grimm, M.; Grube, M.; Schiefelbein, U.; Zühlke, D.; Bernhardt, J.; Riedel, K. The Lichens’ Microbiota, Still a Mystery? Front. Microbiol. 2021, 12, 623839. [Google Scholar] [CrossRef]
- Bungartz, F.; Søchting, U.; Arup, U. Teloschistaceae (Lichenized Ascomycota) from the Galapagos Islands: A Phylogenetic Revision Based on Morphological, Anatomical, Chemical, and Molecular Data. Plant Fungal Syst. 2020, 65, 515–576. [Google Scholar] [CrossRef]
- Caneva, G.; Fidanza, M.R.; Tonon, C.; Favero-Longo, S.E. Biodeterioration Patterns and Their Interpretation for Potential Applications to Stone Conservation: A Hypothesis from Allelopathic Inhibitory Effects of Lichens on the Caestia Pyramid (Rome). Sustainability 2020, 12, 1132. [Google Scholar] [CrossRef]
- Cámara, B.; De Buergo, M.Á.; Fort, R.; Souza-Egipsy, V.; Perez-Ortega, S.; De Los Ríos, A.; Wierzchos, J.; Ascaso, C. Anthropic Effect on the Lichen Colonization in Building Stones from Cultural Heritage. Period. Di Mineral. 2015, 84, 539–552. [Google Scholar] [CrossRef]
- Carter, N.E.A.; Viles, H.A. Lichen Hotspots: Raised Rock Temperatures beneath Verrucaria Nigrescens on Limestone. Geomorphology 2004, 62, 1–16. [Google Scholar] [CrossRef]
- Kamyabi, A.S.; Iranbakhsh, A.; Moniri, M.H.; Asri, Y. New Records of Five Verrucaria Species (Verrucariacaea, Ascomycota) from Iran. Feddes Repert. 2023, 134, 191–199. [Google Scholar] [CrossRef]
- Pinna, D.; Galeotti, M.; Perito, B.; Daly, G.; Salvadori, B. In Situ Long-Term Monitoring of Recolonization by Fungi and Lichens after Innovative and Traditional Conservative Treatments of Archaeological Stones in Fiesole (Italy). Int. Biodeterior. Biodegrad. 2018, 132, 49–58. [Google Scholar] [CrossRef]
- Morando, M.; Matteucci, E.; Nascimbene, J.; Borghi, A.; Piervittori, R.; Favero-Longo, S.E. Effectiveness of Aerobiological Dispersal and Microenvironmental Requirements Together Influence Spatial Colonization Patterns of Lichen Species on the Stone Cultural Heritage. Sci. Total Environ. 2019, 685, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Tonon, C.; Favero-Longo, S.E.; Matteucci, E.; Piervittori, R.; Croveri, P.; Appolonia, L.; Meirano, V.; Serino, M.; Elia, D. Microenvironmental Features Drive the Distribution of Lichens in the House of the Ancient Hunt, Pompeii, Italy. Int. Biodeterior. Biodegrad. 2019, 136, 71–81. [Google Scholar] [CrossRef]
- Favero-Longo, S.E.; Castelli, D.; Fubini, B.; Piervittori, R. Lichens on Asbestos-Cement Roofs: Bioweathering and Biocovering Effects. J. Hazard. Mater. 2009, 162, 1300–1308. [Google Scholar] [CrossRef]
- Nascimbene, J.; Salvadori, O. Lichen Recolonization on Restored Calcareous Statues of Three Venetian Villas. Int. Biodeterior. Biodegrad. 2008, 62, 313–318. [Google Scholar] [CrossRef]
- Altieri, A.; Pietrini, M.; Ricci, S.; Roccardi, A.; Piervittori, R. The Temples of the Archaeological Area of Paestum (Italy): A Case Study on Biodeterioration. In Proceedings of the 9th International Congress on Deterioration and Conservation of Stone, Venice, Italy, 19–24 June 2000; Fassina, V., Ed.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2000; pp. 433–443, ISBN 9780444505170. [Google Scholar]
- de los Ríos, A.; Cámara, B.; García del Cura, M.Á.; Rico, V.J.; Galván, V.; Ascaso, C. Deteriorating Effects of Lichen and Microbial Colonization of Carbonate Building Rocks in the Romanesque Churches of Segovia (Spain). Sci. Total Environ. 2009, 407, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, B.; Yang, X.; Ge, Q. Deterioration-Associated Microbiome of Stone Monuments: Structure, Variation, and Assembly. Appl. Environ. Microbiol. 2018, 84, 1–19. [Google Scholar] [CrossRef]
- Kosecka, M.; Kukwa, M.; Jabłońska, A.; Flakus, A.; Rodriguez-Flakus, P.; Ptach, L.; Guzow-Krzemińska, B. Phylogeny and Ecology of Trebouxia Photobionts From Bolivian Lichens. Front. Microbiol. 2022, 13, 779784. [Google Scholar] [CrossRef] [PubMed]
- Kroken, S.; Taylor, J.W. Phylogenetic Species, Reproductive Mode, and Specificity of the Green Alga Trebouxia Forming Lichens with the Fungal Genus Letharia. Bryologist 2000, 103, 645–660. [Google Scholar] [CrossRef]
- Ding, X.; Lan, W.; Yan, A.; Li, Y.; Katayama, Y.; Gu, J.D. Microbiome Characteristics and the Key Biochemical Reactions Identified on Stone World Cultural Heritage under Different Climate Conditions. J. Environ. Manag. 2022, 302, 114041. [Google Scholar] [CrossRef]
- Ding, Y.; Salvador, C.S.C.; Caldeira, A.T.; Angelini, E.; Schiavon, N. Biodegradation and Microbial Contamination of Limestone Surfaces: An Experimental Study from Batalha Monastery, Portugal. Corros. Mater. Degrad. 2021, 2, 31–45. [Google Scholar] [CrossRef]
- Bates, S.T.; Cropsey, G.W.G.; Caporaso, J.G.; Knight, R.; Fierer, N. Bacterial Communities Associated with the Lichen Symbiosis. Appl. Environ. Microbiol. 2011, 77, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Cernava, T.; Soh, J.; Fuchs, S.; Aschenbrenner, I.; Lassek, C.; Wegner, U.; Becher, D.; Riedel, K.; Sensen, C.W.; et al. Exploring Functional Contexts of Symbiotic Sustain within Lichen-Associated Bacteria by Comparative Omics. ISME J. 2015, 9, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Wedin, M. Lichenized Fungi and the Evolution of Symbiotic Organization. Microbiol. Spectr. 2016, 4, 749–765. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.P.; McDonald, I.R.; Wood, A.P. The Family Mehylobacteriaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 313–340. ISBN 9783642301971. [Google Scholar]
- Xu, F.F.; Morohoshi, T.; Wang, W.Z.; Yamaguchi, Y.; Liang, Y.; Ikeda, T. Evaluation of Intraspecies Interactions in Biofilm Formation by Methylobacterium Species Isolated from Pink-Pigmented Household Biofilms. Microbes Environ. 2014, 29, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.C.; Madureira, A.R.; Pintado, M.; Moreira, P.R. Biocontamination and Diversity of Epilithic Bacteria and Fungi Colonising Outdoor Stone and Mortar Sculptures. Appl. Microbiol. Biotechnol. 2022, 106, 3811–3828. [Google Scholar] [CrossRef] [PubMed]
- Bjelland, T.; Grube, M.; Hoem, S.; Jorgensen, S.L.; Daae, F.L.; Thorseth, I.H.; Øvreås, L. Microbial Metacommunities in the Lichen-Rock Habitat. Environ. Microbiol. Rep. 2011, 3, 434–442. [Google Scholar] [CrossRef]
- Egas, C.; Barroso, C.; Froufe, H.J.C.; Pacheco, J.; Albuquerque, L.; da Costa, M.S. Complete Genome Sequence of the Radiation-Resistant Bacterium Rubrobacter Radiotolerans RSPS-4. Stand. Genom. Sci. 2015, 9, 1062–1075. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Piñar, G.; Vybiral, D.; Lubitz, W.; Rölleke, S. Rubrobacter-Related Bacteria Associated with Rosy Discolouration of Masonry and Lime Wall Paintings. Arch. Microbiol. 2001, 176, 347–354. [Google Scholar] [CrossRef]
- Ettenauer, J.D.; Jurado, V.; Piñar, G.; Miller, A.Z.; Santner, M.; Saiz-Jimenez, C.; Sterflinger, K. Halophilic Microorganisms Are Responsible for the Rosy Discolouration of Saline Environments in Three Historical Buildings with Mural Paintings. PLoS ONE 2014, 9, e103844. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Li, X.; Osuga, Y.; Kawashima, A.; Gu, J.D.; Nasu, M.; Katayama, Y. Bacterial Communities in Pigmented Biofilms Formed on the Sandstone Bas-Relief Walls of the Bayon Temple, Angkor Thom, Cambodia. Microbes Environ. 2013, 28, 422–431. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).