Large Intra-Age Group Variation in Chromosome Abnormalities in Human Blastocysts
Abstract
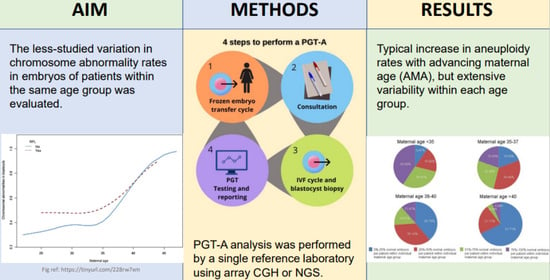
:1. Introduction
2. Materials and Methods
Consent and IRB Approval
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hassold, T.; Hall, H.; Hunt, P. The origin of human aneuploidy: Where we have been. where we are going. Hum. Mol. Genet. 2007, 16, R203–R208. [Google Scholar] [CrossRef]
- Subramaniyam, S.; Pulijaal, V.R.; Mathew, S. Double and multiple chromosomal aneuploidies in spontaneous abortions: A single institutional experience. J. Hum. Reprod. Sci. 2014, 7, 262–268. [Google Scholar] [CrossRef]
- Munne, S.; Lee, A.; Rosenwaks, Z.; Grifo, J.; Cohen, J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum. Reprod. 1993, 8, 2185–2191. Available online: http://www.ncbi.nlm.nih.gov/pubmed/8150922 (accessed on 11 November 2021). [CrossRef]
- Munne, S.; Weier, H.U.; Stein, J.; Grifo, J.; Cohen, J. A fast and efficient method for simultaneous X and Y in situ hybridization of human blastomeres. J. Assist. Reprod. Genet. 1993, 10, 82–90. Available online: http://www.ncbi.nlm.nih.gov/pubmed/8499685 (accessed on 11 November 2021). [CrossRef] [PubMed]
- Gutierrez-Mateo, C.; Colls, P.; Sanchez-Garcia, J.; Escudero, T.; Prates, R.; Ketterson, K.; Wells, D.; Munne, S. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil. Steril. 2011, 95, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.S.; Gemelos, G.; Baner, J.; Ryan, A.; Cinnioglu, C.; Banjevic, M.; Ross, R.; Alper, M.; Barrett, B.; Frederick, J.; et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum. Reprod. 2010, 25, 1066–1075. [Google Scholar] [CrossRef]
- Kung, A.; Munne, S.; Bankowski, B.; Coates, A.; Wells, D. Validation of next-generation sequencing for comprehensive chromosome screening of embryos. Reprod. Biomed. Online 2015, 31, 760–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treff, N.R.; Fedick, A.; Tao, X.; Devkota, B.; Taylor, D.; Scott, R.T., Jr. Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil. Steril. 2013, 99, 1377–1384.e6. [Google Scholar] [CrossRef] [PubMed]
- Treff, N.R.; Tao, X.; Ferry, K.M.; Su, J.; Taylor, D.; Scott, R.T., Jr. Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil. Steril. 2012, 97, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.; Delhanty, J.D. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol. Hum. Reprod. 2000, 6, 1055–1062. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11044470 (accessed on 1 August 2016). [CrossRef]
- Wells, D.; Kaur, K.; Grifo, J.; Glassner, M.; Taylor, J.C.; Fragouli, E.; Munne, S. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J. Med. Genet. 2014, 51, 553–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilton, L.; Williamson, R.; McBain, J.; Edgar, D.; Voullaire, L. Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. N. Engl. J. Med. 2001, 345, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- McArthur, S.J.; Leigh, D.; Marshall, J.T.; de Boer, K.A.; Jansen, R.P. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil. Steril. 2005, 84, 1628–1636. [Google Scholar] [CrossRef] [PubMed]
- Schoolcraft, W.B.; Fragouli, E.; Stevens, J.; Munne, S.; Katz-Jaffe, M.G.; Wells, D. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil. Steril. 2010, 94, 1700–1706. [Google Scholar] [CrossRef] [PubMed]
- Sermon, K.; Capalbo, A.; Cohen, J.; Coonen, E.; De Rycke, M.; De Vos, A.; Delhanty, J.; Fiorentino, F.; Gleicher, N.; Griesinger, G.; et al. The why, the how and the when of PGS 2.0: Current practices and expert opinions of fertility specialists, molecular biologists, and embryologists. MHR Basic Sci. Reprod. Med. 2016, 22, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Ata, B.; Kaplan, B.; Danzer, H.; Glassner, M.; Opsahl, M.; Tan, S.L.; Munne, S. Array CGH analysis shows that aneuploidy is not related to the number of embryos generated. Reprod. Biomed. Online 2012, 24, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Franasiak, J.M.; Forman, E.J.; Hong, K.H.; Werner, M.D.; Upham, K.M.; Treff, N.R.; Scott, R.T., Jr. The nature of aneuploidy with increasing age of the female partner: A review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil. Steril. 2014, 101, 656–663.e1. [Google Scholar] [CrossRef]
- Harton, G.L.; Munne, S.; Surrey, M.; Grifo, J.; Kaplan, B.; McCulloh, D.H.; Griffin, D.K.; Wells, D.; PGD Practitioners Group. Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil. Steril. 2013, 100, 1695–1703. [Google Scholar] [CrossRef]
- Munne, S.; Alikani, M.; Tomkin, G.; Grifo, J.; Cohen, J. Embryo morphology, developmental rates, and maternal age are correlated with chromosome abnormalities. Fertil. Steril. 1995, 64, 382–391. Available online: http://www.ncbi.nlm.nih.gov/pubmed/7615118 (accessed on 11 November 2021). [CrossRef] [PubMed] [Green Version]
- Munne, S.; Chen, S.; Colls, P.; Garrisi, J.; Zheng, X.; Cekleniak, N.; Lenzi, M.; Hughes, P.; Fischer, J.; Garrisi, M.; et al. Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod. Biomed. Online 2007, 14, 628–634. Available online: http://www.ncbi.nlm.nih.gov/pubmed/17509208 (accessed on 1 August 2016). [CrossRef]
- Konstantinidis, M.; Prates, R.; Goodall, N.-N.; Fischer, J.; Tecson, V.; Lemma, T.; Chu, B.; Jordan, A.; Armenti, E.; Wells, D.; et al. Live births following Karyomapping of human blastocysts: Experience from clinical application of the method. Reprod. Biomed. Online 2015, 31, 394–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuliev, A.; Verlinsky, Y. Meiotic and mitotic nondisjunction: Lessons from preimplantation genetic diagnosis. Hum. Reprod. Update 2004, 10, 401–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabinowitz, M.; Ryan, A.; Gemelos, G.; Hill, M.; Baner, J.; Cinnioglu, C.; Banjevic, M.; Potter, D.; Petrov, D.A.; Demko, Z. Origins and rates of aneuploidy in human blastomeres. Fertil. Steril. 2012, 97, 395–401. [Google Scholar] [CrossRef]
- Treff, N.R.; Su, J.; Tao, X.; Miller, K.A.; Levy, B.; Scott, R.T., Jr. A novel single-cell DNA fingerprinting method successfully distinguishes sibling human embryos. Fertil. Steril. 2010, 94, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Bielanska, M.; Tan, S.L.; Ao, A. High rate of mixoploidy among human blastocysts cultured in vitro. Fertil. Steril. 2002, 78, 1248–1253. Available online: http://www.ncbi.nlm.nih.gov/pubmed/12477520 (accessed on 11 November 2021). [CrossRef]
- Campbell, I.M.; Yuan, B.; Robberecht, C.; Pfundt, R.; Szafranski, P.; McEntagart, M.E.; Nagamani, S.C.S.; Erez, A.; Bartnik, M.; Wiśniowiecka-Kowalnik, B.; et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am. J. Hum. Genet. 2014, 95, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Fragouli, E.; Alfarawati, S.; Spath, K.; Wells, D. Morphological and cytogenetic assessment of cleavage and blastocyst stage embryos. Mol. Hum. Reprod. 2014, 20, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Magli, M.C.; Gianaroli, L.; Ferraretti, A.P.; Lappi, M.; Ruberti, A.; Farfalli, V. Embryo morphology and development are dependent on the chromosomal complement. Fertil. Steril. 2007, 87, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Munne, S.; Ary, J.; Zouves, C.; Escudero, T.; Barnes, F.; Cinioglu, C.; Ary, B.; Cohen, J. Wide range of chromosome abnormalities in the embryos of young egg donors. Reprod. Biomed. Online 2006, 12, 340–346. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16569324 (accessed on 11 November 2021). [CrossRef]
- Munne, S.; Grifo, J.; Wells, D. Mosaicism: “survival of the fittest” versus “no embryo left behind”. Fertil. Steril. 2016, 105, 1146–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandalinas, M.; Sadowy, S.; Alikani, M.; Calderon, G.; Cohen, J.; Munne, S. Developmental ability of chromosomally abnormal human embryos to develop to the blastocyst stage. Hum. Reprod. 2001, 16, 1954–1958. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11527904 (accessed on 11 November 2021). [CrossRef] [PubMed] [Green Version]
- Delhanty, J.D.A.; Griffin, D.K.; Handyside, A.H.; Harper, J.; Pieters, M.H.E.C.; Winston, R.M.L. Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination by fluorescent in-situ hybridisation. Hum. Mol. Genet. 1993, 2, 1183–1185. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K.; Handyside, A.H.; Penketh, R.J.A.; Winston, R.M.L.; Delhanty, J.D.A. Fluorescent in-situ hybridisation to interphase nuclei of human preimplantation embryos with X and Y specific probes. Hum. Reprod. 1991, 6, 101–105. [Google Scholar] [CrossRef]
- Munne, S.; Weier, H.U.; Grifo, J.; Cohen, J. Chromosome mosaicism in human embryos. Biol. Reprod. 1994, 51, 373–379. Available online: http://www.ncbi.nlm.nih.gov/pubmed/7803609 (accessed on 11 November 2021). [CrossRef] [PubMed]
- Fragouli, E.; Alfarawati, S.; Spath, K.; Tarozzi, N.; Borini, A.; Wells, D. The developmental potential of mosaic embryos. Fertil. Steril. 2017, 104, e96. [Google Scholar] [CrossRef]
- Greco, E.; Biricik, A.; Cotarelo, R.P.; Iammarone, E.; Rubino, P.; Tesarik, J.; Fiorentino, F.; Minasi, M.G. Successful implantation and live birth of a healthy boy after triple biopsy and double vitrification of oocyte-embryo-blastocyst. SpringerPlus 2015, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Grifo, J.; Colls, P.; Ribustello, L.; Escudero, T.; Liu, E.; Munne, S. Why do array-CGH (ACGH) euploid embryos miscarry? Reanalysis by NGS reveals undetected abnormalities which would have prevented 56% of the miscarriages. Fertil. Steril. 2017, 104, e14. [Google Scholar] [CrossRef]
- Munne, S.; Large, M.; Ribustello, L.; Blazek, J.; Gouw, F.; Grifo, J.; Haddad, G.; Chang, W.; Grunert, G.M.; Huang, A.; et al. PGS analysis of over 33,000 blastocysts using high resolution Next Generation Sequencing (HRNGS) of over 33,000 blastocysts using high resolution Next Generation Sequencing (HRNGS). Fertil. Steril. 2016, 106, e18–e19. [Google Scholar] [CrossRef]
- Fiorentino, F.; Spizzichino, L.; Bono, S.; Biricik, A.; Kokkali, G.; Rienzi, L.; Ubaldi, F.M.; Iammarrone, E.; Gordon, A.; Pantos, K. PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum. Reprod. 2011, 26, 1925–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munne, S.; Magli, C.; Adler, A.; Wright, G.; de Boer, K.; Mortimer, D.; Tucker, M.; Cohen, J.; Gianaroli, L. Treatment-related chromosome abnormalities in human embryos. Hum. Reprod. 1997, 12, 780–784. Available online: http://www.ncbi.nlm.nih.gov/pubmed/9159442 (accessed on 11 November 2021). [CrossRef] [PubMed] [Green Version]
- Munne, S.; Alikani, M. Culture-induced chromosome abnormalities: The canary in the mine. Reprod. Biomed. Online 2011, 22, 506–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munne, S.; Alikani, M.; Barritt, J.; Hesla, J.; Kaplan, B.; Alper, M.; McCulloh, D. Egg donor aneuploidy rates significantly differ between fertility centers. Fertil. Steril. 2014, 102, e121–e122. [Google Scholar] [CrossRef]
- Munne, S.; Held, K.R.; Magli, C.M.; Ata, B.; Wells, D.; Fragouli, E.; Baukloh, V.; Fischer, R.; Gianaroli, L. Intra-age, intercenter, and intercycle differences in chromosome abnormalities in oocytes. Fertil. Steril. 2012, 97, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, A.S.; Thornhill, A.R.; Ottolini, C.S.; Gordon, A.; Brown, A.P.C.; Taylor, J.; Bennett, K.; Handyside, A.; Griffin, D.K. Array comparative genomic hybridisation on first polar bodies suggests that non-disjunction is not the predominant mechanism leading to aneuploidy in humans. J. Med. Genet. 2011, 48, 433–437. Available online: http://jmg.bmj.com/content/48/7/433.abstract (accessed on 11 November 2021). [CrossRef] [PubMed]
- Fiorentino, F.; Biricik, A.; Bono, S.; Spizzichino, L.; Cotroneo, E.; Cottone, G.; Kokocinski, F.; Michel, C.E. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil. Steril. 2014, 101, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Colls, P.; Escudero, T.; Fischer, J.; Cekleniak, N.A.; Ben-Ozer, S.; Meyer, B.; Damien, M.; Grifo, J.A.; Hershlag, A.; Munne, S. Validation of array comparative genome hybridization for diagnosis of translocations in preimplantation human embryos. Reprod. Biomed. Online 2012, 24, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troude, P.; Guibert, J.; Bouyer, J.; de La Rochebrochard, E. Medical factors associated with early IVF discontinuation. Reprod. BioMed. Online 2017, 28, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Forman, E.J.; Hong, K.H.; Ferry, K.M.; Tao, X.; Taylor, D.; Levy, B.; Treff, N.R.; Scott, R.T., Jr. In-vitro fertilization with single euploid blastocyst transfer: A randomized controlled trial. Fertil. Steril. 2017, 100, 100–107.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, C.; Bellver, J.; Rodrigoa, L.; Castillón, G.; Guillén, A.; Vidal, C.; Giles, J.; Ferrando, M.; Cabanillas, S.; Remohí, J.; et al. In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: A randomized controlled study. Fertil. Steril. 2017, 107, 1122–1129. [Google Scholar] [CrossRef] [Green Version]
- Scott, R.T., Jr.; Upham, K.M.; Forman, E.J.; Hong, K.H.; Scott, K.L.; Taylor, D.; Tao, X.; Treff, N.R. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: A randomized controlled trial. Fertil. Steril. 2017, 100, 697–703. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, J.; Collins, G.S.; Salem, S.A.; Liu, X.; Lyle, S.S.; Peck, A.C.; Sills, E.S.; Salem, R.D. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: Results from a randomized pilot study. Mol. Cytogenet. 2012, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz-Jaffe, M.G.; Surrey, E.S.; Minjarez, D.A.; Gustofson, R.L.; Stevens, J.M.; Schoolcraft, W.B. Association of abnormal ovarian reserve parameters with a higher incidence of aneuploid blastocysts. Obstet. Gynecol. 2013, 121, 71–77. [Google Scholar] [CrossRef]
- Miyamoto, T.; Hasuike, S.; Yogev, L.; Maduro, M.R.; Ishikawa, M.; Westphal, H.; Lamb, D.J. Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet 2003, 362, 1714–1719. [Google Scholar] [CrossRef]
- Bannister, L.A.; Pezza, R.J.; Donaldson, J.R.; de Rooij, D.G.; Schimenti, K.J.; Camerini-Otero, R.D.; Schimenti, J.C. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007, 5, e105. [Google Scholar] [CrossRef] [Green Version]
- Cherry, S.M.; Adelman, C.A.; Theunissen, J.W.; Hassold, T.J.; Hunt, P.A.; Petrini, J.H. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr. Biol. 2007, 17, 373–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koehler, K.E.; Schrump, S.E.; Cherry, J.P.; Hassold, T.J.; Hunt, P.A. Near-human aneuploidy levels in female mice with homeologous chromosomes. Curr. Biol. 2006, 16, R579–R580. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, S.; Pellegrini, M.; Shuda, K.; Fernandez-Capetillo, O.; Liu, Y.; Martin, B.K.; Burkett, S.; Southon, E.; Pati, D.; Tessarollo, L.; et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 2007, 176, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Matzuk, M.M. Deconstructing mammalian reproduction: Using knockouts to define fertility pathways. Reproduction 2006, 131, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Schimenti, J.C. The genetics of human infertility by functional interrogation of SNPs in mice. Proc. Natl. Acad. Sci. USA 2015, 112, 10431–10436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munné, S.; Alikani, M.; Ribustello, L.; Colls, P.; Martínez-Ortiz, P.A.; Referring Physician Group; McCulloh, D.H. Euploidy rates in donor egg cycles significantly differ between fertility centers. Human Reprod. 2017, 32, 743–749. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.Y.T.; Mahbub, S.B.; Campbell, J.M.; Habibalahi, A.; Campugan, C.A.; Rose, R.D.; Chow, D.J.X.; Mustafa, S.; Goldys, E.M.; Dunning, K.R. Non-invasive, label-free optical analysis to detect aneuploidy within the inner cell mass of the preimplantation embryo. Hum. Reprod. 2021, deab233. [Google Scholar] [CrossRef] [PubMed]
- Victor, A.R.; Griffin, D.K.; Brake, A.J.; Tyndall, J.C.; Murphy, A.E.; Lepkowsky, L.T.; Lal, A.; Zouves, C.G.; Barnes, F.L.; McCoy, R.C.; et al. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum. Reprod. 2019, 34, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Coorens, T.H.H.; Oliver, T.R.W.; Sanghvi, R.; Sovio, U.; Cook, E.; Vento-Tormo, R.; Haniffa, M.; Young, M.D.; Rahbari, R.; Sebire, N.; et al. Inherent mosaicism and extensive mutation of human placentas. Nature 2021, 592, 80–85. [Google Scholar] [CrossRef]
- Desmyttere, S.; De Schepper, J.; Nekkebroeck, J.; De Vos, A.; De Rycke, M.; Staessen, C.; Liebaers, I.; Bonduelle, M. Two-year auxological and medical outcome of singletons born after embryo biopsy applied in preimplantation genetic diagnosis or preimplantation genetic screening. Hum. Reprod. 2009, 24, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Age | Min. (%) | 1st Quartile | Median | Mean | 3rd Quartile | Max. (%) | IQR | n (Cycles) |
|---|---|---|---|---|---|---|---|---|
| Donor | 0 | 16.67 | 30.77 | 32.64 | 46.15 | 100 | 29.48 | 1538 |
| 17–25 | 0 | 21.43 | 40 | 38.01 | 50 | 100 | 28.57 | 85 |
| 26 | 0 | 17.05 | 33.33 | 36.25 | 50 | 100 | 32.95 | 78 |
| 27 | 0 | 20 | 37.5 | 38.72 | 50 | 100 | 30 | 101 |
| 28 | 0 | 14.29 | 28.57 | 31.96 | 50 | 100 | 35.71 | 192 |
| 29 | 0 | 20 | 33.33 | 36.58 | 50 | 100 | 30 | 251 |
| 30 | 0 | 17.8 | 33.33 | 36.05 | 50 | 100 | 32.2 | 336 |
| 31 | 0 | 16.67 | 33.33 | 36.27 | 50 | 100 | 33.33 | 461 |
| 32 | 0 | 25 | 40 | 41.96 | 60 | 100 | 35 | 529 |
| 33 | 0 | 20 | 37.5 | 39.62 | 55.56 | 100 | 35.56 | 669 |
| 34 | 0 | 25 | 40 | 42.11 | 60 | 100 | 35 | 680 |
| 35 | 0 | 25 | 45.8 | 44.91 | 62.5 | 100 | 37.5 | 816 |
| 36 | 0 | 26.58 | 50 | 46.22 | 66.67 | 100 | 40.09 | 896 |
| 37 | 0 | 33.33 | 50 | 50.93 | 70.72 | 100 | 37.39 | 939 |
| 38 | 0 | 37.5 | 57.14 | 56.81 | 77.78 | 100 | 40.28 | 1043 |
| 39 | 0 | 40 | 62.5 | 59.96 | 83.33 | 100 | 43.33 | 1095 |
| 40 | 0 | 50 | 75 | 68.09 | 100 | 100 | 50 | 1044 |
| 41 | 0 | 57.14 | 80 | 74.01 | 100 | 100 | 42.86 | 938 |
| 42 | 0 | 66.67 | 93.33 | 78.14 | 100 | 100 | 33.33 | 811 |
| 43 | 0 | 75 | 100 | 84.75 | 100 | 100 | 25 | 613 |
| 44 | 0 | 80 | 100 | 87.29 | 100 | 100 | 20 | 358 |
| 45 | 0 | 75 | 100 | 83.5 | 100 | 100 | 25 | 182 |
| a | ||||||||||||||||
| Cohort Size | EGD | <30 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | >42 |
| 1 to 3 | 59.24% | 59.42% | 64.88% | 64.58% | 60.52% | 58.58% | 56.97% | 50.45% | 49.85% | 48.00% | 43.85% | 38.91% | 30.70% | 25.45% | 21.29% | 13.33% |
| 4 to 6 | 64.57% | 61.47% | 58.25% | 59.12% | 53.88% | 58.15% | 53.73% | 53.81% | 52.17% | 46.88% | 39.52% | 38.93% | 30.98% | 26.25% | 20.20% | 15.08% |
| 7 to 9 | 66.31% | 62.56% | 60.42% | 61.04% | 56.24% | 56.96% | 57.29% | 53.50% | 52.89% | 45.77% | 42.91% | 38.33% | 28.10% | 22.90% | 18.28% | 12.71% |
| 10 to 12 | 67.12% | 63.94% | 65.67% | 63.62% | 56.63% | 60.13% | 54.86% | 59.37% | 53.93% | 49.62% | 40.96% | 38.67% | 30.92% | 22.42% | 24.76% | 20.39% |
| 13 to 15 | 68.00% | 63.01% | 66.82% | 65.30% | 58.12% | 60.10% | 61.25% | 52.92% | 53.35% | 56.00% | 43.44% | 41.56% | 35.22% | 29.78% | 25.37% | 24.44% |
| >16 | 67.26% | 64.46% | 66.33% | 70.88% | 60.25% | 60.23% | 61.29% | 59.06% | 57.41% | 48.42% | 44.68% | 43.55% | 38.71% | 29.41% | 17.91% | 11.11% |
| Total | 17,032 | 4662 | 2228 | 2873 | 3199 | 3993 | 3817 | 4523 | 4786 | 4785 | 5144 | 5109 | 4624 | 4036 | 3246 | 4471 |
| b | ||||||||||||||||
| Cohort Size | EGD | 21–31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45+ |
| 1 | 63.64% | 60.00% | 48.57% | 57.14% | 68.18% | 45.71% | 54.79% | 53.75% | 40.00% | 43.48% | 36.43% | 28.69% | 25.16% | 15.52% | 9.64% | 15.48% |
| 2 | 61.93% | 63.30% | 60.00% | 64.66% | 52.24% | 50.51% | 45.10% | 46.85% | 42.04% | 36.25% | 30.66% | 25.15% | 21.23% | 13.54% | 12.21% | 13.04% |
| 3 | 57.43% | 62.33% | 64.29% | 55.00% | 57.41% | 50.91% | 51.42% | 46.96% | 45.53% | 39.53% | 29.31% | 24.84% | 19.73% | 13.57% | 12.28% | 14.29% |
| >3 | 66.53% | 62.39% | 56.05% | 58.49% | 56.11% | 54.72% | 53.11% | 47.66% | 41.33% | 38.92% | 30.74% | 24.96% | 20.17% | 15.12% | 15.29% | 15.23% |
| a | ||||||||||||||||
| Cohort Size | EGD | <30 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | >42 |
| 1 to 3 | 58.59% | 47.92% | 40.54% | 46.09% | 48.28% | 40.00% | 43.69% | 43.25% | 42.86% | 32.26% | 30.03% | 28.81% | 25.00% | 21.91% | 12.28% | 11.69% |
| 4 to 6 | 53.72% | 48.40% | 52.03% | 54.10% | 49.75% | 42.63% | 44.20% | 45.83% | 38.46% | 32.77% | 35.53% | 30.54% | 23.46% | 17.09% | 14.38% | 12.64% |
| 7 to 9 | 57.09% | 47.45% | 53.69% | 49.75% | 43.60% | 45.53% | 42.09% | 43.81% | 39.36% | 37.76% | 36.48% | 27.29% | 24.74% | 19.48% | 12.72% | 11.48% |
| 10 to 12 | 60.42% | 52.38% | 49.48% | 43.75% | 42.46% | 44.95% | 38.67% | 45.56% | 40.52% | 37.50% | 34.97% | 25.91% | 26.58% | 15.79% | 6.58% | 13.79% |
| 13 to 15 | 64.11% | 52.98% | 34.29% | 61.87% | 32.80% | 37.76% | 36.14% | 46.94% | 49.70% | 25.77% | 26.61% | 24.03% | 19.20% | 13.89% | 7.94% | 9.41% |
| >16 | 62.84% | 59.26% | 33.33% | 43.40% | 39.56% | 47.78% | 44.25% | 30.36% | 41.03% | 36.72% | 23.97% | 24.09% | 17.56% | 20.29% | 8.33% | 7.27% |
| Total | 6090 | 1822 | 862 | 1270 | 1615 | 1436 | 1680 | 1959 | 1913 | 1898 | 1964 | 2026 | 1893 | 1570 | 1149 | 1386 |
| b | ||||||||||||||||
| Cohort Size | EGD | 21–31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45+ |
| 1 | 40.91% | 38.19% | 53.33% | 37.50% | 31.58% | 33.33% | 36.11% | 33.33% | 21.95% | 28.57% | 17.74% | 19.05% | 15.52% | 11.11% | 8.82% | 11.11% |
| 2 | 50.00% | 41.35% | 56.52% | 28.85% | 40.22% | 37.93% | 38.30% | 32.00% | 24.60% | 36.96% | 25.27% | 26.71% | 13.11% | 6.67% | 12.00% | 6.90% |
| 3 | 60.45% | 46.24% | 39.58% | 40.15% | 46.94% | 46.30% | 41.83% | 27.32% | 32.28% | 20.29% | 23.50% | 17.95% | 11.90% | 13.33% | 12.00% | 5.88% |
| >3 | 59.98% | 50.81% | 43.96% | 44.36% | 42.22% | 44.80% | 40.31% | 35.96% | 33.68% | 28.35% | 22.74% | 17.56% | 11.20% | 10.68% | 12.62% | 12.10% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawarkar, S.; Griffin, D.K.; Ribustello, L.; Munné, S. Large Intra-Age Group Variation in Chromosome Abnormalities in Human Blastocysts. DNA 2021, 1, 91-104. https://doi.org/10.3390/dna1020010
Sawarkar S, Griffin DK, Ribustello L, Munné S. Large Intra-Age Group Variation in Chromosome Abnormalities in Human Blastocysts. DNA. 2021; 1(2):91-104. https://doi.org/10.3390/dna1020010
Chicago/Turabian StyleSawarkar, Sarthak, Darren K. Griffin, Lia Ribustello, and Santiago Munné. 2021. "Large Intra-Age Group Variation in Chromosome Abnormalities in Human Blastocysts" DNA 1, no. 2: 91-104. https://doi.org/10.3390/dna1020010
APA StyleSawarkar, S., Griffin, D. K., Ribustello, L., & Munné, S. (2021). Large Intra-Age Group Variation in Chromosome Abnormalities in Human Blastocysts. DNA, 1(2), 91-104. https://doi.org/10.3390/dna1020010