Karyotype Organization of the Endangered Species Yellow Cardinal (Gubernatrix cristata)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal
2.2. Cell Culture and Karyotype Description
2.3. The 18S rDNA Clusters in Gubernatrix cristata and in Passeriformes Species
3. Results
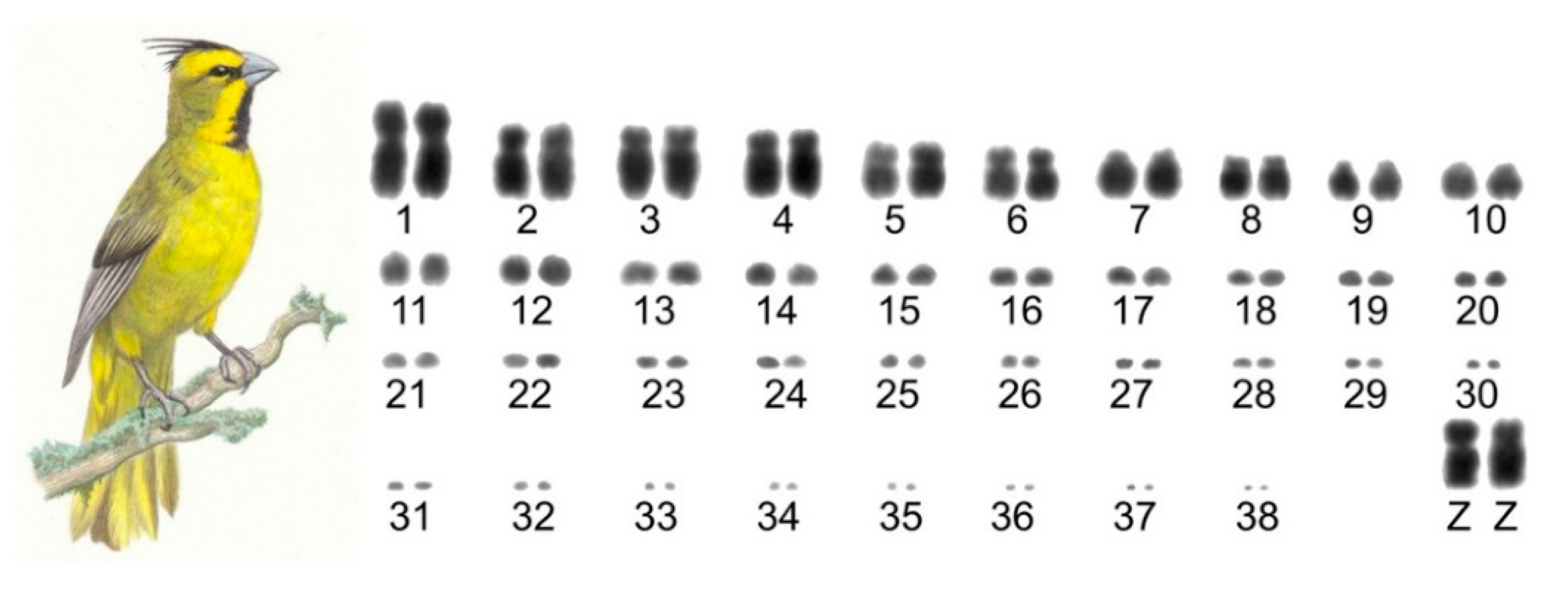
3.1. The Karyotype of G. cristata
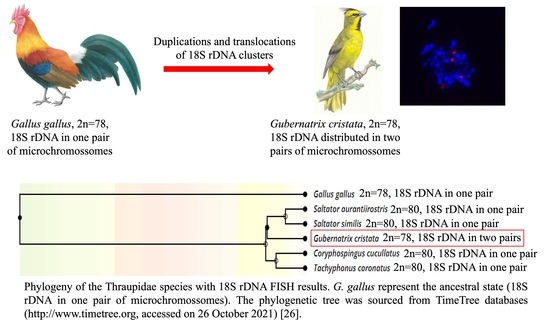
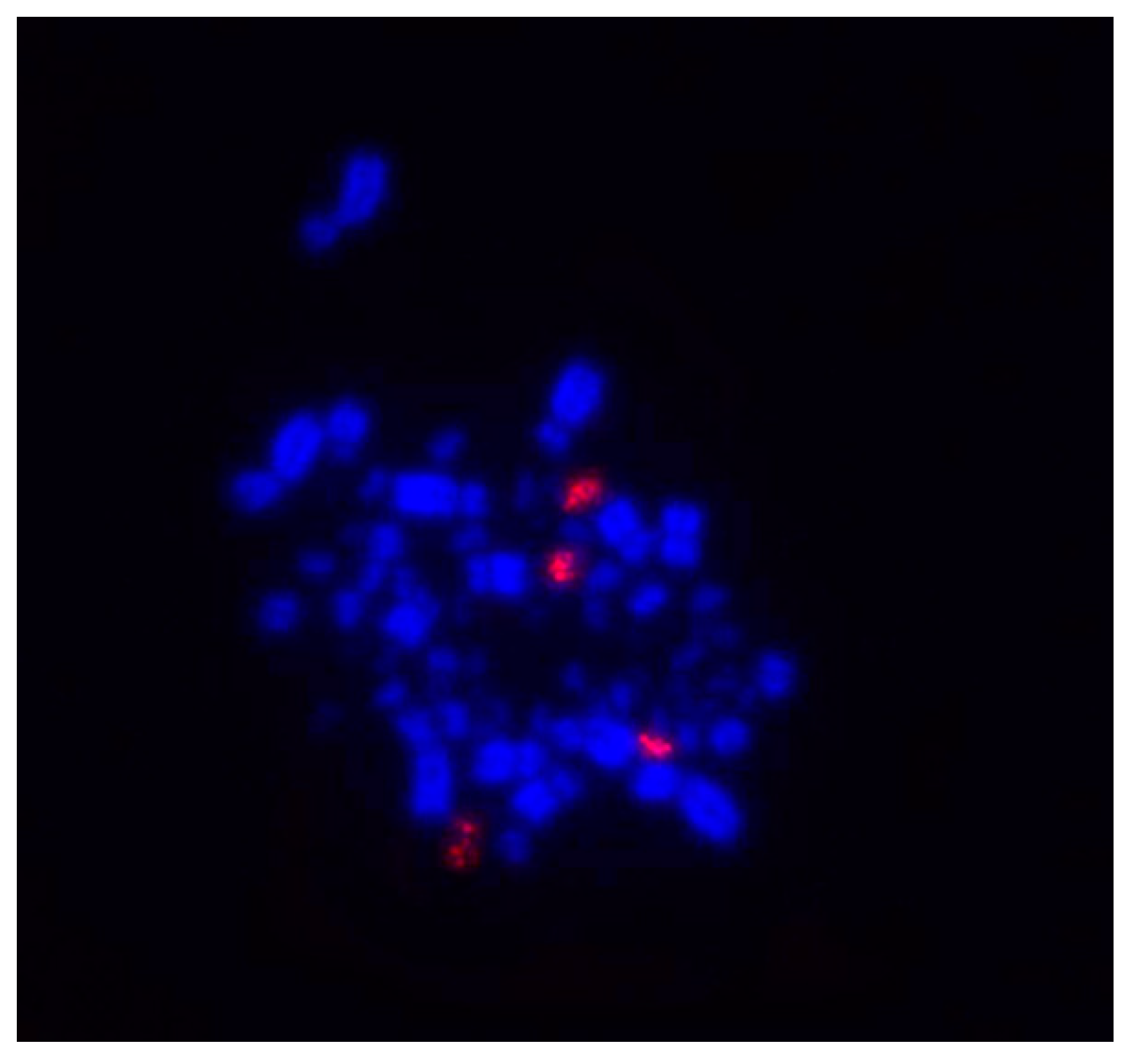
3.2. The 18S rDNA Distribution in G. cristata
3.3. Comparisions of 18S rDNA Distribution among Passeriformes Species
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Degrandi, T.M.; Barcellos, S.; Costa, A.; Garnero, A.; Hass, I.; Gunski, R.J. Introducing the Bird Chromosome Database: An Overview of Cytogenetic Studies in Birds. Cytogenet. Genome Res. 2020, 160, 199–205. [Google Scholar] [CrossRef]
- Griffin, D.; Robertson, L.; Tempest, H.; Skinner, B. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 2007, 117, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; Ferguson-Smith, M.A.; De Oliveira, E.H.C. Karyotype Evolution in Birds: From Conventional Staining to Chromosome Painting. Genes 2018, 9, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankl-Vilches, C.; Kuhl, H.; Werber, M.; Klages, S.; Kerick, M.; Bakker, A.; De Oliveira, E.H.C.; Reusch, C.; Capuano, F.; Vowinckel, J.; et al. Using the canary genome to decipher the evolution of hormone-sensitive gene regulation in seasonal singing birds. Genome Biol. 2015, 16, 19. [Google Scholar] [CrossRef] [Green Version]
- Peona, V.; Weissensteiner, M.H.; Suh, A. How complete are “complete” genome assemblies?-An avian perspective. Mol. Ecol. Resour. 2018, 18, 1188–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauclair, L.; Ramé, C.; Arensburger, P.; Piégu, B.; Guillou, F.; Dupont, J.; Bigot, Y. Sequence properties of certain GC rich avian genes, their origins and absence from genome assemblies: Case studies. BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Birdlife International. Scientists Discover an “Invisible Barrier” that Holds the Answer to One of Nature’s Little Mysteries. Birdlife News Posts. 16 March 2012. Available online: http://www.birdlife.org (accessed on 14 June 2012).
- Pessino, M.E.M. Nuevo registro y descripcion de hibridos entre cardenal amarillo (Gubernatrix cristata) y diuca comun (Diuca diuca) en la provincia de La Pampa, Argentina. Rev. Nuestras Aves 2006, 52, 16–18. [Google Scholar]
- Martins-Ferreira, C.; Repenning, M.; Damiani, R.V. Gubernatrix cristata. In Plano de Ação Nacional Para a Conservação Daos Passeriformes Ameaçados Dos Campos Sulinos E Espinilho; Serafini, P.P., Ed.; Instituto Chico Mendes de Conservação da Biodiversidade ICMBio: Brasília, Brazil, 2013; pp. 116–119. [Google Scholar]
- Domínguez, M.; Tiedemann, R.; Reboreda, J.C.; Segura, L.; Tittarelli, F.; Mahler, B. Genetic structure reveals management units for the yellow cardinal (Gubernatrix cristata), endangered by habitat loss and illegal trapping. Conserv. Genet. 2017, 18, 1131–1140. [Google Scholar] [CrossRef]
- Chebez, J.C. Los Que Se Van: Especies Argentinas En Peligro; Albatros: Buenos Aires, Argentina, 1994; p. 604. [Google Scholar]
- Ortiz, D. Distribucion historica y actual del cardenal amarillo (Gubernatrix cristata) en el litoral luvial argentino. In Temas de la Biodiversidad del Litoral Luvial Argentino, III; Acenolaza, F.G., Ed.; INSUGEO: San Miguel de Tucumán, Argentina, 2008; pp. 121–126. [Google Scholar]
- Ridgely, R.S.; Tudor, G. The Birds of South America; University of Texas Press: Austin, TX, USA, 1989; Volume I, p. 516. [Google Scholar]
- Azpiroz, A.B. Aves del Uruguay: Lista E Introduccion A su Biologia Y Conservacion; Aves Uruguay-GUPECA: Montevideo, Uruguay, 2003; p. 104. [Google Scholar]
- Bencke, G.A. Diversidade e conservação da fauna dos Campos do Sul do Brasil. In Campos Sulinos: Conservação e uso Sustentável da Biodiversidade; Pillar, V.D.P., Muller, S.C., Castilhos, Z.M.d.S., Jacques, A.V.A., Eds.; MMA: Brasília, Brazil, 2009; pp. 101–121. [Google Scholar]
- Martins-Ferreira, C.; Santos, M.O.; Hadddrath, O.; Baker, A.J.; de Freitas, T.R.O. Isolation and characterization of 10 mi-crosatellite loci in the Yellow Cardinal Gubernatrix cristata En. Mol. Ecol. Resour. 2010, 10, 751–754. [Google Scholar]
- Fontana, C.S.; Bencke, G.A.; Reis, R.E. Livro Vermelho da Fauna Ameaçada de Exatianção No Rio Grande Do Sul; EDIPUCRS: Porto Alegre, Brazil, 2003; p. 632. [Google Scholar]
- Di Giacomo, A.S. Areas Importantes Para La Conservacion de Las Aves En Argentina: Sitios Prioritarios Para la Conservacion de la Biodiversidad. TEMAS De Naturaleza Y Conservacion, 5th ed.; Aves Argentinas/Asociacion Ornitologica del Plata: Buenos Aires, Argentina, 2005. [Google Scholar]
- Dominguez, M.; Lapido, R.; Gorrindo, A.; Archuby, D.; Correa, E.; Llanos, F.; Reales, F.; Piantanida, F.; Marateo, G.; Meriggi, J.; et al. A citizen science survey discloses the current distribution of the endangered Yellow Cardinal Gubernatrix cristata in Argentina. Bird Conserv. Int. 2020, 31, 139–150. [Google Scholar] [CrossRef]
- Martins-Ferreira, C.; Bencke, G.A.; Fontana, C.S.; Dias, R.A.; Repenning, M.; Damiani, R.V.; Mauricio, G.N.; Gianuca, A.T.; Franz, I.; Rovedder, C.E.; et al. Plano de Ação Nacional Para a Conservação Dos Passeriformes Ameaçados dos Campos Sulinos e Espinilho; Instituto chico mendes de Conservação da Biodiversidade, ICMBio: Brasília, Brazil, 2013; p. 212. [Google Scholar]
- Beier, C.; Repenning, M.; Da Pereira, M.S.; Pereira, A.; Fontana, C.S. Cooperative breeding and demography of Yellow Cardinal Gubernatrix cristata in Brazil. Rev. Bras. Ornitol. 2017, 25, 12–19. [Google Scholar] [CrossRef]
- Furo, I.D.O.; Kretschmer, R.; Dos Santos, M.S.; Carvalho, C.A.D.L.; Gunski, R.J.; O’Brien, P.C.; Ferguson-Smith, M.A.; Cioffi, M.B.; De Oliveira, E.H. Chromosomal Mapping of Repetitive DNAs in Myiopsitta monachus and Amazona aestiva (Psittaciformes, Psittacidae) with Emphasis on the Sex Chromosomes. Cytogenet. Genome Res. 2017, 151, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.S. Reviewing the chromosome nomenclature of Levan et al. Rev. Bras. Genet. 1986, 9, 741–743. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenet-ics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Shinsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Daniels, L.M. Molecular and cytogenetic organization of the 5S ribosomal DNA array in chicken (Gallus gallus). Chromosome Res. 2003, 11, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Degrandi, T.M.; Gunski, R.J.; Garnero, A.D.V.; De Oliveira, E.H.C.; Kretschmer, R.; De Souza, M.S.; Barcellos, S.A.; Hass, I. The distribution of 45S rDNA sites in bird chromosomes suggests multiple evolutionary histories. Genet. Mol. Biol. 2020, 43, e20180331. [Google Scholar] [CrossRef] [PubMed]
- Ribas, T.; Nagamachi, C.Y.; Aleixo, A.; Pinheiro, M.L.S.; O’brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Suarez, P.; Pieczarka, J.C. Chromosome painting in Glyphorynchus spirurus (Vieillot, 1819) detects a new fission in Passeriformes. PLoS ONE 2018, 13, e0202040. [Google Scholar] [CrossRef]
- Rodrigues, B.S.; Kretschmer, R.; Gunski, R.J.; Garnero, A.; O’Brien, P.C.; Ferguson-Smith, M.; De Oliveira, E.H. Chromosome Painting in Tyrant Flycatchers Confirms a Set of Inversions Shared by Oscines and Suboscines (Aves, Passeriformes). Cytogenet. Genome Res. 2017, 153, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; De Oliveira, E.H.C.; Dos Santos, M.S.; Furo, I.D.O.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Garnero, A.; Gunski, R.J. Chromosome mapping of the large elaenia (Elaenia spectabilis): Evidence for a cytogenetic signature for passeriform birds? Biol. J. Linn. Soc. 2015, 115, 391–398. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, T.D.; Kretschmer, R.; Bertocchi, N.; O’Brien, P.C.; Ferguson-Smith, M.A.; Garnero, A.D.V.; De Oliveira, E.H.C.; Gunski, R.J. The molecular cytogenetic characterization of Conopophaga lineata indicates a common chromosome rearrangement in the Parvorder Furnariida (Aves, Passeriformes). Genet. Mol. Biol. 2020, 43, e20200018. [Google Scholar] [CrossRef]
- Santos, M.D.S.D.; Kretschmer, R.; Frankl-Vilches, C.; Bakker, A.; Gahr, M.; O’brien, P.C.M.; Ferguson-Smith, M.A.; De Oliveira, E.H.C. Comparative cytogenetics between two important songbird, models: The zebra finch and the canary. PLoS ONE 2017, 12, e0170997. [Google Scholar] [CrossRef]
- Kretschmer, R.; Gunski, R.J.; Garnero, A.; Furo, I.D.O.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; De Oliveira, E.H.C. Molecular Cytogenetic Characterization of Multiple Intrachromosomal Rearrangements in Two Representatives of the Genus Turdus (Turdidae, Passeriformes). PLoS ONE 2014, 9, e103338. [Google Scholar] [CrossRef] [PubMed]
- Bülau, S.E.; Kretschmer, R.; Gunski, R.J.; Garnero, A.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; De Oliveira, E.H.C.; De Freitas, T.R.O. Chromosomal polymorphism and comparative chromosome painting in the rufous-collared sparrow (Zonotrichia capensis). Genet. Mol. Biol. 2018, 41, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.D.S.D.; Kretschmer, R.; Silva, F.A.O.; Ledesma, M.A.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Garnero, A.; De Oliveira, E.H.C.; Gunski, R.J. Intrachromosomal rearrangements in two representatives of the genus Saltator (Thraupidae, Passeriformes) and the occurrence of heteromorphic Z chromosomes. Genetica 2015, 143, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Potter, S.; O’Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.; Fukui, K.; Graves, J.A.M.; Griffin, D.; Grutzner, F.; et al. Chromosomics: Bridging the Gap between Genomes and Chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, V.C.S.; Garnero, A.D.V.; dos Santos, L.P.; Silva, R.R.; Barbosa, M.; Bonifácio, H.L.; Gunski, R.J. Alta similaridade cariotípica na família Emberezidae (Aves: Passeriformes). Biosci. J. 2009, 25, 99–111. [Google Scholar]
- Völker, M.; Backström, N.; Skinner, B.; Langley, E.J.; Bunzey, S.K.; Ellegren, H.; Griffin, D.K. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 2010, 20, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Hooper, D.M.; Price, T.D. Chromosomal inversion differences correlate with range overlap in passerine birds. Nat. Ecol. Evol. 2017, 1, 1526–1534. [Google Scholar] [CrossRef]
- Nishida-Umehara, C.; Tsuda, Y.; Ishijima, J.; Ando, J.; Fujiwara, A.; Matsuda, Y.; Griffin, D.K. The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res. 2007, 15, 721–734. [Google Scholar] [CrossRef]
- Stitou, S.; Burgos, M.; Zurita, F.; Jimenez, R.; Sanchez, A.; De La Guardia, R.D. Recent evolution of NOR-bearing and sex chromosomes of the North African rodent Lemniscomys barbarus. Chromosome Res. 1997, 5, 481–485. [Google Scholar] [CrossRef]

| Species | Family | Suborder | No 1 | Diploid Number | Reference |
|---|---|---|---|---|---|
| Syndactila rufosuperciliata | Furnariidae | Suboscines | 2 micros | 82 | [27] |
| Cranioleuca obsoleta | Furnariidae | Suboscines | 2 micros | 82 | [27] |
| Furnarius rufus | Furnariidae | Suboscines | 2 micros | 82 | [27] |
| Synallaxis albescens | Furnariidae | Suboscines | 2 micros | 82 | [27] |
| Anumbius annumbi | Furnariidae | Suboscines | 2 micros | 82 | [27] |
| Dendrocolaptes platyrostris | Furnariidae | Suboscines | 2 macros | 82 | [27] |
| Glyphorynchus spirurus | Furnariidae | Suboscines | 2 macros | 80 | [28] |
| Schiffornis virescens | Tityridae | Suboscines | 2 micros | 82 | [27] |
| Myiarchus ferox | Tyrannidae | Suboscines | 2 micros | 76 | [27] |
| Pitangus sulphuratus | Tyrannidae | Suboscines | 2 micros | 80 | [29] |
| Satrapa icterophrys | Tyrannidae | Suboscines | 2 micros | 82 | [29] |
| Serpophaga subcristata | Tyrannidae | Suboscines | 4 micros | 82 | [29] |
| Elaenia spectabilis | Tyrannidae | Suboscines | 4 micros | 80 | [30] |
| Conopophaga lineata | Conopophagidae | Suboscines | 2 micros | 78 | [31] |
| Taeniopygia guttata | Estrildidae | Oscines | 2 micros | 80 | [32] |
| Basileuterus culicivorus | Parulidae | Oscines | 2 micros | 80 | [27] |
| Serinus canaria | Fringillidae | Oscines | 4 micros | 80 | [32] |
| Agelaioides badius | Icteridae | Oscines | 4 micros | 80 | [27] |
| Molothrus bonariensis | Icteridae | Oscines | 2 micros | 80 | [27] |
| Turdus rufiventris | Turdidae | Oscines | 6 micros | 78 | [33] |
| Turdus albicollis | Turdidae | Oscines | 4 micros | 78 | [33] |
| Zonotrichia capensis | Passerellidae | Oscines | 2 micros | 80 | [34] |
| Saltator similis | Thraupidae | Oscines | 2 micros | 80 | [35] |
| Saltator aurantiirostris | Thraupidae | Oscines | 2 micros | 80 | [35] |
| Tachyphonus coronatus | Thraupidae | Oscines | 2 micros | 80 | [27] |
| Coryphospingus cucullatus | Thraupidae | Oscines | 2 micros | 80 | [27] |
| Gubernatrix cristata | Thraupidae | Oscines | 4 micros | 78 | Present study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bülau, S.E.; Kretschmer, R.; Furo, I.d.O.; de Oliveira, E.H.C.; de Freitas, T.R.O. Karyotype Organization of the Endangered Species Yellow Cardinal (Gubernatrix cristata). DNA 2021, 1, 77-83. https://doi.org/10.3390/dna1020008
Bülau SE, Kretschmer R, Furo IdO, de Oliveira EHC, de Freitas TRO. Karyotype Organization of the Endangered Species Yellow Cardinal (Gubernatrix cristata). DNA. 2021; 1(2):77-83. https://doi.org/10.3390/dna1020008
Chicago/Turabian StyleBülau, Sandra Eloisa, Rafael Kretschmer, Ivanete de Oliveira Furo, Edivaldo Herculano Correa de Oliveira, and Thales Renato Ochotorena de Freitas. 2021. "Karyotype Organization of the Endangered Species Yellow Cardinal (Gubernatrix cristata)" DNA 1, no. 2: 77-83. https://doi.org/10.3390/dna1020008
APA StyleBülau, S. E., Kretschmer, R., Furo, I. d. O., de Oliveira, E. H. C., & de Freitas, T. R. O. (2021). Karyotype Organization of the Endangered Species Yellow Cardinal (Gubernatrix cristata). DNA, 1(2), 77-83. https://doi.org/10.3390/dna1020008