Kinetic Studies on the 2-Oxoglutarate/Fe(II)-Dependent Nucleic Acid Modifying Enzymes from the AlkB and TET Families
Abstract
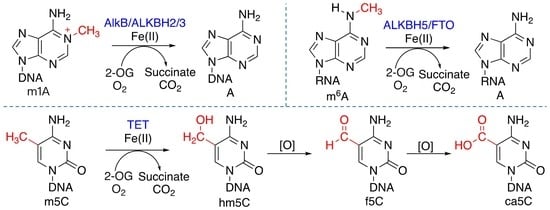
1. Introduction
2. Kinetic Studies of the AlkB and TET Family Enzymes
2.1. ALKBH1
2.2. ALKBH2
2.3. ALKBH3
2.4. ALKBH5
2.5. FTO
2.6. Ten-Eleven Translocation and J-Binding Protein (TET/JBP) Proteins
2.7. AlkB
2.8. Other Mammalian Homologs of AlkB
3. Conclusions
Funding
Conflicts of Interest
References
- Rydberg, B.; Lindahl, T. Nonenzymatic Methylation of DNA by the Intracellular Methyl Group Donor S-Adenosyl-L-Methionine Is a Potentially Mutagenic Reaction. EMBO J. 1982, 1, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Mathison, B.H.; Frame, S.R.; Bogdanffy, M.S. DNA Methylation, Cell Proliferation, and Histopathology in Rats Following Repeated Inhalation Exposure to Dimethyl Sulfate. Inhal. Toxicol. 2004, 16, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.P.; Cho, Y.-J.; Huang, H.; Kim, H.-Y.; Kozekov, I.D.; Kozekova, A.; Wang, H.; Minko, I.G.; Lloyd, R.S.; Harris, T.M.; et al. Interstrand DNA Cross-Links Induced by α,β-Unsaturated Aldehydes Derived from Lipid Peroxidation and Environmental Sources. Acc. Chem. Res. 2008, 41, 793–804. [Google Scholar] [CrossRef]
- Marnett, L. Endogenous DNA Damage and Mutation. Trends Genet. 2001, 17, 214–221. [Google Scholar] [CrossRef]
- Bordin, D.L.; Lirussi, L.; Nilsen, H. Cellular Response to Endogenous DNA Damage: DNA Base Modifications in Gene Expression Regulation. DNA Repair 2021, 99, 103051. [Google Scholar] [CrossRef]
- Barrows, L.R.; Magee, P.N. Nonenzymatic Methylation of DNA by S-Adenosylmethionine in Vitro. Carcinogenesis 1982, 3, 349–351. [Google Scholar] [CrossRef]
- Sedgwick, B.; Lindahl, T. Recent Progress on the Ada Response for Inducible Repair of DNA Alkylation Damage. Oncogene 2002, 21, 8886–8894. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Sedgwick, B.; Sekiguchi, M.; Nakabeppu, Y. Regulation and Expression of the Adaptive Response to Alkylating Agents. Annu. Rev. Biochem. 1988, 57, 133–157. [Google Scholar] [CrossRef]
- Mielecki, D.; Grzesiuk, E. Ada Response—A Strategy for Repair of Alkylated DNA in Bacteria. FEMS Microbiol. Lett. 2014, 355, 1–11. [Google Scholar] [CrossRef]
- Mielecki, D.; Wrzesiński, M.; Grzesiuk, E. Inducible Repair of Alkylated DNA in Microorganisms. Mutat. Res./Rev. Mutat. Res. 2015, 763, 294–305. [Google Scholar] [CrossRef]
- Aas, P.A.; Otterlei, M.; Falnes, P.Ø.; Vågbø, C.B.; Skorpen, F.; Akbari, M.; Sundheim, O.; Bjørås, M.; Slupphaug, G.; Seeberg, E.; et al. Human and Bacterial Oxidative Demethylases Repair Alkylation Damage in Both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. The DNA-Repair Protein AlkB, EGL-9, and Leprecan Define New Families of 2-Oxoglutarate- and Iron-Dependent Dioxygenases. Genome Biol 2001, 2, research0007.1. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.Ø.; Johansen, R.F.; Seeberg, E. AlkB-Mediated Oxidative Demethylation Reverses DNA Damage in Escherichia coli. Nature 2002, 419, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative Demethylation by Escherichia coli AlkB Directly Reverts DNA Base Damage. Nature 2002, 419, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Clifton, I.J.; McDonough, M.A.; Ehrismann, D.; Kershaw, N.J.; Granatino, N.; Schofield, C.J. Structural Studies on 2-Oxoglutarate Oxygenases and Related Double-Stranded β-Helix Fold Proteins. J. Inorg. Biochem. 2006, 100, 644–669. [Google Scholar] [CrossRef]
- Hausinger, R.P. Fe(II)/α-Ketoglutarate-Dependent Hydroxylases and Related Enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 21–68. [Google Scholar] [CrossRef] [PubMed]
- Welford, R.W.D.; Kirkpatrick, J.M.; McNeill, L.A.; Puri, M.; Oldham, N.J.; Schofield, C.J. Incorporation of Oxygen into the Succinate Co-Product of Iron(II) and 2-Oxoglutarate Dependent Oxygenases from Bacteria, Plants and Humans. FEBS Lett. 2005, 579, 5170–5174. [Google Scholar] [CrossRef]
- Hutton, J.J.; Kaplan, A.; Udenfriend, S. Conversion of the Amino Acid Sequence Gly-Pro-Pro in Protein to Gly-Pro-Hyp by Collagen Proline Hydroxylase. Arch. Biochem. Biophys. 1967, 121, 384–391. [Google Scholar] [CrossRef]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef]
- Waheed, S.O.; Ramanan, R.; Chaturvedi, S.S.; Lehnert, N.; Schofield, C.J.; Christov, C.Z.; Karabencheva-Christova, T.G. Role of Structural Dynamics in Selectivity and Mechanism of Non-Heme Fe(II) and 2-Oxoglutarate-Dependent Oxygenases Involved in DNA Repair. ACS Cent. Sci. 2020, 6, 795–814. [Google Scholar] [CrossRef]
- Delaney, J.C.; Essigmann, J.M. Mutagenesis, Genotoxicity, and Repair of 1-Methyladenine, 3-Alkylcytosines, 1-Methylguanine, and 3-Methylthymine in AlkB Escherichia coli. Proc. Natl. Acad. Sci. USA 2004, 101, 14051–14056. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, P.; Robins, P.; Lindahl, T.; Sedgwick, B. Demethylation of 3-Methylthymine in DNA by Bacterial and Human DNA Dioxygenases. J. Biol. Chem. 2004, 279, 40470–40474. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fedeles, B.I.; Shrivastav, N.; Delaney, J.C.; Yang, X.; Wong, C.; Drennan, C.L.; Essigmann, J.M. Removal of N-Alkyl Modifications from N(2)-Alkylguanine and N(4)-Alkylcytosine in DNA by the Adaptive Response Protein AlkB. Chem. Res. Toxicol. 2013, 26, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Delaney, J.C.; Page, C.M.; Yang, X.; Chen, A.S.; Wong, C.; Drennan, C.L.; Essigmann, J.M. Exocyclic Carbons Adjacent to the N6 of Adenine Are Targets for Oxidation by the Escherichia coli Adaptive Response Protein AlkB. J. Am. Chem. Soc. 2012, 134, 8896–8901. [Google Scholar] [CrossRef]
- Delaney, J.C.; Smeester, L.; Wong, C.; Frick, L.E.; Taghizadeh, K.; Wishnok, J.S.; Drennan, C.L.; Samson, L.D.; Essigmann, J.M. AlkB Reverses Etheno DNA Lesions Caused by Lipid Oxidation in Vitro and in Vivo. Nat. Struct. Mol. Biol. 2005, 12, 855–860. [Google Scholar] [CrossRef]
- Mishina, Y.; Yang, C.-G.; He, C. Direct Repair of the Exocyclic DNA Adduct 1,N6-Ethenoadenine by the DNA Repair AlkB Proteins. J. Am. Chem. Soc. 2005, 127, 14594–14595. [Google Scholar] [CrossRef]
- Maciejewska, A.M.; Poznański, J.; Kaczmarska, Z.; Krowisz, B.; Nieminuszczy, J.; Polkowska-Nowakowska, A.; Grzesiuk, E.; Kuśmierek, J.T. AlkB Dioxygenase Preferentially Repairs Protonated Substrates: Specificity Against Exocyclic Adducts and Molecular Mechanism of Action. J. Biol. Chem. 2013, 288, 432–441. [Google Scholar] [CrossRef]
- Maciejewska, A.M.; Ruszel, K.P.; Nieminuszczy, J.; Lewicka, J.; Sokołowska, B.; Grzesiuk, E.; Kuśmierek, J.T. Chloroacetaldehyde-Induced Mutagenesis in Escherichia coli: The Role of AlkB Protein in Repair of 3,N4-Ethenocytosine and 3,N4-α-Hydroxyethanocytosine. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2010, 684, 24–34. [Google Scholar] [CrossRef]
- Kurowski, M.A.; Bhagwat, A.S.; Papaj, G.; Bujnicki, J.M. Phylogenomic Identification of Five New Human Homologs of the DNA Repair Enzyme AlkB. BMC Genom. 2003, 4, 48. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Andrade-Navarro, M.A. The FTO (Fat Mass and Obesity Associated) Gene Codes for a Novel Member of the Non-Heme Dioxygenase Superfamily. BMC Biochem. 2007, 8, 23. [Google Scholar] [CrossRef]
- Duncan, T.; Trewick, S.C.; Koivisto, P.; Bates, P.A.; Lindahl, T.; Sedgwick, B. Reversal of DNA Alkylation Damage by Two Human Dioxygenases. Proc. Natl. Acad. Sci. USA 2002, 99, 16660–16665. [Google Scholar] [CrossRef]
- Loos, R.J.F.; Bouchard, C. FTO: The First Gene Contributing to Common Forms of Human Obesity. Obes. Rev. 2008, 9, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Crawford, D.J.; Liu, M.Y.; Nabel, C.S.; Cao, X.-J.; Garcia, B.A.; Kohli, R.M. Tet2 Catalyzes Stepwise 5-Methylcytosine Oxidation by an Iterative and de Novo Mechanism. J. Am. Chem. Soc. 2016, 138, 730–733. [Google Scholar] [CrossRef]
- Tamanaha, E.; Guan, S.; Marks, K.; Saleh, L. Distributive Processing by the Iron(II)/α-Ketoglutarate-Dependent Catalytic Domains of the TET Enzymes Is Consistent with Epigenetic Roles for Oxidized 5-Methylcytosine Bases. J. Am. Chem. Soc. 2016, 138, 9345–9348. [Google Scholar] [CrossRef]
- Yu, M.; Hon, G.C.; Szulwach, K.E.; Song, C.-X.; Jin, P.; Ren, B.; He, C. Tet-Assisted Bisulfite Sequencing of 5-Hydroxymethylcytosine. Nat. Protoc. 2012, 7, 2159–2170. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.; Iyer, L.M.; Hu, J.; Wang, G.; Fu, Y.; Yu, M.; Dai, Q.; Aravind, L.; He, C. A TET Homologue Protein from Coprinopsis Cinerea (CcTET) That Biochemically Converts 5-Methylcytosine to 5-Hydroxymethylcytosine, 5-Formylcytosine, and 5-Carboxylcytosine. J. Am. Chem. Soc. 2014, 136, 4801–4804. [Google Scholar] [CrossRef]
- Yu, Z.; Genest, P.-A.; ter Riet, B.; Sweeney, K.; DiPaolo, C.; Kieft, R.; Christodoulou, E.; Perrakis, A.; Simmons, J.M.; Hausinger, R.P.; et al. The Protein That Binds to DNA Base J in Trypanosomatids Has Features of a Thymidine Hydroxylase. Nucleic Acids Res. 2007, 35, 2107–2115. [Google Scholar] [CrossRef]
- Cliffe, L.J.; Kieft, R.; Southern, T.; Birkeland, S.R.; Marshall, M.; Sweeney, K.; Sabatini, R. JBP1 and JBP2 Are Two Distinct Thymidine Hydroxylases Involved in J Biosynthesis in Genomic DNA of African Trypanosomes. Nucleic Acids Res. 2009, 37, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Sabatini, R. Base J: Discovery, Biosynthesis, and Possible Functions. Annu. Rev. Microbiol. 2008, 62, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB Family of Fe(II)/α-Ketoglutarate-Dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Kanazhevskaya, L.Y.; Fedorova, O.S. DNA Demethylation in the Processes of Repair and Epigenetic Regulation Performed by 2-Ketoglutarate-Dependent DNA Dioxygenases. Int. J. Mol. Sci. 2021, 22, 10540. [Google Scholar] [CrossRef]
- Perry, G.S.; Das, M.; Woon, E.C.Y. Inhibition of AlkB Nucleic Acid Demethylases: Promising New Epigenetic Targets. J. Med. Chem. 2021, 64, 16974–17003. [Google Scholar] [CrossRef] [PubMed]
- Alemu, E.A.; He, C.; Klungland, A. ALKBHs-Facilitated RNA Modifications and de-Modifications. DNA Repair 2016, 44, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Kanazhevskaya, L.Y.; Smyshlyaev, D.A.; Alekseeva, I.V.; Fedorova, O.S. Conformational Dynamics of Dioxygenase AlkB and DNA in the Course of Catalytically Active Enzyme–Substrate Complex Formation. Russ. J. Bioorg. Chem. 2019, 45, 630–640. [Google Scholar] [CrossRef]
- Ma, L.; Lu, H.; Tian, Z.; Yang, M.; Ma, J.; Shang, G.; Liu, Y.; Xie, M.; Wang, G.; Wu, W.; et al. Structural Insights into the Interactions and Epigenetic Functions of Human Nucleic Acid Repair Protein ALKBH6. J. Biol. Chem. 2022, 298, 101671. [Google Scholar] [CrossRef]
- Kanazhevskaya, L.Y.; Alekseeva, I.V.; Fedorova, O.S. A Single-Turnover Kinetic Study of DNA Demethylation Catalyzed by Fe(II)/α-Ketoglutarate-Dependent Dioxygenase AlkB. Molecules 2019, 24, 4576. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA Methylation: The Mark and Its Mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Kinde, B.; Gabel, H.W.; Gilbert, C.S.; Griffith, E.C.; Greenberg, M.E. Reading the Unique DNA Methylation Landscape of the Brain: Non-CpG Methylation, Hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. USA 2015, 112, 6800–6806. [Google Scholar] [CrossRef]
- Dubin, D.T.; Taylor, R.H. The Methylation State of Poly A-Containing Messenger RNA from Cultured Hamster Cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, H.; Nair, J. New DNA-Based Biomarkers for Oxidative Stress and Cancer Chemoprevention Studies. Eur. J. Cancer 2000, 36, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Zdżalik, D.; Domańska, A.; Prorok, P.; Kosicki, K.; van den Born, E.; Falnes, P.Ø.; Rizzo, C.J.; Guengerich, F.P.; Tudek, B. Differential Repair of Etheno-DNA Adducts by Bacterial and Human AlkB Proteins. DNA Repair 2015, 30, 1–10. [Google Scholar] [CrossRef]
- Nair, J.; Godschalk, R.W.; Nair, U.; Owen, R.W.; Hull, W.E.; Bartsch, H. Identification of 3, N4-Etheno-5-Methyl-2′-Deoxycytidine in Human DNA: A New Modified Nucleoside Which May Perturb Genome Methylation. Chem. Res. Toxicol. 2012, 25, 162–169. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, S.; Nelakanti, R.; Zhao, W.; Liu, G.; Li, Z.; Liu, X.; Wu, T.; Xiao, A.; Li, H. Mammalian ALKBH1 Serves as an N6-MA Demethylase of Unpairing DNA. Cell Res. 2020, 30, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-H.; Jin, S.-G.; Cai, S.; Chen, Y.; Pfeifer, G.P.; O’Connor, T.R. Repair of Methylation Damage in DNA and RNA by Mammalian AlkB Homologues. J. Biol. Chem. 2005, 280, 39448–39459. [Google Scholar] [CrossRef]
- Yang, T.; Cheong, A.; Mai, X.; Zou, S.; Woon, E.C.Y. A Methylation-Switchable Conformational Probe for the Sensitive and Selective Detection of RNA Demethylase Activity. Chem. Commun. 2016, 52, 6181–6184. [Google Scholar] [CrossRef]
- Chen, F.; Bian, K.; Tang, Q.; Fedeles, B.I.; Singh, V.; Humulock, Z.T.; Essigmann, J.M.; Li, D. Oncometabolites D- and L-2-Hydroxyglutarate Inhibit the AlkB Family DNA Repair Enzymes under Physiological Conditions. Chem. Res. Toxicol. 2017, 30, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Yang, T.; Dong, J.; Prasetya, F.; Xie, Y.; Wong, K.H.Q.; Cheong, A.; Woon, E.C.Y. Multiprotein Dynamic Combinatorial Chemistry: A Strategy for the Simultaneous Discovery of Subfamily-Selective Inhibitors for Nucleic Acid Demethylases FTO and ALKBH3. Chem. Asian J. 2018, 13, 2854–2867. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Lu, J.; Cheng, J.; Rao, Q.; Li, Z.; Hou, H.; Lou, Z.; Zhang, L.; Li, W.; Gong, W.; et al. Structural Insight into Substrate Preference for TET-Mediated Oxidation. Nature 2015, 527, 118–122. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yi, C. Switching Demethylation Activities between AlkB Family RNA/DNA Demethylases through Exchange of Active-Site Residues. Angew. Chem. Int. Ed. 2014, 53, 3659–3662. [Google Scholar] [CrossRef]
- Li, F.; Kennedy, S.; Hajian, T.; Gibson, E.; Seitova, A.; Xu, C.; Arrowsmith, C.H.; Vedadi, M. A Radioactivity-Based Assay for Screening Human M6A-RNA Methyltransferase, METTL3-METTL14 Complex, and Demethylase ALKBH5. SLAS Discov. 2016, 21, 290–297. [Google Scholar] [CrossRef]
- Zou, S.; Toh, J.D.W.; Wong, K.H.Q.; Gao, Y.-G.; Hong, W.; Woon, E.C.Y. N6-Methyladenosine: A Conformational Marker That Regulates the Substrate Specificity of Human Demethylases FTO and ALKBH5. Sci. Rep. 2016, 6, 25677. [Google Scholar] [CrossRef]
- Wang, L.; Song, C.; Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.-C.; Yang, Q. NADP Modulates RNA M6A Methylation and Adipogenesis via Enhancing FTO Activity. Nat. Chem. Biol. 2020, 16, 1394–1402. [Google Scholar] [CrossRef]
- Khatiwada, B.; Purslow, J.A.; Underbakke, E.S.; Venditti, V. N-Terminal Fusion of the N-Terminal Domain of Bacterial Enzyme I Facilitates Recombinant Expression and Purification of the Human RNA Demethylases FTO and Alkbh5. Protein Expr. Purif. 2020, 167, 105540. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible Methylation of M6Am in the 5′ Cap Controls MRNA Stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Ma, M.; Harding, H.P.; O’Rahilly, S.; Ron, D.; Yeo, G.S.H. Kinetic Analysis of FTO (Fat Mass and Obesity-Associated) Reveals That It Is Unlikely to Function as a Sensor for 2-Oxoglutarate. Biochem. J. 2012, 444, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Yang, C.-G.; Yang, S.; Jian, X.; Yi, C.; Zhou, Z.; He, C. Oxidative Demethylation of 3-Methylthymine and 3-Methyluracil in Single-Stranded DNA and RNA by Mouse and Human FTO. FEBS Lett. 2008, 582, 3313–3319. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, P.; Duncan, T.; Lindahl, T.; Sedgwick, B. Minimal Methylated Substrate and Extended Substrate Range of Escherichia coli AlkB Protein, a 1-Methyladenine-DNA Dioxygenase. J. Biol. Chem. 2003, 278, 44348–44354. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Edstrom, W.C.; Benach, J.; Hamuro, Y.; Weber, P.C.; Gibney, B.R.; Hunt, J.F. Crystal Structures of Catalytic Complexes of the Oxidative DNA/RNA Repair Enzyme AlkB. Nature 2006, 439, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Roy, T.W.; Bhagwat, A.S. Kinetic Studies of Escherichia coli AlkB Using a New Fluorescence-Based Assay for DNA Demethylation. Nucleic Acids Res. 2007, 35, e147. [Google Scholar] [CrossRef]
- Yu, B.; Hunt, J.F. Enzymological and Structural Studies of the Mechanism of Promiscuous Substrate Recognition by the Oxidative DNA Repair Enzyme AlkB. Proc. Natl. Acad. Sci. USA 2009, 106, 14315–14320. [Google Scholar] [CrossRef]
- Ergel, B.; Gill, M.L.; Brown, L.; Yu, B.; Palmer, A.G.; Hunt, J.F. Protein Dynamics Control the Progression and Efficiency of the Catalytic Reaction Cycle of the Escherichia coli DNA-Repair Enzyme AlkB. J. Biol. Chem. 2014, 289, 29584–29601. [Google Scholar] [CrossRef]
- Baldwin, M.R.; Admiraal, S.J.; O’Brien, P.J. Transient Kinetic Analysis of Oxidative Dealkylation by the Direct Reversal DNA Repair Enzyme AlkB. J. Biol. Chem. 2020, 295, 7317–7326. [Google Scholar] [CrossRef]
- Wang, Y.; Katanski, C.D.; Watkins, C.; Pan, J.N.; Dai, Q.; Jiang, Z.; Pan, T. A High-Throughput Screening Method for Evolving a Demethylase Enzyme with Improved and New Functionalities. Nucleic Acids Res. 2021, 49, e30. [Google Scholar] [CrossRef]
- Karkhanina, A.A.; Mecinović, J.; Musheev, M.U.; Krylova, S.M.; Petrov, A.P.; Hewitson, K.S.; Flashman, E.; Schofield, C.J.; Krylov, S.N. Direct Analysis of Enzyme-Catalyzed DNA Demethylation. Anal. Chem. 2009, 81, 5871–5875. [Google Scholar] [CrossRef] [PubMed]
- Nigam, R.; Anindya, R. Escherichia coli Single-Stranded DNA Binding Protein SSB Promotes AlkB-Mediated DNA Dealkylation Repair. Biochem. Biophys. Res. Commun. 2018, 496, 274–279. [Google Scholar] [CrossRef]
- Shivange, G.; Kodipelli, N.; Anindya, R. 2-Hydrazinobenzothiazole-Based Etheno-Adduct Repair Protocol (HERP): A Method for Quantitative Determination of Direct Repair of Etheno-Bases. DNA Repair 2015, 28, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-F.; Carter, K.C.; Wang, R.-P.; Shell, B.K. Molecular Cloning and Functional Analysis of a Human CDNA Encoding an Escherichia coli AlkB Homolog, a Protein Involved in DNA Alkylation Damage Repair. Nucleic Acids Res. 1996, 24, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; He, C. DNA Repair by Reversal of DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012575. [Google Scholar] [CrossRef]
- Ougland, R.; Lando, D.; Jonson, I.; Dahl, J.A.; Moen, M.N.; Nordstrand, L.M.; Rognes, T.; Lee, J.T.; Klungland, A.; Kouzarides, T.; et al. ALKBH1 Is a Histone H2A Dioxygenase Involved in Neural Differentiation. Stem Cells 2012, 30, 2672–2682. [Google Scholar] [CrossRef]
- Ma, C.-J.; Ding, J.-H.; Ye, T.-T.; Yuan, B.-F.; Feng, Y.-Q. AlkB Homologue 1 Demethylates N3-Methylcytidine in MRNA of Mammals. ACS Chem. Biol. 2019, 14, 1418–1425. [Google Scholar] [CrossRef]
- Westbye, M.P.; Feyzi, E.; Aas, P.A.; Vågbø, C.B.; Talstad, V.A.; Kavli, B.; Hagen, L.; Sundheim, O.; Akbari, M.; Liabakk, N.-B.; et al. Human AlkB Homolog 1 Is a Mitochondrial Protein That Demethylates 3-Methylcytosine in DNA and RNA. J. Biol. Chem. 2008, 283, 25046–25056. [Google Scholar] [CrossRef]
- Haag, S.; Sloan, K.E.; Ranjan, N.; Warda, A.S.; Kretschmer, J.; Blessing, C.; Hübner, B.; Seikowski, J.; Dennerlein, S.; Rehling, P.; et al. NSUN3 and ABH1 Modify the Wobble Position of Mt-TRNAMet to Expand Codon Recognition in Mitochondrial Translation. EMBO J. 2016, 35, 2104–2119. [Google Scholar] [CrossRef]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Kenjyo, M.; Suzuki, T. ALKBH1 Is an RNA Dioxygenase Responsible for Cytoplasmic and Mitochondrial TRNA Modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA Methylation on N6-Adenine in Mammalian Embryonic Stem Cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated TRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e16. [Google Scholar] [CrossRef]
- Yang, C.-G.; Yi, C.; Duguid, E.M.; Sullivan, C.T.; Jian, X.; Rice, P.A.; He, C. Crystal Structures of DNA/RNA Repair Enzymes AlkB and ABH2 Bound to DsDNA. Nature 2008, 452, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Monsen, V.T.; Sundheim, O.; Aas, P.A.; Westbye, M.P.; Sousa, M.M.L.; Slupphaug, G.; Krokan, H.E. Divergent β-Hairpins Determine Double-Strand versus Single-Strand Substrate Recognition of Human AlkB-Homologues 2 and 3. Nucleic Acids Res. 2010, 38, 6447–6455. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liu, H.; Sun, X.; Yang, C.-G. Mechanistic Insight into the Recognition of Single-Stranded and Double-Stranded DNA Substrates by ABH2 and ABH3. Mol. BioSyst. 2010, 6, 2143. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tang, Q.; Bian, K.; Humulock, Z.T.; Yang, X.; Jost, M.; Drennan, C.L.; Essigmann, J.M.; Li, D. Adaptive Response Enzyme AlkB Preferentially Repairs 1-Methylguanine and 3-Methylthymine Adducts in Double-Stranded DNA. Chem. Res. Toxicol. 2016, 29, 687–693. [Google Scholar] [CrossRef]
- Bian, K.; Lenz, S.A.P.; Tang, Q.; Chen, F.; Qi, R.; Jost, M.; Drennan, C.L.; Essigmann, J.M.; Wetmore, S.D.; Li, D. DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB Oxidize 5-Methylcytosine to 5-Hydroxymethylcytosine, 5-Formylcytosine and 5-Carboxylcytosine in Vitro. Nucleic Acids Res. 2019, 47, 5522–5529. [Google Scholar] [CrossRef]
- You, C.; Wang, P.; Nay, S.L.; Wang, J.; Dai, X.; O’Connor, T.R.; Wang, Y. Roles of Aag, Alkbh2, and Alkbh3 in the Repair of Carboxymethylated and Ethylated Thymidine Lesions. ACS Chem. Biol. 2016, 11, 1332–1338. [Google Scholar] [CrossRef]
- Sundheim, O.; Vågbø, C.B.; Bjørås, M.; Sousa, M.M.L.; Talstad, V.; Aas, P.A.; Drabløs, F.; Krokan, H.E.; Tainer, J.A.; Slupphaug, G. Human ABH3 Structure and Key Residues for Oxidative Demethylation to Reverse DNA/RNA Damage. EMBO J. 2006, 25, 3389–3397. [Google Scholar] [CrossRef]
- Chen, Z.; Qi, M.; Shen, B.; Luo, G.; Wu, Y.; Li, J.; Lu, Z.; Zheng, Z.; Dai, Q.; Wang, H. Transfer RNA Demethylase ALKBH3 Promotes Cancer Progression via Induction of TRNA-Derived Small RNAs. Nucleic Acids Res. 2019, 47, 2533–2545. [Google Scholar] [CrossRef]
- Dango, S.; Mosammaparast, N.; Sowa, M.E.; Xiong, L.-J.; Wu, F.; Park, K.; Rubin, M.; Gygi, S.; Harper, J.W.; Shi, Y. DNA Unwinding by ASCC3 Helicase Is Coupled to ALKBH3-Dependent DNA Alkylation Repair and Cancer Cell Proliferation. Mol. Cell 2011, 44, 373–384. [Google Scholar] [CrossRef]
- Brickner, J.R.; Soll, J.M.; Lombardi, P.M.; Vågbø, C.B.; Mudge, M.C.; Oyeniran, C.; Rabe, R.; Jackson, J.; Sullender, M.E.; Blazosky, E.; et al. A Ubiquitin-Dependent Signalling Axis Specific for ALKBH-Mediated DNA Dealkylation Repair. Nature 2017, 551, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6A RNA Methylomes Revealed by M6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ensfelder, T.T.; Kurz, M.Q.; Iwan, K.; Geiger, S.; Matheisl, S.; Müller, M.; Beckmann, R.; Carell, T. ALKBH5-Induced Demethylation of Mono- and Dimethylated Adenosine. Chem. Commun. 2018, 54, 8591–8593. [Google Scholar] [CrossRef]
- Takahashi, H.; Hase, H.; Yoshida, T.; Tashiro, J.; Hirade, Y.; Kitae, K.; Tsujikawa, K. Discovery of Two Novel ALKBH5 Selective Inhibitors That Exhibit Uncompetitive or Competitive Type and Suppress the Growth Activity of Glioblastoma Multiforme. Chem. Biol. Drug Des. 2022, 100, 1–12. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Gerken, T.; Girard, C.A.; Tung, Y.-C.L.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.H.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate-Dependent Nucleic Acid Demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential M6A, M6Am, and M1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-Mediated Formation of N6-Hydroxymethyladenosine and N6-Formyladenosine in Mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, L.; Zheng, G.; Fu, Y.; Ji, Q.; Liu, F.; Chen, H.; He, C. Crystal Structure of the RNA Demethylase ALKBH5 from Zebrafish. FEBS Lett. 2014, 588, 892–898. [Google Scholar] [CrossRef]
- Waheed, S.O.; Ramanan, R.; Chaturvedi, S.S.; Ainsley, J.; Evison, M.; Ames, J.M.; Schofield, C.J.; Christov, C.Z.; Karabencheva-Christova, T.G. Conformational Flexibility Influences Structure–Function Relationships in Nucleic Acid N-Methyl Demethylases. Org. Biomol. Chem. 2019, 17, 2223–2231. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cao, Z.; Sharon, D.A.; Shaik, S. Computations Reveal a Rich Mechanistic Variation of Demethylation of N-Methylated DNA/RNA Nucleotides by FTO. ACS Catal. 2015, 5, 7077–7090. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. Modified Nucleosides and Bizarre 5′-Termini in Mouse Myeloma MRNA. Nature 1975, 255, 28–33. [Google Scholar] [CrossRef]
- Wei, C.-M.; Gershowitz, A.; Moss, B. N6, O2′-Dimethyladenosine a Novel Methylated Ribonucleoside next to the 5′ Terminal of Animal Cell and Virus MRNAs. Nature 1975, 257, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Cliffe, L.J.; Siegel, T.N.; Marshall, M.; Cross, G.A.M.; Sabatini, R. Two Thymidine Hydroxylases Differentially Regulate the Formation of Glucosylated DNA at Regions Flanking Polymerase II Polycistronic Transcription Units throughout the Genome of Trypanosoma Brucei. Nucleic Acids Res. 2010, 38, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Vainio, S.; Genest, P.-A.; ter Riet, B.; van Luenen, H.; Borst, P. Evidence That J-Binding Protein 2 Is a Thymidine Hydroxylase Catalyzing the First Step in the Biosynthesis of DNA Base J. Mol. Biochem. Parasitol. 2009, 164, 157–161. [Google Scholar] [CrossRef]
- van Leeuwen, F.; Taylor, M.C.; Mondragon, A.; Moreau, H.; Gibson, W.; Kieft, R.; Borst, P. β-D-Glucosyl-Hydroxymethyluracil Is a Conserved DNA Modification in Kinetoplastid Protozoans and Is Abundant in Their Telomeres. Proc. Natl. Acad. Sci. USA 1998, 95, 2366–2371. [Google Scholar] [CrossRef]
- Toaldo, C.B.; Kieft, R.; Dirks-Mulder, A.; Sabatini, R.; van Luenen, H.G.A.M.; Borst, P. A Minor Fraction of Base J in Kinetoplastid Nuclear DNA Is Bound by the J-Binding Protein 1. Mol. Biochem. Parasitol. 2005, 143, 111–115. [Google Scholar] [CrossRef]
- Torabifard, H.; Cisneros, G.A. Insight into Wild-Type and T1372E TET2-Mediated 5hmC Oxidation Using Ab Initio QM/MM Calculations. Chem. Sci. 2018, 9, 8433–8445. [Google Scholar] [CrossRef]
- Waheed, S.O.; Chaturvedi, S.S.; Karabencheva-Christova, T.G.; Christov, C.Z. Catalytic Mechanism of Human Ten-Eleven Translocation-2 (TET2) Enzyme: Effects of Conformational Changes, Electric Field, and Mutations. ACS Catal. 2021, 11, 3877–3890. [Google Scholar] [CrossRef]
- Waheed, S.O.; Varghese, A.; Chaturvedi, S.S.; Karabencheva-Christova, T.G.; Christov, C.Z. How Human TET2 Enzyme Catalyzes the Oxidation of Unnatural Cytosine Modifications in Double-Stranded DNA. ACS Catal. 2022, 12, 5327–5344. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Torabifard, H.; Crawford, D.J.; DeNizio, J.E.; Cao, X.-J.; Garcia, B.A.; Cisneros, G.A.; Kohli, R.M. Mutations along a TET2 Active Site Scaffold Stall Oxidation at 5-Hydroxymethylcytosine. Nat. Chem. Biol. 2017, 13, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Yamamoto, Y.; Sekiguchi, M. A New Gene (AlkB) of Escherichia coli That Controls Sensitivity to Methyl Methane Sulfonate. J. Bacteriol. 1983, 153, 1301–1307. [Google Scholar] [CrossRef]
- Wang, J.; Qi, R.; Li, H.; Christov, C.; Lehnert, N.; Li, D. Genetic and Epigenetic Biomarkers Related to 2-Oxoglutarate/Fe(II)-Dependent Oxygenases and Implications for Disease and Toxicology. In Biomarkers in Toxicology; Patel, V.B., Preedy, V.R., Rajendram, R., Eds.; Biomarkers in Disease: Methods, Discoveries and Applications; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–28. [Google Scholar] [CrossRef]
- Fang, D.; Cisneros, G.A. Alternative Pathway for the Reaction Catalyzed by DNA Dealkylase AlkB from Ab Initio QM/MM Calculations. J. Chem. Theory Comput. 2014, 10, 5136–5148. [Google Scholar] [CrossRef] [PubMed]
- Quesne, M.G.; Latifi, R.; Gonzalez-Ovalle, L.E.; Kumar, D.; de Visser, S.P. Quantum Mechanics/Molecular Mechanics Study on the Oxygen Binding and Substrate Hydroxylation Step in AlkB Repair Enzymes. Chem.–A Eur. J. 2014, 20, 435–446. [Google Scholar] [CrossRef]
- Fang, D.; Lord, R.L.; Cisneros, G.A. Ab Initio QM/MM Calculations Show an Intersystem Crossing in the Hydrogen Abstraction Step in Dealkylation Catalyzed by AlkB. J. Phys. Chem. B 2013, 117, 6410–6420. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Usharani, D.; Li, C.; Shaik, S. Theory Uncovers an Unusual Mechanism of DNA Repair of a Lesioned Adenine by AlkB Enzymes. J. Am. Chem. Soc. 2014, 136, 13895–13901. [Google Scholar] [CrossRef]
- Dai, Q.; Zheng, G.; Schwartz, M.H.; Clark, W.C.; Pan, T. Selective Enzymatic Demethylation of N2,N2-Dimethylguanosine in RNA and Its Application in High-Throughput TRNA Sequencing. Angew. Chem. Int. Ed. 2017, 56, 5017–5020. [Google Scholar] [CrossRef]
- Li, M.-M.; Nilsen, A.; Shi, Y.; Fusser, M.; Ding, Y.-H.; Fu, Y.; Liu, B.; Niu, Y.; Wu, Y.-S.; Huang, C.-M.; et al. ALKBH4-Dependent Demethylation of Actin Regulates Actomyosin Dynamics. Nat. Commun. 2013, 4, 1832. [Google Scholar] [CrossRef]
- Mielecki, D.; Zugaj, D.Ł.; Muszewska, A.; Piwowarski, J.; Chojnacka, A.; Mielecki, M.; Nieminuszczy, J.; Grynberg, M.; Grzesiuk, E. Novel AlkB Dioxygenases—Alternative Models for In Silico and In Vivo Studies. PLoS ONE 2012, 7, e30588. [Google Scholar] [CrossRef]
- Tsujikawa, K.; Koike, K.; Kitae, K.; Shinkawa, A.; Arima, H.; Suzuki, T.; Tsuchiya, M.; Makino, Y.; Furukawa, T.; Konishi, N.; et al. Expression and Sub-Cellular Localization of Human ABH Family Molecules. J. Cell. Mol. Med. 2007, 11, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-S.; Xiong, Q.-P.; Peña Perez, S.; Liu, C.; Wei, J.; Le, C.; Zhang, L.; Harada, B.T.; Dai, Q.; Feng, X.; et al. ALKBH7-Mediated Demethylation Regulates Mitochondrial Polycistronic RNA Processing. Nat Cell Biol 2021, 23, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Bobiak, M.L. Biochemical Characterization of Human AlkBH7. Doctoral Dissertation, Stony Brook University, Stony Brook, NY, USA, 2009. [Google Scholar]
- Pastore, C.; Topalidou, I.; Forouhar, F.; Yan, A.C.; Levy, M.; Hunt, J.F. Crystal Structure and RNA Binding Properties of the RNA Recognition Motif (RRM) and AlkB Domains in Human AlkB Homolog 8 (ABH8), an Enzyme Catalyzing TRNA Hypermodification. J. Biol. Chem. 2012, 287, 2130–2143. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Cai, A.; Bian, K.; Chen, F.; Delaney, J.C.; Adusumalli, S.; Bach, A.C.; Akhlaghi, F.; Cho, B.P.; Li, D. Characterization of Byproducts from Chemical Syntheses of Oligonucleotides Containing 1-Methyladenine and 3-Methylcytosine. ACS Omega 2017, 2, 8205–8212. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | Substrate |
|---|---|
| AlkB | DNA: m1A, m3C, m1G, m3T, m4C, m2G, m22G, m5C, e1A, εA, εC, 1,N2-εG, e2G, EA, FF, HF, αHOPG, γHOPG, M1G, HEC, HPC |
| RNA: m1A, m3C, m1G | |
| ALKBH1 | DNA: m3C, m6A |
| RNA: m3C, m5C, m1A | |
| ALKBH2 | DNA: m1A, m3C, m1G, m3T, m5C, e1A, e3T, εA, εC, 1,N2-εG |
| ALKBH3 | DNA: m1A, m3C, m3T, m5C, e1A, e3T, εC, εA |
| RNA: m1A, m3C, m6A | |
| ALKBH4 | DNA: m6A |
| ALKBH5 | RNA: m6A, m66A |
| ALKBH6 | -- |
| ALKBH7 | RNA: m1A, m3C, m22G, εA |
| ALKBH8 | RNA: mc5mU |
| FTO | DNA: m3T, m6A |
| RNA: m3U, m6A, m1A, m3C | |
| TET1-3 | DNA: m5C, T |
| RNA: m5C |
| Enzyme | DNA/ RNA | Substrate | DNA/RNA Sequence 5′-3′ | Kcat (min−1) | Km (µm) | Kcat/Km (min−1 µm−1) | Reference |
|---|---|---|---|---|---|---|---|
| ALKBH1 | DNA | m6A | ACCTTATGGAXAGCATGCTTG in ds-DNA | 0.136 ± 0.0036 | 3.18 ± 0.28 | 0.04 | [58] |
| DNA | ACCTTATGGAXAGCATGCTTG | 0.076 ± 0.0012 | 2.79 ± 0.18 | 0.03 | |||
| ALKBH2 | DNA | m1A | AAAGCAGXATTCGAAAAAGCGAAA in ds-DNA | 823.2 ± 120 | 0.320 ± 0.073 | 2573 | [59] |
| DNA | AAAGCAGXATTCGAAAAAGCGAAA | 198 ± 16.2 | 0.183 ± 0.023 | 1082 | |||
| RNA | AAAGCAGXAUUCGAA in ds-RNA | 2.19 ± 0.05 | 0.30 ± 0.07 | 7.4 | [60] | ||
| RNA | CGCGXAUUCGCG | 3.67 ± 0.35 | 1.09 ± 0.14 | 3.4 | |||
| RNA | AAAGCAGXAUUCGAA | 4.07 ± 0.15 | 0.95 ± 0.11 | 4.3 | |||
| DNA | GAAGACCTXGGCGTCC in ds-DNA | 2.5 ± 0.1 | 7.3 ± 0.9 | 0.34 | [61] | ||
| DNA | GAAGACCTXGGCGTCC | 1.1 ± 0.1 | 4.1 ± 0.9 | 0.27 | |||
| DNA | m3C | AAAGCACXGGTCGAAAAAGCGAAA in ds-DNA | 530.4±52.8 | 0.167 ± 0.027 | 3176 | [59] | |
| DNA | AAAGCACXGGTCGAAAAAGCGAAA | 63.6±7.2 | 0.0822 ± 0.022 | 774 | |||
| DNA | GAAGACCTXGGCGTCC in ds-DNA | 2.6 ± 0.1 | 1.9 ± 0.4 | 1.3 | [61] | ||
| DNA | GAAGACCTXGGCGTCC | 1.7 ± 0.1 | 1.4 ± 0.2 | 1.2 | |||
| ALKBH3 | DNA | m1A | AAAGCAGXATTCGAAAAAGCGAAA in ds-DNA | 109.8±2.28 | 0.263 ± 0.110 | 418 | [59] |
| DNA | AAAGCAGXATTCGAAAAAGCGAAA | 178.8±44.4 | 0.182 ± 0.140 | 982 | |||
| RNA | AAAGCAGXAUUCGAA in ds-RNA | 2.57 ± 0.27 | 6.60 ± 0.19 | 0.39 | [60] | ||
| RNA | AAAGCAGXAUUCGAA | 3.56 ± 0.31 | 1.12 ± 0.16 | 3.2 | |||
| RNA | CGCGXAUUCGCG | 3.13 ± 0.22 | 1.47 ± 0.08 | 2.1 | |||
| DNA | GAAGACCTXGGCGTCC | 1.2 ± 0.0 | 2.3 ± 0.1 | 0.51 | [61] | ||
| DNA | AAAGCAGXATTCGAA | 3.04 ± 0.22 | 0.97 ± 0.07 | 3.1 | [62] | ||
| DNA | m3C | AAAGCACXGGTCGAAAAAGCGAAA in ds-DNA | 2.268±0.462 | 0.0084 ± 0.016 | 270 | [59] | |
| DNA | AAAGCACXGGTCGAAAAAGCGAAA | 123.6±19.2 | 0.162 ± 0.048 | 763 | |||
| DNA | GAAGACCTXGGCGTCC | 1.7 ± 0.0 | 1.9 ± 0.4 | 0.87 | [61] | ||
| TET2 | DNA | m5C | ACCACXGGTGGT | 0.127 ± 0.019 | 0.48 ± 0.19 | 0.27 | [63] |
| DNA | hm5C | ACCACXGGTGGT | 0.038 ± 0.005 | 0.90 ± 0.30 | 0.04 | ||
| DNA | f5C | ACCACXGGTGGT | 0.0276 ± 0.002 | 1.30 ± 0.27 | 0.02 |
| Enzyme | DNA/ RNA | Substrate | DNA/RNA Sequence 5′-3′ | Kcat (min−1) | Km (µm) | Kcat/Km (min−1 µm−1) | Reference |
|---|---|---|---|---|---|---|---|
| ALKBH5 | RNA | m6A | AUUGUCAXCAGCAGC | 0.169 ± 0.0106 | 1.38 ± 0.2653 | 0.12 | [64] |
| DNA | ATTGTCAXCAGCAGA | 0.174 ± 0.008 | 1.66 ± 0.16 | 0.11 | [65] | ||
| RNA | UACACUCGAUCUGGXCUAAAGCU GCUC-biotin-3′ | 0.3 ± 0.067 | 2.5 ± 0.5 | 0.12 | [66] | ||
| RNA | UACACUCGAUCUGGXCUAAAGCU GCUC-biotin-3′ | 0.192 | |||||
| RNA | GGXCU | 0.140 ± 0.013 | 2.344 ± 0.140 | 0.06 | [67] | ||
| DNA | GAXCA | 0.162 ± 0.014 | 2.251 ± 0.042 | 0.07 | |||
| RNA | GCGGXCUCCAGAUG | 0.172 ± 0.010 | 1.755 ± 0.088 | 0.1 | |||
| RNA | CCCCXCCCCCCCCC | 0.137 ± 0.021 | 2.583 ± 0.256 | 0.05 | |||
| RNA | GGXCU | 0.16 ± 0.02 | 1.64 ± 0.05 | 0.1 | [62] | ||
| RNA | AUUGUCAXCAGCAG | 0.306 ± 0.034 | 1.335 ± 0.213 | 0.23 | [68] | ||
| DNA | GGXCT | 2.6 ± 0.6 | 1.6 ± 0.1 | 1.6 | [69] | ||
| FTO | RNA | m6A | AUUGUCAXCAGCAGC | 0.296 ± 0.004 | 0.409 ± 0.023 | 0.72 | [33] |
| RNA | AUUGUCAXCAGCAGC | 0.381 ± 0.114 | 0.6 ± 0.12 | 0.63 | [65] | ||
| RNA | m7GpppXCA | 7.77 | 16.09 | 0.48 | [70] | ||
| RNA | m7GpppACX | 0.46 | 6.4 | 0.07 | |||
| RNA | GGXCU | 0.54 | 9.29 | 0.06 | |||
| RNA | GGXCU | 0.347 ± 0.015 | 0.508 ± 0.126 | 0.68 | [67] | ||
| DNA | GGXCT | 0.334 ± 0.57 | 0.586 ± 0.137 | 0.57 | |||
| DNA | GCGGXCUCCAGAUG | 0.376 ± 0.009 | 0.488 ± 0.074 | 0.77 | |||
| RNA | CCCCXCCCCCCCCC | 0.268 ± 0.012 | 0.688 ± 0.025 | 0.39 | |||
| RNA | GGXCU | 0.35 ± 0.03 | 0.51 ± 0.06 | 0.69 | [62] | ||
| RNA | AUUGUCAXCAGCAG | 0.46 ± 0.055 | 0.59 ± 0.094 | 0.78 | [68] | ||
| RNA | containing 50 μM NADPH | 0.406 ± 0.0467 | 0.401 ± 0.0521 | 1.01 | |||
| 50 μM NADH | 0.290 ± 0.0311 | 0.528 ± 0.0660 | 0.55 | ||||
| 50 μM NADP+ | 0.282 ± 0.0340 | 0.961 ± 0.127 | 0.29 | ||||
| 50 μM NAD+ | 0.224 ± 0.0291 | 1.125 ± 0.158 | 0.20 | ||||
| 50 μM Vc | 0.136 ± 0.0258 | 3.015 ± 0.572 | 0.045 | ||||
| DNA | GGXCT | 0.015 ± 0.005 | 12 ± 2 | 0.001 | [69] | ||
| RNA | m6Am | m7GpppX | 8.78 | 1.34 | 6.55 | [70] | |
| RNA | m3U | CTGACGGAGAXGAACGTCAG | 2.88 | [71] | |||
| RNA | CUUGUCAXCAGCAGA | 0.115 ± 0.022 | 8.51 ± 3.13 | 0.014 ± 0.007 | [72] | ||
| DNA | m3T | CTTGTCAXCAGCAGA | 0.007 ± 0.0002 | 0.95 ± 0.12 | 0.007 ± 0.002 | [72] |
| Enzyme | DNA/ RNA | Substrate | DNA/RNA Sequence 5′-3′ | Kcat (min−1) | Km (µm) | Kcat/Km (min−1 µm−1) | Reference |
|---|---|---|---|---|---|---|---|
| AlkB | DNA | m1A | poly(dA) methylated with [14C]MeI | 11.7 ± 0.2 | 1.4 ± 0.2 | 8.6 | [73] |
| DNA | TXT | 7.4 ± 0.6 | 2.8 ± 0.9 | 2.6 | |||
| DNA | TX | 3.7 | 4.4 | 0.8 | |||
| DNA | TXT | 2.7 ± 0.8 | 1.4 ± 0.5 | 1.9 | [74] | ||
| DNA | CGTCGXATTCTAGAGCCCC | 3.7 ± 0.2 | 5.4 ± 0.9 | 0.68 | [75] | ||
| DNA | CGTCGXATTCTAGAGCCCC in ds-DNA | 3.1 ± 0.2 | 6.2 ± 1.3 | 0.48 | |||
| DNA | TXT | 2.7 ± 0.8 | 1.4 ± 0.9 | 1.9 | [76] | ||
| DNA | CAXAT | 5.4 ± 1.3 | 0.06 ± 0.01 | 97 | |||
| DNA | TXT | 5.2 ± 0.2 | 3.2 ± 0.4 | 1.6 | [77] | ||
| DNA | ATTGTCAXCAGCAGA | 7.41 ± 0.47 | 2.00 ± 0.35 | 3.7 | [65] | ||
| RNA | AUUGUCAXCAGCAGC | 3.72 ± 0.19 | 2.32 ± 0.31 | 1.6 | |||
| RNA | AAAGCAGXAUUCGAA in ds-DNA | 2.25 ± 0.18 | 3.56 ± 0.24 | 0.63 | [60] | ||
| RNA | r(CGCGXAUUCGCG) probe | 4.10 ± 0.29 | 1.30 ± 0.12 | 3.2 | |||
| RNA | AAAGCAGXAUUCGAA | 3.75 ± 0.12 | 1.44 ± 0.25 | 2.6 | |||
| DNA | GAAGACCTXGGCGTCC | 4.2 ± 0.2 | 7.1 ± 1.1 | 0.59 | [61] | ||
| DNA | GAAGACCTXGGCGTCC in ds-RNA | 4.8 ± 0.2 | 12.7 ± 1.3 | 0.38 | |||
| DNA | CGATAGCATCCTXCCTTCTCTCCAT | 54 ± 1.8 | 0.041 ± 0.007 | 1317 | [78] | ||
| DNA | CGATAGCATCCTXCCTTCTCTCCAT in ds-DNA | 46.2 ± 1.2 | 0.65 ± 0.05 | 71.1 | |||
| DNA | m6A | ATTGTCAXCAGCAGA | 0.107 ± 0.013 | 14.93 ± 2.46 | 0.01 | [65] | |
| D135S | RNA | m1G | GAGCXUUAG | 2.2 | [79] | ||
| D135T | RNA | GAGCXUUAG | 0.052 ± 0.008 | 3.3 ± 1.3 | 15.7 ± 3.7 | ||
| DNA | m3C | CGTCGAATTXTA GAGCCCC | 2.2 ± 0.1 | 3.4 ± 0.6 | 0.65 | [75] | |
| DNA | CGTCGAATTXTA GAGCCCC in ds-DNA | 3.3 ± 0.2 | 9.3 ± 2.4 | 0.35 | |||
| DNA | TXT | 21 ± 4 | 24 ± 5 | 0.9 | [76] | ||
| DNA | CAXAT | 23 + 10 | 0.29 ± 0.03 | 78.3 | |||
| DNA | TTXTTTTTTTTTTTT | 2.6 ± 0.3 | 0.0353 ± 0.0066 | 73.6 | [80] | ||
| DNA | CAXAT | 21.2 ± 1.1 | 0.4 ± 0.1 | 53 | [77] | ||
| DNA | GAAGACCTXGGCGTCC in ds-DNA | 8.2 ± 0.4 | 10.8 ± 1.9 | 0.76 | [61] | ||
| DNA | GAAGACCTXGGCGTCC | 24.5 ± 0.7 | 19.9 ± 1.3 | 1.2 | |||
| DNA | 70-mer Poly T with X at position 1 | 2.4 | [81] | ||||
| DNA | 70-mer Poly T with X at position 35 | 6.7 | |||||
| DNA | 70-mer Poly T with X at position 15 | 8.2 | |||||
| DNA | εA | TXT | 0.06 | [25] | |||
| DNA | GAAGACCTXGGCGTCC | 1.8 | [26] | ||||
| DNA | TXT | 0.13 ± 0.05 | 60 ± 14 | 0.002 | [76] | ||
| DNA | 40-mer containing A treated by chloroacetaldehyde | 0.134 | 67.4 | 0.0019 | [82] | ||
| DNA | CGATAGCATCCTXCCTTCTCTCCAT | 45 ± 6.6 | 5.3 ± 1.3 | 8.5 | [78] | ||
| DNA | CGATAGCATCCTXCCTTCTCTCCAT in ds-DNA | 102 ± 30 | 8.4 ± 3.4 | 12.1 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Ma, J.; Christov, C.Z.; Karabencheva-Christova, T.; Lehnert, N.; Li, D. Kinetic Studies on the 2-Oxoglutarate/Fe(II)-Dependent Nucleic Acid Modifying Enzymes from the AlkB and TET Families. DNA 2023, 3, 65-84. https://doi.org/10.3390/dna3020005
Peng Z, Ma J, Christov CZ, Karabencheva-Christova T, Lehnert N, Li D. Kinetic Studies on the 2-Oxoglutarate/Fe(II)-Dependent Nucleic Acid Modifying Enzymes from the AlkB and TET Families. DNA. 2023; 3(2):65-84. https://doi.org/10.3390/dna3020005
Chicago/Turabian StylePeng, Zhiyuan, Jian Ma, Christo Z. Christov, Tatyana Karabencheva-Christova, Nicolai Lehnert, and Deyu Li. 2023. "Kinetic Studies on the 2-Oxoglutarate/Fe(II)-Dependent Nucleic Acid Modifying Enzymes from the AlkB and TET Families" DNA 3, no. 2: 65-84. https://doi.org/10.3390/dna3020005
APA StylePeng, Z., Ma, J., Christov, C. Z., Karabencheva-Christova, T., Lehnert, N., & Li, D. (2023). Kinetic Studies on the 2-Oxoglutarate/Fe(II)-Dependent Nucleic Acid Modifying Enzymes from the AlkB and TET Families. DNA, 3(2), 65-84. https://doi.org/10.3390/dna3020005