8-OxodG: A Potential Biomarker for Chronic Oxidative Stress Induced by High-LET Radiation
Abstract
:1. Background
2. Human Exposure to High-LET Radiation
3. Potential Sources of 8-OxodG
3.1. Cellular Sources of 8-OxodG
3.2. DNA Trapped in Vesicles
3.3. Dietary and Microbiome Released 8-OxodG
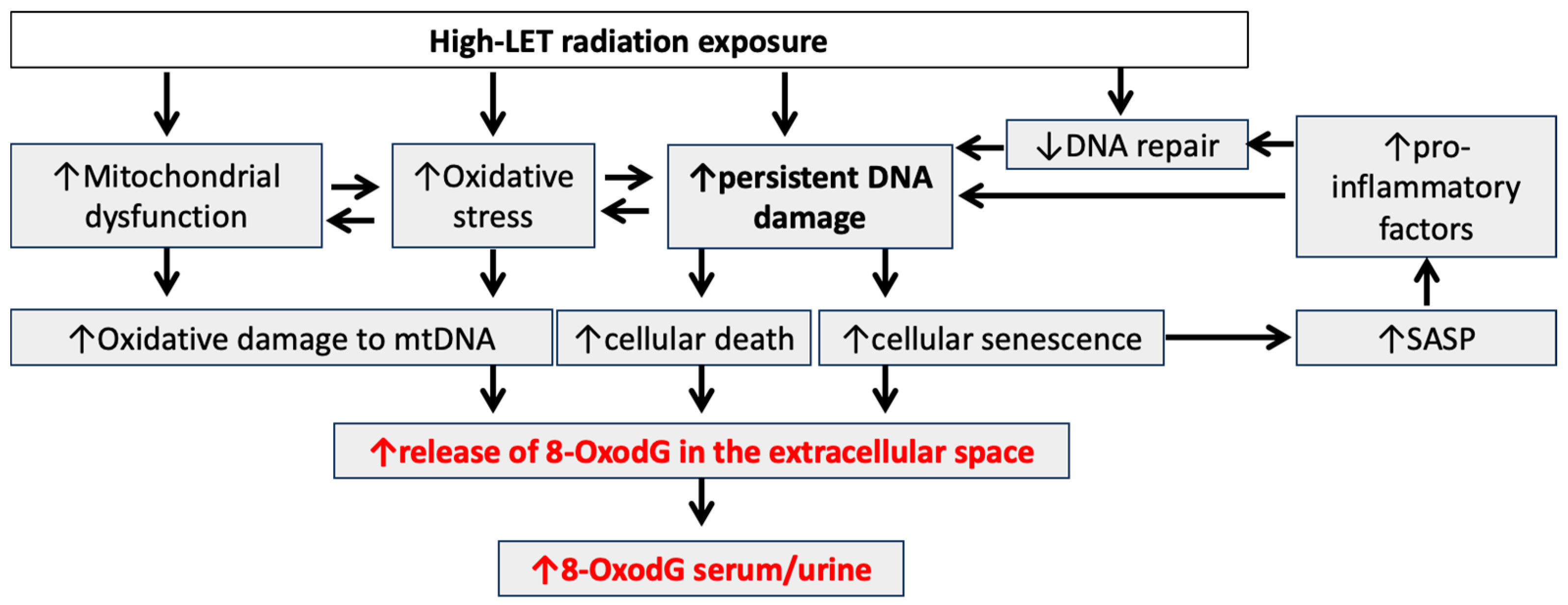
4. High-LET Radiation-Induced Chronic Oxidative Stress
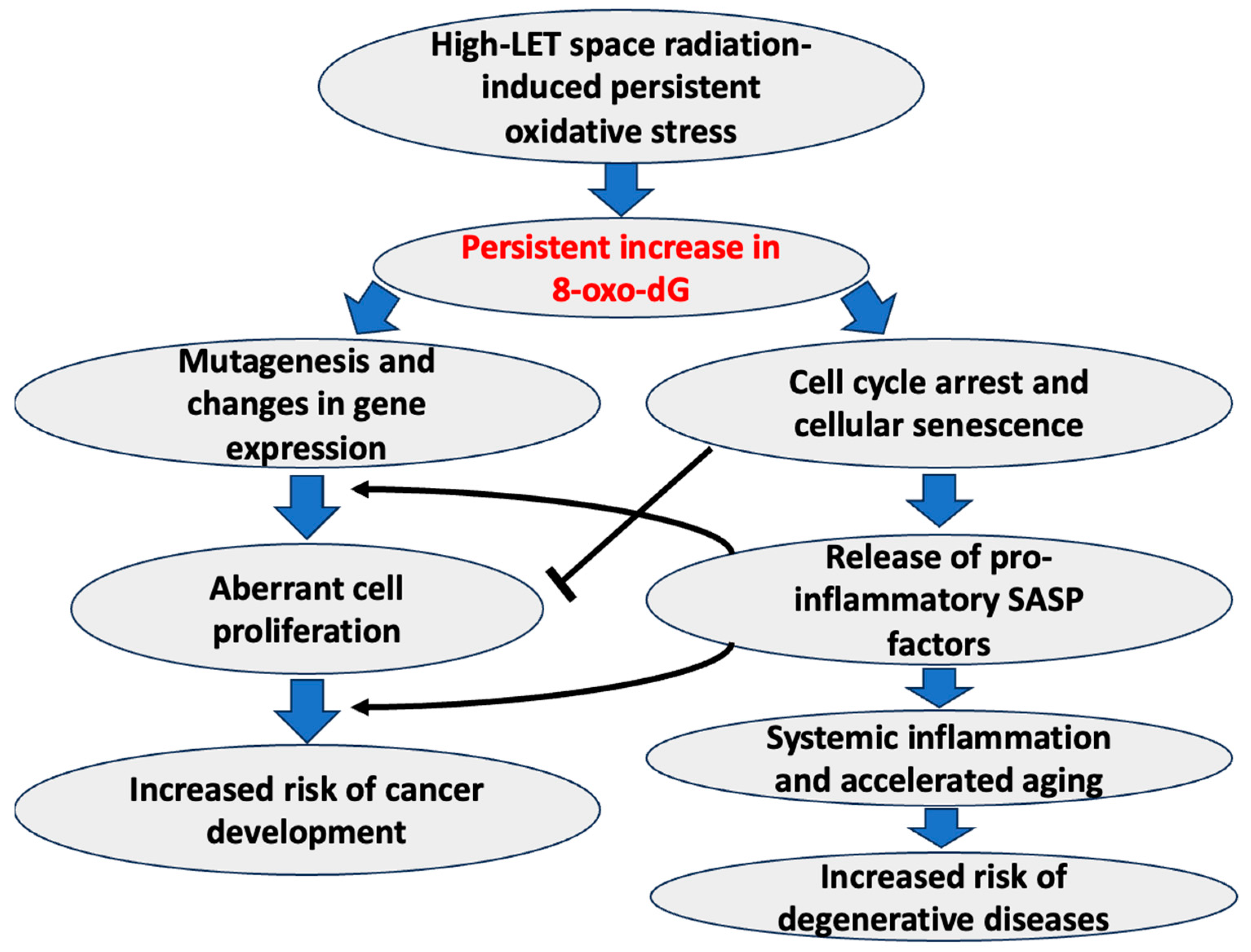
5. 8-OxodG as a Biomarker of High-LET Radiation-Induced Chronic Oxidative Stress and Associated Adverse Health Effects
6. Analytical Methods for Quantitative Assessment of 8-OxodG
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bailey, D.L.; Humm, J.L.; Todd-Pokropek, A.; Aswegen, A.V. (Eds.) Nuclear Medicine Physics: A Handbook for Teachers and Students; International Atomic Energy Agency, Division of Human Health: Vienna, Austria, 2014. [Google Scholar]
- National Research Council. Health Risks from Exposure to Low Levels of Ionizing Radiation: BEIR VII Phase 2; The National Academies Press: Washington DC, USA, 2006; p. 422. [Google Scholar]
- Kumar, K.; Kumar, S.; Datta, K.; Fornace, A.J.; Suman, S. High-LET-Radiation-Induced Persistent DNA Damage Response Signaling and Gastrointestinal Cancer Development. Curr. Oncol. 2023, 30, 5497–5514. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2012; p. 908. [Google Scholar]
- Ray, S.; Cekanaviciute, E.; Lima, I.P.; Sørensen, B.S.; Costes, S.V. Comparing Photon and Charged Particle Therapy Using DNA Damage Biomarkers. Int. J. Part Ther. 2018, 5, 15–24. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Ionizing Radiation, Part 1: X- and Gamma (γ)-radiation, and Neutrons; International Agency for Research on Cancer: Lyon, France, 2000; p. 491. [Google Scholar]
- Henriksen, T.; Maillie, D. Radiation and Health; CRC Press: London, UK, 2002; p. 241. [Google Scholar]
- Chatterjee, A.; Schaefer, H.J. Microdosimetric structure of heavy ion tracks in tissue. Radiat. Environ. Biophys. 1976, 13, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.L.; Chatterjee, A. Radiation chemistry of heavy-particle tracks. 1. General considerations. J. Phys. Chem. 1980, 84, 3529–3536. [Google Scholar] [CrossRef]
- Swarts, S.G.; Gilbert, D.C.; Sharma, K.K.; Razskazovskiy, Y.; Purkayastha, S.; Naumenko, K.A.; Bernhard, W.A. Mechanisms of direct radiation damage in DNA, based on a study of the yields of base damage, deoxyribose damage, and trapped radicals in d(GCACGCGTGC)(2). Radiat. Res. 2007, 168, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Kant, M.; Jaruga, P.; Coskun, E.; Ward, S.; Stark, A.D.; Baumann, T.; Becker, D.; Adhikary, A.; Sevilla, M.D.; Dizdaroglu, M. Ne-22 Ion-Beam Radiation Damage to DNA: From Initial Free Radical Formation to Resulting DNA-Base Damage. ACS Omega 2021, 6, 16600–16611. [Google Scholar] [CrossRef]
- Alizadeh, E.; Sanz, A.G.; García, G.; Sanche, L. Radiation Damage to DNA: The Indirect Effect of Low Energy Electrons. J. Phys. Chem. Lett. 2013, 4, 820–825. [Google Scholar] [CrossRef]
- D’Auria Vieira de Godoy, P.R.; Nakamura, A.; Khavari, A.P.; Sangsuwan, T.; Haghdoost, S. Effect of dose and dose rate of gamma irradiation on the formation of micronuclei in bone marrow cells isolated from whole-body-irradiated mice. Environ. Mol. Mutagen. 2021, 62, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Asaithamby, A.; Chen, D.J. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat. Res. 2011, 711, 87–99. [Google Scholar] [CrossRef]
- de Almeida, A.J.P.O.; de Oliveira, J.C.P.L.; da Silva Pontes, L.V.; de Souza Júnior, J.F.; Gonçalves, T.A.F.; Dantas, S.H.; de Almeida Feitosa, M.S.; Silva, A.O.; de Medeiros, I.A. ROS: Basic Concepts, Sources, Cellular Signaling, and its Implications in Aging Pathways. Oxid. Med. Cell Longev. 2022, 2022, 1225578. [Google Scholar] [CrossRef]
- Suman, S.; Seth, R.K.; Chandna, S. Mitochondrial antioxidant defence in radio-resistant Lepidopteran insect cells. Bioinformation 2009, 4, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Seth, R.K.; Chandna, S. A calcium-insensitive attenuated nitrosative stress response contributes significantly in the radioresistance of Sf9 insect cells. Int. J. Biochem. Cell Biol. 2011, 43, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Checa, J.; Aran, J.M. Reactive Oxygen Species: Drivers of Physiological and Pathological Processes. J. Inflamm. Res. 2020, 13, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suman, S.; Fornace, A.J.; Datta, K. Space radiation triggers persistent stress response, increases senescent signaling, and decreases cell migration in mouse intestine. Proc. Natl. Acad. Sci. USA 2018, 115, E9832–E9841. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Rodriguez, O.C.; Winters, T.A.; Fornace, A.J.; Albanese, C.; Datta, K. Therapeutic and space radiation exposure of mouse brain causes impaired DNA repair response and premature senescence by chronic oxidant production. Aging 2013, 5, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Kumar, S.; Fornace, A.J.; Datta, K. The effect of carbon irradiation is associated with greater oxidative stress in mouse intestine and colon relative to γ-rays. Free Radic. Res. 2018, 52, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suman, S.; Moon, B.H.; Fornace, A.J.; Datta, K. Low dose radiation upregulates Ras/p38 and NADPH oxidase in mouse colon two months after exposure. Mol. Biol. Rep. 2023, 50, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Zeliger, H. Oxidative Stress: Its Mechanisms and Impacts on Human Health and Disease Onset; Elsevier Science: Amsterdam, The Netherlands, 2022; p. 502. [Google Scholar]
- Chao, M.R.; Evans, M.D.; Hu, C.W.; Ji, Y.; Møller, P.; Rossner, P.; Cooke, M.S. Biomarkers of nucleic acid oxidation—A summary state-of-the-art. Redox. Biol. 2021, 42, 101872. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Jaruga, P.; Dizdaroglu, M.; Fornace, A.J.; Datta, K. Heavy ion space radiation triggers ongoing DNA base damage by downregulating DNA repair pathways. Life Sci. Space Res. 2020, 27, 27–32. [Google Scholar] [CrossRef]
- Hada, M.; Georgakilas, A.G. Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef]
- Hagiwara, Y.; Oike, T.; Niimi, A.; Yamauchi, M.; Sato, H.; Limsirichaikul, S.; Held, K.D.; Nakano, T.; Shibata, A. Clustered DNA double-strand break formation and repair pathway following heavy-ion irradiation. J. Radiat. Res. 2019, 60, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.M.; Nennett, P.V.; Sidorkina, O.; Laval, J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. USA 2000, 97, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Nikitaki, Z.; Nikolov, V.; Mavragani, I.V.; Mladenov, E.; Mangelis, A.; Laskaratou, D.A.; Fragkoulis, G.I.; Hellweg, C.E.; Martin, O.A.; Emfietzoglou, D.; et al. Measurment of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET). Free Radic. Res. 2016, 50, S64–S78. [Google Scholar] [CrossRef] [PubMed]
- Sage, E.; Shikazono, N. Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 2017, 107, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Hahm, J.Y.; Park, J.; Jang, E.S.; Chi, S.W. 8-Oxoguanine: From oxidative damage to epigenetic and epitranscriptional modification. Exp. Mol. Med. 2022, 54, 1626–1642. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, M.K.; Aboujaoude, E.N.; Chen, Q.; Ames, B.N. Assays of oxidative DNA damage biomarkers 8-oxo-2’-deoxyguanosine and 8-oxoguanine in nuclear DNA and biological fluids by high-performance liquid chromatography with electrochemical detection. Methods Enzymol. 1994, 234, 16–33. [Google Scholar] [PubMed]
- Thanan, R.; Oikawa, S.; Hiraku, Y.; Ohnishi, S.; Ma, N.; Pinlaor, S.; Yongvanit, P.; Kawanishi, S.; Murata, M. Oxidative stress and its significant roles in neurodegenerative diseases and cancer. Int. J. Mol. Sci. 2014, 16, 193–217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, L. The Significance of 8-oxoGsn in Aging-Related Diseases. Aging Dis. 2020, 11, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Chiorcea-Paquim, A.M. 8-oxoguanine and 8-oxodeoxyguanosine Biomarkers of Oxidative DNA Damage: A Review on HPLC-ECD Determination. Molecules 2022, 27, 1620. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, P.; Wang, Z.; Han, L.; Li, J.; Tian, C.; Zhao, F.; Wang, J.; Zhao, F.; Zhang, Q.; et al. Serum 8-Hydroxy-2’-Deoxyguanosine Level as a Potential Biomarker of Oxidative DNA Damage Induced by Ionizing Radiation in Human Peripheral Blood. Dose Response 2019, 17, 1559325818820649. [Google Scholar] [CrossRef]
- Ribeiro, A.; Husson, O.; Drey, N.; Murray, I.; May, K.; Thurston, J.; Oyen, W. Ionising radiation exposure from medical imaging—A review of Patient’s (un) awareness. Radiography 2020, 26, e25–e30. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Shukla, S.K.; Suman, S. Editorial: Multifaceted Approaches Combining Low or High LET Radiation and Pharmacological Interventions in Cancer and Radioprotection: From Bench to Bedside. Front. Oncol. 2022, 12, 880607. [Google Scholar] [CrossRef] [PubMed]
- Baudin, C.; Vacquier, B.; Thin, G.; Chenene, L.; Guersen, J.; Partarrieu, I.; Louet, M.; Ducou Le Pointe, H.; Mora, S.; Verdun-Esquer, C.; et al. Occupational exposure to ionizing radiation in medical staff: Trends during the 2009-2019 period in a multicentric study. Eur. Radiol. 2023, 33, 5675–5684. [Google Scholar] [CrossRef] [PubMed]
- Huff, J.L.; Poignant, F.; Rahmanian, S.; Khan, N.; Blakely, E.A.; Britten, R.A.; Chang, P.; Fornace, A.J.; Hada, M.; Kronenberg, A.; et al. Galactic cosmic ray simulation at the NASA space radiation laboratory—Progress, challenges and recommendations on mixed-field effects. Life Sci. Space Res. 2023, 36, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, L.C.; Slaba, T.C.; Guida, P.; Rusek, A. NASA’s first ground-based Galactic Cosmic Ray Simulator: Enabling a new era in space radiobiology research. PLoS Biol. 2020, 18, e3000669. [Google Scholar] [CrossRef] [PubMed]
- Helm, A.; Fournier, C. High-LET charged particles: Radiobiology and application for new approaches in radiotherapy. Strahlenther. Onkol. 2023, 199, 1225–1241. [Google Scholar] [CrossRef] [PubMed]
- Ekendahl, D.; Rubovič, P.; Žlebčík, P.; Hupka, I.; Huml, O.; Bečková, V.; Malá, H. NEUTRON DOSE ASSESSMENT USING SAMPLES OF HUMAN BLOOD AND HAIR. Radiat. Prot. Dosim. 2019, 186, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Akashi, M.; Maekawa, K. Medical management of heavily exposed victims: An experience at the Tokaimura criticality accident. J. Radiol. Prot. 2021, 41, S391. [Google Scholar] [CrossRef] [PubMed]
- Leonard, B.E. The radon inverse dose rate effect and high-LET galactic hazards. Radiat. Prot. Dosim. 2005, 115, 310–315. [Google Scholar] [CrossRef]
- Suman, S.; Kumar, S.; Moon, B.H.; Strawn, S.J.; Thakor, H.; Fan, Z.; Shay, J.W.; Fornace, A.J.; Datta, K. Relative Biological Effectiveness of Energetic Heavy Ions for Intestinal Tumorigenesis Shows Male Preponderance and Radiation Type and Energy Dependence in APC(1638N/+) Mice. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 131–138. [Google Scholar] [CrossRef]
- Datta, K.; Suman, S.; Kallakury, B.V.; Fornace, A.J. Exposure to heavy ion radiation induces persistent oxidative stress in mouse intestine. PLoS ONE 2012, 7, e42224. [Google Scholar] [CrossRef]
- Suman, S.; Datta, K.; Trani, D.; Laiakis, E.C.; Strawn, S.J.; Fornace, A.J. Relative biological effectiveness of 12C and 28Si radiation in C57BL/6J mice. Radiat. Environ. Biophys. 2012, 51, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Moon, B.H.; Datta, K.; Kallakury, B.V.S.; Fornace, A.J. Heavy-ion radiation-induced colitis and colorectal carcinogenesis in Il10-/- mice display co-activation of β-catenin and NF-κB signaling. PLoS ONE 2022, 17, e0279771. [Google Scholar] [CrossRef] [PubMed]
- Shuryak, I.; Fornace, A.J.; Datta, K.; Suman, S.; Kumar, S.; Sachs, R.K.; Brenner, D.J. Scaling Human Cancer Risks from Low LET to High LET when Dose-Effect Relationships are Complex. Radiat. Res. 2017, 187, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Datta, K.; Fornace, A.J.; Suman, S. Total body proton and heavy-ion irradiation causes cellular senescence and promotes pro-osteoclastogenic activity in mouse bone marrow. Heliyon 2022, 8, e08691. [Google Scholar] [CrossRef] [PubMed]
- Restier-Verlet, J.; El-Nachef, L.; Ferlazzo, M.L.; Al-Choboq, J.; Granzotto, A.; Bouchet, A.; Foray, N. Radiation on Earth or in Space: What Does It Change. Int. J. Mol. Sci. 2021, 22, 3739. [Google Scholar] [CrossRef]
- Norbury, J.W.; Schimmerling, W.; Slaba, T.C.; Azzam, E.I.; Badavi, F.F.; Baiocco, G.; Benton, E.; Bindi, V.; Blakely, E.A.; Blattnig, S.R.; et al. Galactic cosmic ray simulation at the NASA Space Radiation Laboratory. Life Sci. Space Res. 2016, 8, 38–51. [Google Scholar] [CrossRef]
- Hart, D.A. Homo sapiens-A Species Not Designed for Space Flight: Health Risks in Low Earth Orbit and Beyond, Including Potential Risks When Traveling beyond the Geomagnetic Field of Earth. Life 2023, 13, 757. [Google Scholar] [CrossRef]
- Sishc, B.J.; Zawaski, J.; Saha, J.; Carnell, L.S.; Fabre, K.M.; Elgart, S.R. The Need for Biological Countermeasures to Mitigate the Risk of Space Radiation-Induced Carcinogenesis, Cardiovascular Disease, and Central Nervous System Deficiencies. Life Sci. Space Res. 2022, 35, 4–8. [Google Scholar] [CrossRef]
- Kumar, K.; Moon, B.H.; Datta, K.; Fornace, A.J.; Suman, S. Simulated galactic cosmic radiation (GCR)-induced expression of Spp1 coincide with mammary ductal cell proliferation and preneoplastic changes in ApcMin/+ mouse. Life Sci. Space Res. 2023, 36, 116–122. [Google Scholar] [CrossRef]
- Pariset, E.; Bertucci, A.; Petay, M.; Malkani, S.; Lopez Macha, A.; Paulino Lima, I.G.; Gomez Gonzalez, V.; Tin, A.S.; Tang, J.; Plante, I.; et al. DNA Damage Baseline Predicts Resilience to Space Radiation and Radiotherapy. Cell Rep. 2020, 33, 108434. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, D.M.; Asaithamby, A.; Blattnig, S.R.; Costes, S.V.; Doetsch, P.W.; Dynan, W.S.; Hahnfeldt, P.; Hlatky, L.; Kidane, Y.; Kronenberg, A.; et al. Evaluating biomarkers to model cancer risk post cosmic ray exposure. Life Sci. Space Res. 2016, 9, 19–47. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Burrows, C.J. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: Role and Response of Short Guanine Tracts at Genomic Locations. Int. J. Mol. Sci. 2019, 20, 4258. [Google Scholar] [CrossRef] [PubMed]
- Kino, K.; Hirao-Suzuki, M.; Morikawa, M.; Sakaga, A.; Miyazawa, H. Generation, repair and replication of guanine oxidation products. Genes Environ. 2017, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Rokhlenko, Y.; Cadet, J.; Geacintov, N.E.; Shafirovich, V. Mechanistic aspects of hydration of guanine radical cations in DNA. J. Am. Chem. Soc. 2014, 136, 5956–5962. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, M.D.; Becker, D.; Kumar, A.; Adhikary, A. Gamma and ion-beam irradiation of DNA: Free radical mechanisms, electron effects, and radiation chemical track structure. Radiat. Phys. Chem. Oxf. Engl. 1993 2016, 128, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Balanikas, E.; Banyasz, A.; Baldacchino, G.; Markovitsi, D. Populations and dynamics of guanine radicals in DNA strands-direct versus indirect generation. Molecules 2019, 24, 2347. [Google Scholar] [CrossRef]
- Guo, C.; Ding, P.; Xie, C.; Ye, C.; Ye, M.; Pan, C.; Cao, X.; Zhang, S.; Zheng, S. Potential application of the oxidative nucleic acid damage biomarkers in detection of diseases. Oncotarget 2017, 8, 75767–75777. [Google Scholar] [CrossRef]
- Helbock, H.J.; Beckman, K.B.; Shigenaga, M.K.; Walter, P.B.; Woodall, A.A.; Yeo, H.C.; Ames, B.N. DNA oxidation matters: The HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc. Natl. Acad. Sci. USA 1998, 95, 288–293. [Google Scholar] [CrossRef]
- Hu, C.W.; Chao, M.R.; Sie, C.H. Urinary analysis of 8-oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2’-deoxyguanosine by isotope-dilution LC-MS/MS with automated solid-phase extraction: Study of 8-oxo-7,8-dihydroguanine stability. Free Radic. Biol. Med. 2010, 48, 89–97. [Google Scholar] [CrossRef] [PubMed]
- AbuArrah, M.; Yuli Setianto, B.; Faisal, A.; Hamim Sadewa, A. 8-Hydroxy-2-Deoxyguanosine as Oxidative DNA Damage Biomarker of Medical Ionizing Radiation: A Scoping Review. J. Biomed. Phys. Eng. 2021, 11, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Maynard, S.; Schurman, S.H.; Harboe, C.; de Souza-Pinto, N.C.; Bohr, V.A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 2009, 30, 2–10. [Google Scholar] [CrossRef]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-excision repair of oxidative DNA damage. Nature 2007, 447, 941–950. [Google Scholar] [CrossRef]
- Ba, X.; Boldogh, I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox. Biol. 2018, 14, 669–678. [Google Scholar] [CrossRef]
- Santos, R.X.; Correia, S.C.; Zhu, X.; Smith, M.A.; Moreira, P.I.; Castellani, R.J.; Nunomura, A.; Perry, G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer’s disease. Antioxid. Redox. Signal. 2013, 18, 2444–2457. [Google Scholar] [CrossRef]
- Alexeyev, M.; Shokolenko, I.; Wilson, G.; LeDoux, S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013, 5, a012641. [Google Scholar] [CrossRef]
- Richter, C.; Park, J.W.; Ames, B.N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA 1988, 85, 6465–6467. [Google Scholar] [CrossRef]
- Singh, G.; Pachouri, U.C.; Khaidem, D.C.; Kundu, A.; Chopra, C.; Singh, P. Mitochondrial DNA Damage and Diseases. F1000Res 2015, 4, 176. [Google Scholar] [CrossRef] [PubMed]
- Shokolenko, I.N.; Wilson, G.L.; Alexeyev, M.F. Aging: A mitochondrial DNA perspective, critical analysis and an update. World J. Exp. Med. 2014, 4, 46–57. [Google Scholar] [CrossRef]
- Hastak, K.; Paul, R.K.; Agarwal, M.K.; Thakur, V.S.; Amin, A.R.; Agrawal, S.; Sramkoski, R.M.; Jacobberger, J.W.; Jackson, M.W.; Stark, G.R.; et al. DNA synthesis from unbalanced nucleotide pools causes limited DNA damage that triggers ATR-CHK1-dependent p53 activation. Proc. Natl. Acad. Sci. USA 2008, 105, 6314–6319. [Google Scholar] [CrossRef] [PubMed]
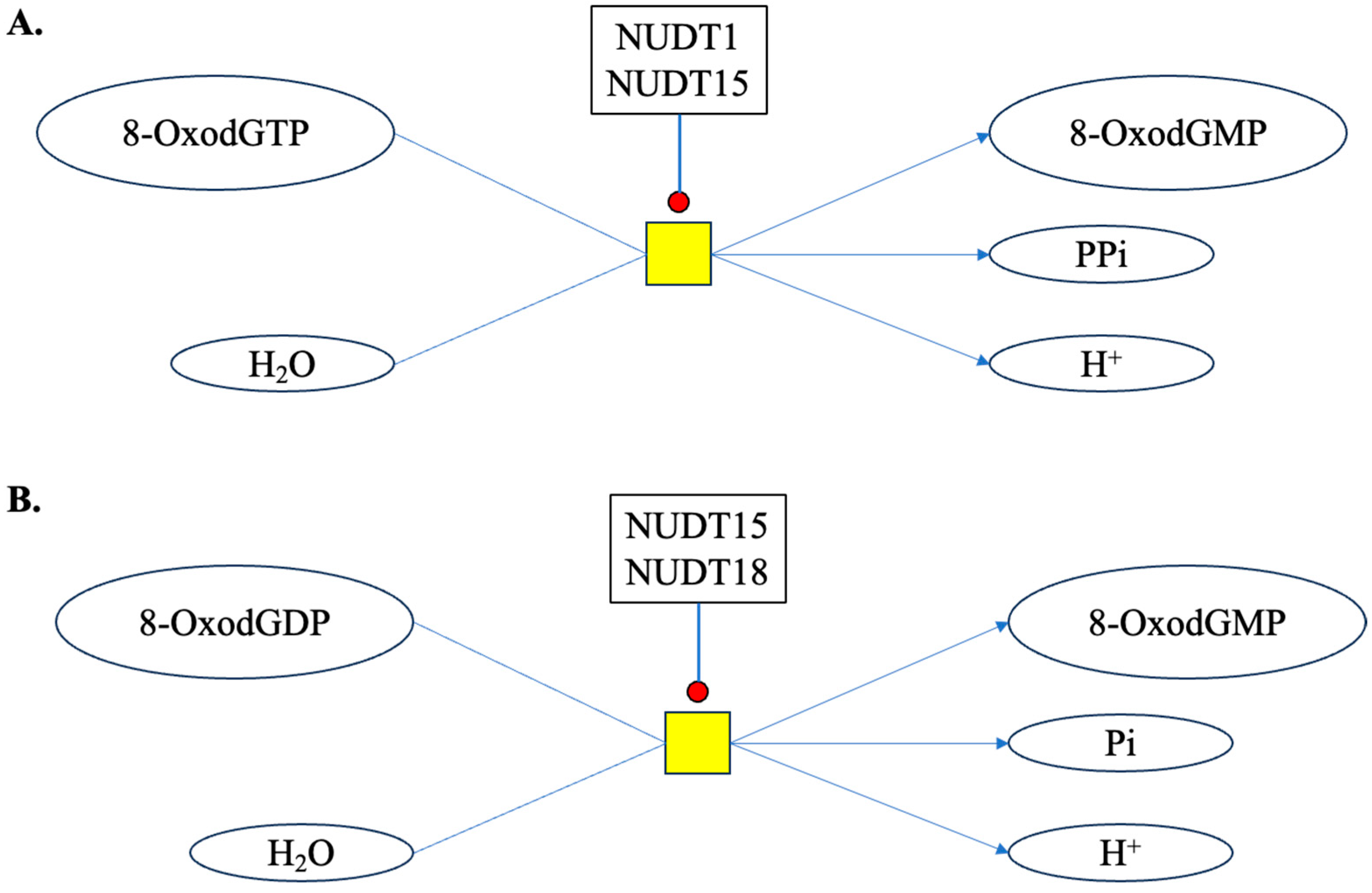
- Broderick, K.; Moutaoufik, M.T.; Aly, K.A.; Babu, M. Sanitation enzymes: Exquisite surveillance of the noncanonical nucleotide pool to safeguard the genetic blueprint. Semin. Cancer Biol. 2023, 94, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, D.; Sakumi, K.; Ohno, M.; Sakai, Y.; Furuichi, M.; Iwai, S.; Nakabeppu, Y. An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress. J. Biol. Chem. 2003, 278, 37965–37973. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Gui, X.; Chu, X.; Sun, Y.; Zhang, S.; Tong, H.; Ju, W.; Li, Y.; Sun, Z.; Xu, M.; et al. MTH1 protects platelet mitochondria from oxidative damage and regulates platelet function and thrombosis. Nat. Commun. 2023, 14, 4829. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, A.V.; Konkova, M.S.; Kostyuk, S.V.; Izevskaya, V.L.; Baranova, A.; Veiko, N.N. Oxidized extracellular DNA as a stress signal in human cells. Oxid. Med. Cell Longev. 2013, 2013, 649747. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, V.A.; Ershova, E.S.; Veiko, N.N.; Malinovskaya, E.M.; Kalyanov, A.A.; Kameneva, L.V.; Stukalov, S.V.; Dolgikh, O.A.; Konkova, M.S.; Ermakov, A.V.; et al. Low-Dose Ionizing Radiation Affects Mesenchymal Stem Cells via Extracellular Oxidized Cell-Free DNA: A Possible Mediator of Bystander Effect and Adaptive Response. Oxid. Med. Cell Longev. 2017, 2017, 9515809. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.G.; Zilhão, R.; Thorsteinsdóttir, S.; Carlos, A.R. Linking Oxidative Stress and DNA Damage to Changes in the Expression of Extracellular Matrix Components. Front. Genet 2021, 12, 673002. [Google Scholar] [CrossRef] [PubMed]
- Chiaradia, E.; Tancini, B.; Emiliani, C.; Delo, F.; Pellegrino, R.M.; Tognoloni, A.; Urbanelli, L.; Buratta, S. Extracellular Vesicles under Oxidative Stress Conditions: Biological Properties and Physiological Roles. Cells 2021, 10, 1763. [Google Scholar] [CrossRef] [PubMed]
- Elzanowska, J.; Semira, C.; Costa-Silva, B. DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 2021, 15, 1701–1714. [Google Scholar] [CrossRef]
- Yahata, T.; Takanashi, T.; Muguruma, Y.; Ibrahim, A.A.; Matsuzawa, H.; Uno, T.; Sheng, Y.; Onizuka, M.; Ito, M.; Kato, S.; et al. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood 2011, 118, 2941–2950. [Google Scholar] [CrossRef]
- Dizdaroglu, M. Oxidatively induced DNA damage: Mechanisms, repair and disease. Cancer Lett. 2012, 327, 26–47. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Henderson, P.T.; Evans, M.D. Sources of extracellular, oxidatively-modified DNA lesions: Implications for their measurement in urine. J. Clin. Biochem. Nutr. 2009, 45, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Garte, S.; Sram, R.J.; Binkova, B.; Kalina, I.; Lyubomirova, K.; Taioli, E.; Singh, R.; Farmer, P.B. Effects of diet on biomarkers of exposure and effects, and on oxidative damage. Mutat. Res. 2007, 620, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Quetglas-Llabrés, M.M.; Monserrat-Mesquida, M.; Bouzas, C.; Mateos, D.; Ugarriza, L.; Gómez, C.; Tur, J.A.; Sureda, A. Oxidative Stress and Inflammatory Biomarkers Are Related to High Intake of Ultra-Processed Food in Old Adults with Metabolic Syndrome. Antioxidants 2023, 12, 1532. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.W.; Kant, M.; Coskun, E.; Kato, T.A.; Jaruga, P.; Palafox, E.; Dizdaroglu, M.; Kool, E.T. Possible Genetic Risks from Heat-Damaged DNA in Food. ACS Cent. Sci. 2023, 9, 1170–1179. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.C.; Liu, C.W.; Chi, L.; Yang, Y.; Lu, K. Effects of Gut Microbiome on Carcinogenic DNA Damage. Chem. Res. Toxicol. 2020, 33, 2130–2138. [Google Scholar] [CrossRef] [PubMed]
- Kunst, C.; Schmid, S.; Michalski, M.; Tümen, D.; Buttenschön, J.; Müller, M.; Gülow, K. The Influence of Gut Microbiota on Oxidative Stress and the Immune System. Biomedicines 2023, 11, 1388. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi SM, J.; Said-Salman, I.; Mortazavi, A.R.; El Khatib, S.; Sihver, L. How the adaptation of the human microbiome to harsh space environment can determine the chances of success for a space mission to Mars and beyond. Front. Microbiol. 2023, 14, 1237564. [Google Scholar] [CrossRef]
- Li, Z.; Ke, X.; Zuo, D.; Wang, Z.; Fang, F.; Li, B. New Insights into the Relationship between Gut Microbiota and Radiotherapy for Cancer. Nutrients 2022, 15, 48. [Google Scholar] [CrossRef]
- Fernandes, A.; Oliveira, A.; Soares, R.; Barata, P. The Effects of Ionizing Radiation on Gut Microbiota: What Can Animal Models Tell Us?-A Systematic Review. Curr. Issues Mol. Biol. 2023, 45, 3877–3910. [Google Scholar] [CrossRef]
- Bardaweel, S.K.; Gul, M.; Alzweiri, M.; Ishaqat, A.; ALSalamat, H.A.; Bashatwah, R.M. Reactive Oxygen Species: The Dual Role in Physiological and Pathological Conditions of the Human Body. Eurasian J. Med. 2018, 50, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.C.; Zhang, Y.; Wang, Y.; Wei, W.; Zhou, Y.X.; Xie, Y.; Wang, X.; Qi, Y.Z.; Chang, L.; Jia, Z.P.; et al. Quantitative proteomics reveals mitochondrial respiratory chain as a dominant target for carbon ion radiation: Delayed reactive oxygen species generation caused DNA damage. Free Radic. Biol. Med. 2019, 130, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.S.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox. Signal. 2014, 21, 396–413. [Google Scholar] [CrossRef] [PubMed]
- Nuszkiewicz, J.; Woźniak, A.; Szewczyk-Golec, K. Ionizing Radiation as a Source of Oxidative Stress-The Protective Role of Melatonin and Vitamin D. Int. J. Mol. Sci. 2020, 21, 5804. [Google Scholar] [CrossRef] [PubMed]
- Karabulutoglu, M.; Finnon, R.; Cruz-Garcia, L.; Hill, M.A.; Badie, C. Oxidative Stress and X-ray Exposure Levels-Dependent Survival and Metabolic Changes in Murine HSPCs. Antioxidants 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Inaba, Y.; Sogo, Y.; Ito, A.; Bekal, M.; Chida, K.; Moritake, T. Total body irradiation causes a chronic decrease in antioxidant levels. Sci. Rep. 2021, 11, 6716. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Choi, J.; Cho, I.; Sheen, Y.Y. Radiotherapy-induced oxidative stress and fibrosis in breast cancer are suppressed by vactosertib, a novel, orally bioavailable TGF-β/ALK5 inhibitor. Sci. Rep. 2022, 12, 16104. [Google Scholar] [CrossRef] [PubMed]
- Chibaya, L.; Karim, B.; Zhang, H.; Jones, S.N. Mdm2 phosphorylation by Akt regulates the p53 response to oxidative stress to promote cell proliferation and tumorigenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2003193118. [Google Scholar] [CrossRef]
- Gonçalves, R.V.; Costa AM, A.; Grzeskowiak, L. Oxidative Stress and Tissue Repair: Mechanism, Biomarkers, and Therapeutics. Oxid. Med. Cell Longev. 2021, 2021, 6204096. [Google Scholar] [CrossRef]
- Datta, K.; Suman, S.; Fornace, A.J. Radiation persistently promoted oxidative stress, activated mTOR via PI3K/Akt, and downregulated autophagy pathway in mouse intestine. Int. J. Biochem. Cell Biol. 2014, 57, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suman, S.; Fornace, A.J.; Datta, K. Intestinal stem cells acquire premature senescence and senescence associated secretory phenotype concurrent with persistent DNA damage after heavy ion radiation in mice. Aging 2019, 11, 4145–4158. [Google Scholar] [CrossRef] [PubMed]
- Tseng, B.P.; Giedzinski, E.; Izadi, A.; Suarez, T.; Lan, M.L.; Tran, K.K.; Acharya, M.M.; Nelson, G.A.; Raber, J.; Parihar, V.K.; et al. Functional consequences of radiation-induced oxidative stress in cultured neural stem cells and the brain exposed to charged particle irradiation. Antioxid. Redox. Signal. 2014, 20, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Oyefeso, F.A.; Goldberg, G.; Opoku, N.Y.P.S.; Vazquez, M.; Bertucci, A.; Chen, Z.; Wang, C.; Muotri, A.R.; Pecaut, M.J. Effects of acute low-moderate dose ionizing radiation to human brain organoids. PLoS ONE 2023, 18, e0282958. [Google Scholar] [CrossRef] [PubMed]
- Laurent, C.; Leduc, A.; Pottier, I.; Prévost, V.; Sichel, F.; Lefaix, J.L. Dramatic increase in oxidative stress in carbon-irradiated normal human skin fibroblasts. PLoS ONE 2013, 8, e85158. [Google Scholar] [CrossRef] [PubMed]
- Alwood, J.S.; Tran, L.H.; Schreurs, A.S.; Shirazi-Fard, Y.; Kumar, A.; Hilton, D.; Tahimic CG, T.; Globus, R.K. Dose- and Ion-Dependent Effects in the Oxidative Stress Response to Space-Like Radiation Exposure in the Skeletal System. Int. J. Mol. Sci. 2017, 18, 2117. [Google Scholar] [CrossRef]
- Suman, S.; Kumar, S.; Moon, B.H.; Angdisen, J.; Kallakury, B.V.S.; Datta, K.; Fornace, A.J. Effects of dietary aspirin on high-LET radiation-induced prostaglandin E2 levels and gastrointestinal tumorigenesis in Apc1638N/+ mice. Life Sci. Space Res. 2021, 31, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Prevost, V.; Sichel, F.; Pottier, I.; Leduc, A.; Lagadu, S.; Laurent, C. Production of early and late nuclear damage and extracellular 8-oxodG in normal skin fibroblasts after carbon ion irradiation compared to X-rays. Toxicol. In Vitro 2018, 52, 116–121. [Google Scholar] [CrossRef]
- Gorini, F.; Scala, G.; Cooke, M.S.; Majello, B.; Amente, S. Towards a comprehensive view of 8-oxo-7,8-dihydro-2’-deoxyguanosine: Highlighting the intertwined roles of DNA damage and epigenetics in genomic instability. DNA Repair. 2021, 97, 103027. [Google Scholar] [CrossRef]
- Roszkowski, K.; Jozwicki, W.; Blaszczyk, P.; Mucha-Malecka, A.; Siomek, A. Oxidative damage DNA: 8-oxoGua and 8-oxodG as molecular markers of cancer. Med. Sci. Monit. 2011, 17, CR329–CR333. [Google Scholar] [CrossRef]
- Moretton, A.; Loizou, J.I. Interplay between Cellular Metabolism and the DNA Damage Response in Cancer. Cancers 2020, 12, 2051. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S.; Murphy, D.L.; Sweasy, J.B. Base excision repair and cancer. Cancer Lett. 2012, 327, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Hanna BM, F.; Michel, M.; Helleday, T.; Mortusewicz, O. NEIL1 and NEIL2 Are Recruited as Potential Backup for OGG1 upon OGG1 Depletion or Inhibition by TH5487. Int. J. Mol. Sci. 2021, 22, 4542. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhou, G.; Hu, W. Carcinogenesis induced by space radiation: A systematic review. Neoplasia 2022, 32, 100828. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Fornace, A.J. Countermeasure development against space radiation-induced gastrointestinal carcinogenesis: Current and future perspectives. Life Sci. Space Res. 2022, 35, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Poetsch, A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020, 18, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.; Takeshita, M.; Grollman, A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 1991, 349, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y.; Sakumi, K.; Sakamoto, K.; Tsuchimoto, D.; Tsuzuki, T.; Nakatsu, Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol. Chem. 2006, 387, 373–379. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Konovalov, K.A.; Pardo-Avila, F.; Tse CK, M.; Oh, J.; Wang, D.; Huang, X. 8-Oxo-guanine DNA damage induces transcription errors by escaping two distinct fidelity control checkpoints of RNA polymerase II. J. Biol. Chem. 2019, 294, 4924–4933. [Google Scholar] [CrossRef] [PubMed]
- Brégeon, D.; Doetsch, P.W. Transcriptional mutagenesis: Causes and involvement in tumour development. Nat. Rev. Cancer 2011, 11, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Song, M.; Adams, L.; Jeong, I.; Je, G.; Guhathakurta, S.; Jiang, J.; Boparai, N.; Dai, W.; Cardozo-Pelaez, F.; et al. Transcriptional mutagenesis of α-synuclein caused by DNA oxidation in Parkinson’s disease pathogenesis. Acta Neuropathol. 2023, 146, 685–705. [Google Scholar] [CrossRef] [PubMed]
- Hailer-Morrison, M.K.; Kotler, J.M.; Martin, B.D.; Sugden, K.D. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2’-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry 2003, 42, 9761–9770. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, M.; Dellino, G.I.; Gambino, V.; Roda, N.; Pelicci, P.G. On the epigenetic role of guanosine oxidation. Redox. Biol. 2020, 29, 101398. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Hickson, L.J.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular senescence: The good, the bad and the unknown. Nat. Rev. Nephrol. 2022, 18, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Basisty, N.; Kale, A.; Jeon, O.H.; Kuehnemann, C.; Payne, T.; Rao, C.; Holtz, A.; Shah, S.; Sharma, V.; Ferrucci, L.; et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020, 18, e3000599. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Prattichizzo, F.; Grillari, J.; Balistreri, C.R. Cellular Senescence and Inflammaging in Age-Related Diseases. Mediators Inflamm. 2018, 2018, 9076485. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suman, S.; Datta, K.; Fornace, A.J. Effects of heavy ion 28Si exposure on mouse natural killer (NK) cells: An important effector for targeting senescent cells. In Proceedings of the 2022 NASA Human Research Program Investigators’ Workshop, Virtual Event, 7–10 February 2022. [Google Scholar]
- Barnes, R.P.; de Rosa, M.; Thosar, S.A.; Detwiler, A.C.; Roginskaya, V.; Van Houten, B.; Bruchez, M.P.; Stewart-Ornstein, J.; Opresko, P.L. Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening. Nat. Struct. Mol. Biol. 2022, 29, 639–652. [Google Scholar] [CrossRef]
- Oikawa, S.; Tada-Oikawa, S.; Kawanishi, S. Site-specific DNA damage at the GGG sequence by UVA involves acceleration of telomere shortening. Biochemistry 2001, 40, 4763–4768. [Google Scholar] [CrossRef]
- Oikawa, S. Sequence-specific DNA damage by reactive oxygen species: Implications for carcinogenesis and aging. Environ. Health Prev. Med. 2005, 10, 65–71. [Google Scholar] [CrossRef]
- Kawanishi, S.; Oikawa, S. Mechanism of telomere shortening by oxidative stress. Ann. N. Y. Acad. Sci. 2004, 1019, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Wilson, V.L.; Taffe, B.G.; Shields, P.G.; Povey, A.C.; Harris, C.C. Detection and quantification of 8-hydroxydeoxyguanosine adducts in peripheral blood of people exposed to ionizing radiation. Environ. Health Perspect. 1993, 99, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Povey, A.C.; Wilson, V.L.; Weston, A.; Doan, V.T.; Wood, M.L.; Essigmann, J.M.; Shields, P.G. Detection of oxidative damage by 32P-postlabelling: 8-hydroxydeoxyguanosine as a marker of exposure. IARC Sci. Publ. 1993, 124, 105–114. [Google Scholar]
- Thomas, M.C.; Woodward, M.; Li, Q.; Pickering, R.; Tikellis, C.; Poulter, N.; Cooper, M.E.; Marre, M.; Zoungas, S.; Chalmers, J.; et al. Relationship Between Plasma 8-OH-Deoxyguanosine and Cardiovascular Disease and Survival in Type 2 Diabetes Mellitus: Results From the ADVANCE Trial. J. Am. Heart Assoc. 2018, 7, e008226. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Li, X.; Wang, R.; Yu, J.; Ye, M.; Mao, L.; Zhang, S.; Zheng, S. Association between Oxidative DNA Damage and Risk of Colorectal Cancer: Sensitive Determination of Urinary 8-Hydroxy-2’-deoxyguanosine by UPLC-MS/MS Analysis. Sci. Rep. 2016, 6, 32581. [Google Scholar] [CrossRef]
- Doroshow, J.H.; Synold, T.W.; Somlo, G.; Akman, S.A.; Gajewski, E. Oxidative DNA base modifications in peripheral blood mononuclear cells of patients treated with high-dose infusional doxorubicin. Blood 2001, 97, 2839–2845. [Google Scholar] [CrossRef] [PubMed]
- Breton, J.; Sichel, F.; Pottier, D.; Prevost, V. Measurement of 8-oxo-7,8-dihydro-2’-deoxyguanosine in peripheral blood mononuclear cells: Optimisation and application to samples from a case-control study on cancers of the oesophagus and cardia. Free Radic. Res. 2005, 39, 21–30. [Google Scholar] [CrossRef]
- Roszkowski, K.; Olinski, R. Urinary 8-oxoguanine as a predictor of survival in patients undergoing radiotherapy. Cancer Epidemiol. Biomark. Prev. 2012, 21, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Fraga, C.G.; Shigenaga, M.K.; Park, J.W.; Degan, P.; Ames, B.N. Oxidative damage to DNA during aging: 8-hydroxy-2’-deoxyguanosine in rat organ DNA and urine. Proc. Natl. Acad. Sci. USA 1990, 87, 4533–4537. [Google Scholar] [CrossRef]
- Evans, M.D.; Saparbaev, M.; Cooke, M.S. DNA repair and the origins of urinary oxidized 2’-deoxyribonucleosides. Mutagenesis 2010, 25, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Evans, M.D.; Dove, R.; Rozalski, R.; Gackowski, D.; Siomek, A.; Lunec, J.; Olinski, R. DNA repair is responsible for the presence of oxidatively damaged DNA lesions in urine. Mutat. Res. 2005, 574, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.D.; Olinski, R.; Loft, S.; Cooke, M.S.; Rossner, J.P.; Sram, R.; Henriksen, T.; Poulsen, H.E.; Weimann, A.; Barbieri, A.; et al. Toward consensus in the analysis of urinary 8-oxo-7,8-dihydro-2’-deoxyguanosine as a noninvasive biomarker of oxidative stress. FASEB J. 2010, 24, 1249–1260. [Google Scholar]
- Lin, R.R.; Li, X.Y.; Weng, Q.H.; Zhou, X.X.; Zheng, F.Y.; Cai, J.P. A study on UHPLC-MS/MS analyses of DNA and RNA oxidative damage metabolites in patients with cervical carcinoma: 8-oxoG in urine as a potential biomarker of cervical carcinoma. Heliyon 2022, 8, e09321. [Google Scholar] [CrossRef]
- Schei, J.; Fuskevåg, O.M.; Stefansson, V.T.N.; Solbu, M.D.; Jenssen, T.G.; Eriksen, B.O.; Melsom, T. Urinary Markers of Oxidative Stress Are Associated with Albuminuria But Not GFR Decline. Kidney Int. Rep. 2018, 3, 573–582. [Google Scholar] [CrossRef]
- Graille, M.; Wild, P.; Sauvain, J.J.; Hemmendinger, M.; Guseva Canu, I.; Hopf, N.B. Urinary 8-OHdG as a Biomarker for Oxidative Stress: A Systematic Literature Review and Meta-Analysis. Int. J. Mol. Sci. 2020, 21, 3743. [Google Scholar] [CrossRef]
- Martinez-Moral, M.P.; Kannan, K. How stable is oxidative stress level? An observational study of intra- and inter-individual variability in urinary oxidative stress biomarkers of DNA, proteins, and lipids in healthy individuals. Environ. Int. 2019, 123, 382–389. [Google Scholar] [CrossRef]
- Silva, R.; Folgosa, F.; Soares, P.; Pereira, A.S.; Garcia, R.; Gestal-Otero, J.J.; Tavares, P.; Gomes da Silva, M.D. Occupational cosmic radiation exposure in Portuguese airline pilots: Study of a possible correlation with oxidative biological markers. Radiat. Environ. Biophys. 2013, 52, 211–220. [Google Scholar] [CrossRef]
- Strigari, L.; Strolin, S.; Morganti, A.G.; Bartoloni, A. Dose-Effects Models for Space Radiobiology: An Overview on Dose-Effect Relationships. Front. Public Health 2021, 9, 733337. [Google Scholar] [CrossRef]
- Foelsche, T. Estimates of radiation doses in space on the basis of current data. Life Sci. Space Res. 1963, 1, 48–94. [Google Scholar] [PubMed]
- Luxton, J.J.; McKenna, M.J.; Taylor, L.E.; George, K.A.; Zwart, S.R.; Crucian, B.E.; Drel, V.R.; Garrett-Bakelman, F.E.; Mackay, M.J.; Butler, D.; et al. Temporal Telomere and DNA Damage Responses in the Space Radiation Environment. Cell Rep. 2020, 33, 108435. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, C.A.; Khalid, R.; Cristea, O.; Greenberger, J.S.; Epperly, M.W.; Lemon, J.A.; Boreham, D.R.; Popov, D.; Gorthi, G.; Ramkumar, N.; et al. Space Radiation Protection Countermeasures in Microgravity and Planetary Exploration. Life 2021, 11, 829. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.P. Space flight and oxidative stress. Nutrition 2002, 18, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.P.; Leskiw, M.J. Oxidant damage during and after spaceflight. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E375–E382. [Google Scholar] [CrossRef] [PubMed]
- Weimann, A.; Belling, D.; Poulsen, H.E. Quantification of 8-oxo-guanine and guanine as the nucleobase, nucleoside and deoxynucleoside forms in human urine by high-performance liquid chromatography-electrospray tandem mass spectrometry. Nucleic. Acids Res. 2002, 30, E7. [Google Scholar] [CrossRef] [PubMed]
- Ravanat, J.L.; Turesky, R.J.; Gremaud, E.; Trudel, L.J.; Stadler, R.H. Determination of 8-oxoguanine in DNA by gas chromatography--mass spectrometry and HPLC--electrochemical detection: Overestimation of the background level of the oxidized base by the gas chromatography--mass spectrometry assay. Chem. Res. Toxicol. 1995, 8, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Hwa Yun, B.; Guo, J.; Bellamri, M.; Turesky, R.J. DNA adducts: Formation, biological effects, and new biospecimens for mass spectrometric measurements in humans. Mass Spectrom. Rev. 2020, 39, 55–82. [Google Scholar] [CrossRef] [PubMed]
- Rossner, P.; Orhan, H.; Koppen, G.; Sakai, K.; Santella, R.M.; Ambroz, A.; Rossnerova, A.; Sram, R.J.; Ciganek, M.; Neca, J.; et al. Urinary 8-oxo-7,8-dihydro-2’-deoxyguanosine analysis by an improved ELISA: An inter-laboratory comparison study. Free Radic. Biol. Med. 2016, 95, 169–179. [Google Scholar] [CrossRef]
- An, N.; Fleming, A.M.; White, H.S.; Burrows, C.J. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano 2015, 9, 4296–4307. [Google Scholar] [CrossRef]
- Pan, L.; Xue, Y.; Wang, K.; Zheng, X.; Boldogh, I. Detection of Oxidatively Modified Base Lesion(s) in Defined DNA Sequences by FLARE Quantitative PCR. Methods Mol. Biol. 2023, 2701, 115–134. [Google Scholar]
- Sakonsinsiri, C.; Puangmali, T.; Sreejivungsa, K.; Koowattanasuchat, S.; Thanan, R.; Chompoosor, A.; Kulchat, S.; Sithithaworn, P. Aptamer-based colorimetric detection of the DNA damage marker 8-oxo-dG using cysteamine-stabilised gold nanoparticles. RSC Adv. 2022, 12, 25478–25486. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, K.; Fornace, A.J., Jr.; Suman, S. 8-OxodG: A Potential Biomarker for Chronic Oxidative Stress Induced by High-LET Radiation. DNA 2024, 4, 221-238. https://doi.org/10.3390/dna4030015
Kumar K, Fornace AJ Jr., Suman S. 8-OxodG: A Potential Biomarker for Chronic Oxidative Stress Induced by High-LET Radiation. DNA. 2024; 4(3):221-238. https://doi.org/10.3390/dna4030015
Chicago/Turabian StyleKumar, Kamendra, Albert J. Fornace, Jr., and Shubhankar Suman. 2024. "8-OxodG: A Potential Biomarker for Chronic Oxidative Stress Induced by High-LET Radiation" DNA 4, no. 3: 221-238. https://doi.org/10.3390/dna4030015
APA StyleKumar, K., Fornace, A. J., Jr., & Suman, S. (2024). 8-OxodG: A Potential Biomarker for Chronic Oxidative Stress Induced by High-LET Radiation. DNA, 4(3), 221-238. https://doi.org/10.3390/dna4030015






