GAS2 Upregulation Is a Targetable Vulnerability in Chronic Myeloid Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Medium
2.2. Single-Cell Transcriptomics Dataset
2.3. Differential Expression Analysis
2.4. Cell Viability Quantification
2.5. Apoptosis Assay by Flow Cytometry
2.6. Cell Morphology Visualization
2.7. RNA Extraction
2.8. cDNA Preparation and qPCR
2.9. Colony-Forming Cell (CFC) Assay
2.10. Synergy Calculation
2.11. Statistical Analysis
3. Results
3.1. GAS2 Is Significantly Upregulated in Persistent BCR::ABL1+ LSCs after TKI Treatment
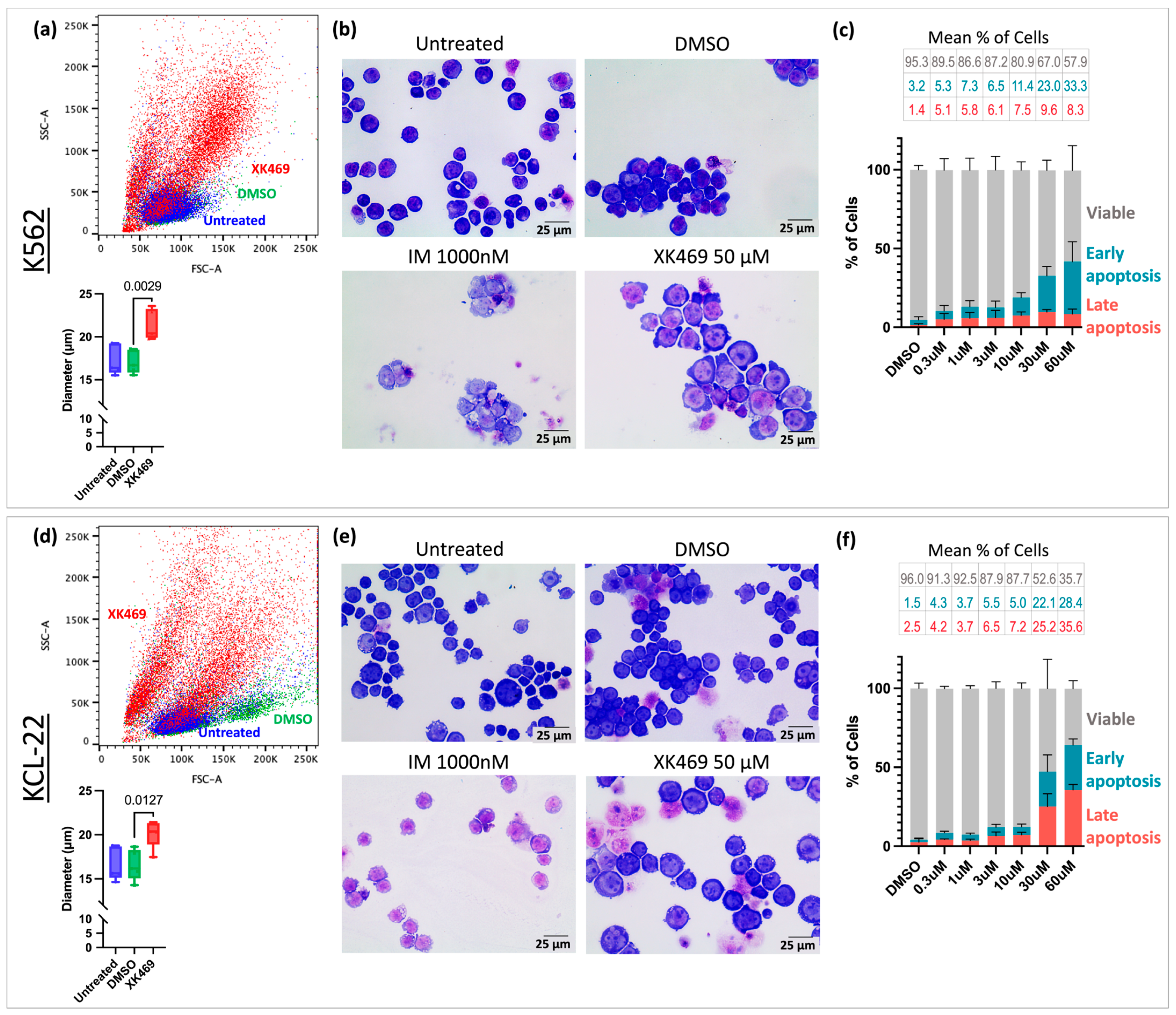
3.2. XK469 Potentially Targets GAS2 and Increases Cell Size
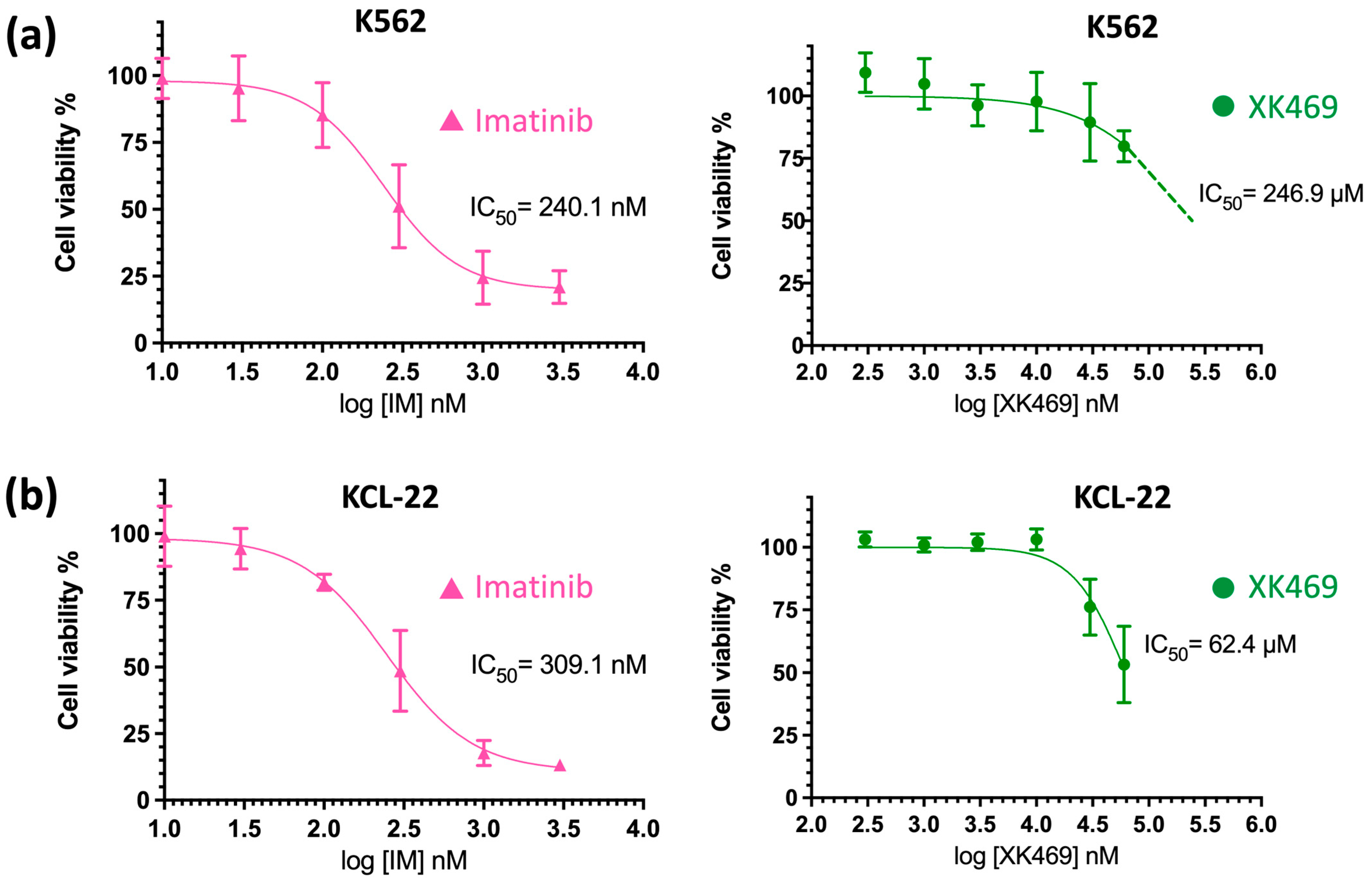
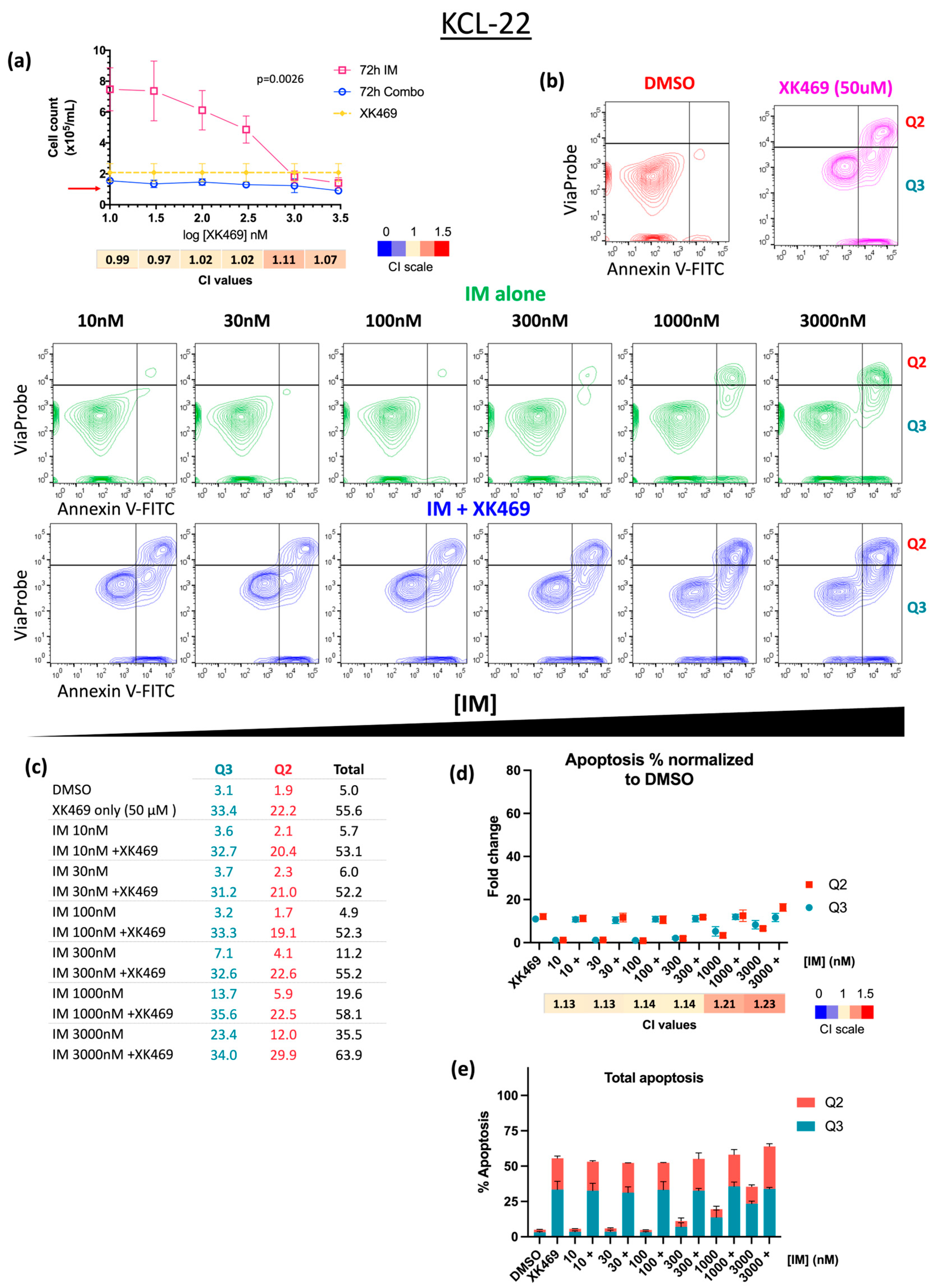
3.3. XK469 Sensitises CML Cells to IM In Vitro
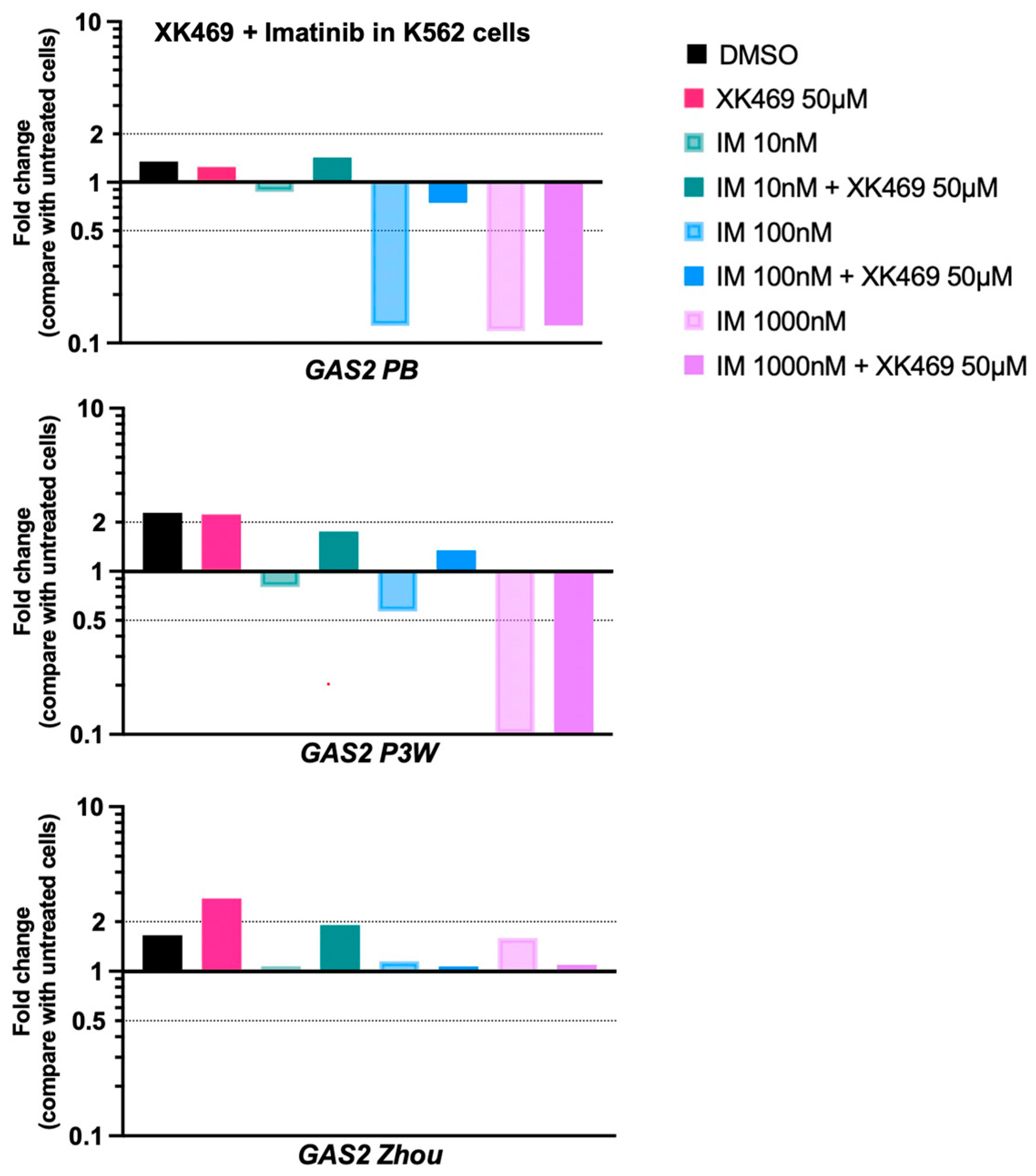
3.4. IM at High Concentration in Combination with XK469 Diminishes GAS2 mRNA Expression in CML Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vetrie, D.; Helgason, G.V.; Copland, M. The leukaemia stem cell: Similarities, differences and clinical prospects in CML and AML. Nat. Rev. Cancer 2020, 20, 158–173. [Google Scholar] [CrossRef] [PubMed]
- Redner, R.L. Why doesn’t imatinib cure chronic myeloid leukemia? Oncologist 2010, 15, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Zhu, X.; Jia, H.; Zhou, L.; Liu, H. Combination Therapies in Chronic Myeloid Leukemia for Potential Treatment-Free Remission: Focus on Leukemia Stem Cells and Immune Modulation. Front. Oncol. 2021, 11, 643382. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Xu, H.; Shah, N.P.; Snyder, D.S.; Forman, S.J.; Sawyers, C.L.; Bhatia, R. Detection of BCR::ABL kinase mutations in CD34+ cells from chronic myelogenous leukemia patients in complete cytogenetic remission on imatinib mesylate treatment. Blood 2005, 105, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, H.G.; Allan, E.K.; Jordanides, N.E.; Mountford, J.C.; Holyoake, T.L. Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood 2007, 109, 4016–4019. [Google Scholar] [CrossRef] [PubMed]
- Giustacchini, A.; Thongjuea, S.; Barkas, N.; Woll, P.S.; Povinelli, B.J.; Booth, C.A.G.; Sopp, P.; Norfo, R.; Rodriguez-Meira, A.; Ashley, N.; et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017, 23, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Warfvinge, R.; Geironson, L.; Sommarin, M.N.E.; Lang, S.; Karlsson, C.; Roschupkina, T.; Stenke, L.; Stentoft, J.; Olsson-Strömberg, U.; Hjorth-Hansen, H.; et al. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood 2017, 129, 2384–2394. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ge, Y.; Sun, L.; Ma, W.; Wu, J.; Zhang, X.; Hu, X.; Eaves, C.J.; Wu, D.; Zhao, Y. Growth arrest specific 2 is up-regulated in chronic myeloid leukemia cells and required for their growth. PLoS ONE 2014, 9, e86195. [Google Scholar] [CrossRef] [PubMed]
- Benetti, R.; Copetti, T.; Dell’Orso, S.; Melloni, E.; Brancolini, C.; Monte, M.; Schneider, C. The Calpain System Is Involved in the Constitutive Regulation of -Catenin Signaling Functions. J. Biol. Chem. 2005, 280, 22070–22080. [Google Scholar] [CrossRef]
- Sun, L.; Zhou, H.; Liu, H.; Ge, Y.; Zhang, X.; Ma, W.; Wu, D.; Zhao, Y. GAS2–Calpain2 axis contributes to the growth of leukemic cells. Acta Biochim. Biophys. Sin. 2015, 47, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Lee, C.L.; Yang, S.H.; Chien, C.C.; Huang, C.C.; Yang, R.N.; Chang, C.-C. Upregulation of the growth arrest-specific-2 in recurrent colorectal cancers, and its susceptibility to chemotherapy in a model cell system. Biochim. Biophys. Acta 2016, 1862, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Alousi, A.; Parchment, R.; Boinpally, R.; Gadgeel, S.; Weigand, R.; McCormick, J.; Lorusso, P. Phase I clinical trial of XK469 in patients with chemo-refractory solid tumors. J. Clin. Oncol. 2004, 22, 2020. [Google Scholar] [CrossRef]
- Undevia, S.D.; Innocenti, F.; Ramirez, J.; House, L.; Desai, A.A.; Skoog, L.A.; Singh, D.A.; Karrison, T.; Kindler, H.L.; Ratain, M.J. A phase I and pharmacokinetic study of the quinoxaline antitumour Agent R(+)XK469 in patients with advanced solid tumours. Eur. J. Cancer 2008, 44, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Stock, W.; Undevia, S.D.; Bivins, C.; Ravandi, F.; Odenike, O.; Faderl, S.; Rich, E.; Borthakur, G.; Godley, L.; Verstovsek, S.; et al. A phase I and pharmacokinetic study of XK469R (NSC 698215), a quinoxaline phenoxypropionic acid derivative, in patients with refractory acute leukemia. Investig. New Drugs 2008, 26, 331–338. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.J.; Faydaci, B.A.; McGuinness, D.; Shaw, R.; Maciewicz, R.A.; Robertson, N.A.; Goodyear, C.S. Searchlight: Automated bulk RNA-seq exploration and visualisation using dynamically generated R scripts. BMC Bioinform. 2021, 22, 411. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Castañeda, E.; Hopcroft, L.E.M.; Rogers, S.; Munje, C.; Bittencourt-Silvestre, J.; Copland, M.; Vetrie, D.; Holyoake, T.; Jørgensen, H.G. Tyrosine Kinase Inhibitor Independent Gene Expression Signature in CML Offers New Targets for LSPC Eradication Therapy. Cancers 2022, 14, 5253. [Google Scholar] [CrossRef] [PubMed]
- Darici, S.; Zavatti, M.; Braglia, L.; Accordi, B.; Serafin, V.; Horne, G.A.; Manzoli, L.; Palumbo, C.; Huang, X.; Jørgensen, H.G.; et al. Synergistic cytotoxicity of dual PI3K/mTOR and FLT3 inhibition in FLT3-ITD AML cells. Adv. Biol. Regul. 2021, 82, 100830. [Google Scholar] [CrossRef]
- Scott, M.T.; Liu, W.; Mitchell, R.; Clarke, C.J.; Kinstrie, R.; Warren, F.; Almasoudi, H.; Stevens, T.; Dunn, K.; Pritchard, J.; et al. Activating p53 abolishes self-renewal of quiescent leukaemic stem cells in residual CML disease. Nat. Commun. 2024, 15, 651. [Google Scholar] [CrossRef] [PubMed]
- Kessel, D.; Reiners, J.J.; Hazeldine, S.T.; Polin, L.; Horwitz, J.P. The role of autophagy in the death of L1210 leukemia cells initiated by the new antitumor agents, XK469 and SH80. Mol. Cancer Ther. 2007, 6, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Parchment, R.E.; LoRusso, P.M.; Zhou, J.Y.; Li, J.; Lawrence, T.S.; Sun, Y.; Wu, G.S. The investigational new drug XK469 induces G(2)-M cell cycle arrest by p53-dependent and -independent pathways. Clin. Cancer Res. 2001, 7, 3336–3342. [Google Scholar] [PubMed]
- Lin, H.; Subramanian, B.; Nakeff, A.; Chen, B.D. XK469, a novel antitumor agent, inhibits signaling by the MEK/MAPK signaling pathway. Cancer Chemother. Pharmacol. 2002, 49, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Foucquier, J.; Guedj, M. Analysis of drug combinations: Current methodological landscape. Pharmacol. Res. Perspect. 2015, 3, e00149. [Google Scholar] [CrossRef] [PubMed]
- Bliss, C.I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- Roell, K.R.; Reif, D.M.; Motsinger-Reif, A.A. An Introduction to Terminology and Methodology of Chemical Synergy-Perspectives from Across Disciplines. Front. Pharmacol. 2017, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Belle, L.; Bruck, F.; Foguenne, J.; Gothot, A.; Beguin, Y.; Baron, F.; Briquet, A. Imatinib and nilotinib inhibit hematopoietic progenitor cell growth, but do not prevent adhesion, migration and engraftment of human cord blood CD34+ cells. PLoS ONE 2012, 7, e52564. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Huang, K.C.; Yamasaki, E.F.; Chan, K.K.; Chohan, L.; Snapka, R.M. XK469, a selective topoisomerase IIbeta poison. Proc. Natl. Acad. Sci. USA 1999, 96, 12168–12173. [Google Scholar] [CrossRef] [PubMed]
- Kakodkar, N.C.; Peddinti, R.; Kletzel, M.; Tian, Y.; Guerrero, L.J.; Undevia, S.D.; Geary, D.; Chlenski, A.; Yang, Q.; Salwen, H.R.; et al. The quinoxaline anti-tumor agent (R+)XK469 inhibits neuroblastoma tumor growth. Pediatr. Blood Cancer 2011, 56, 164–167. [Google Scholar] [CrossRef]
- Sgorbissa, A.; Benetti, R.; Marzinotto, S.; Schneider, C.; Brancolini, C. Caspase-3 and caspase-7 but not caspase-6 cleave Gas2 in vitro: Implications for microfilament reorganization during apoptosis. J. Cell Sci. 1999, 112, 4475–4482. [Google Scholar] [CrossRef] [PubMed]
- Sadaf, S.; Awasthi, D.; Singh, A.K.; Nagarkoti, S.; Kumar, S.; Barthwal, M.K.; Dikshit, M. Pyroptotic and apoptotic cell death in iNOS and nNOS overexpressing K562 cells: A mechanistic insight. Biochem. Pharmacol. 2020, 176, 113779. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Holyoake, T.L. Preclinical approaches in chronic myeloid leukemia: From cells to systems. Exp. Hematol. 2017, 47, 13–23. [Google Scholar] [CrossRef] [PubMed]


| Comparison | Significant Genes | p Adjusted | ||
|---|---|---|---|---|
| Upregulated | Downregulated | Total | ||
| 44 | 18 | 62 | <0.05 |
| 13 | 30 | 43 | <0.05 |
| 1 | 8 | 9 | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramirez-Guzman, L.A.; Huang, W.; Cole, J.J.; Jørgensen, H.G. GAS2 Upregulation Is a Targetable Vulnerability in Chronic Myeloid Leukemia. Int. J. Transl. Med. 2024, 4, 354-368. https://doi.org/10.3390/ijtm4020023
Ramirez-Guzman LA, Huang W, Cole JJ, Jørgensen HG. GAS2 Upregulation Is a Targetable Vulnerability in Chronic Myeloid Leukemia. International Journal of Translational Medicine. 2024; 4(2):354-368. https://doi.org/10.3390/ijtm4020023
Chicago/Turabian StyleRamirez-Guzman, Lizbeth A., Wenjing Huang, John J. Cole, and Heather G. Jørgensen. 2024. "GAS2 Upregulation Is a Targetable Vulnerability in Chronic Myeloid Leukemia" International Journal of Translational Medicine 4, no. 2: 354-368. https://doi.org/10.3390/ijtm4020023
APA StyleRamirez-Guzman, L. A., Huang, W., Cole, J. J., & Jørgensen, H. G. (2024). GAS2 Upregulation Is a Targetable Vulnerability in Chronic Myeloid Leukemia. International Journal of Translational Medicine, 4(2), 354-368. https://doi.org/10.3390/ijtm4020023







