Novel Natural and Synthetic Anticandidal Therapeutic Peptides to Combat Drug-Resistant Infections †
Abstract
:1. Introduction
2. Methodology
2.1. Peptide Design and Synthesis
2.2. Test Organisms
2.3. Biofilm Formation
2.4. Purification and Characterization
2.5. Determination of Antimicrobial Activity and MIC Values of Peptides
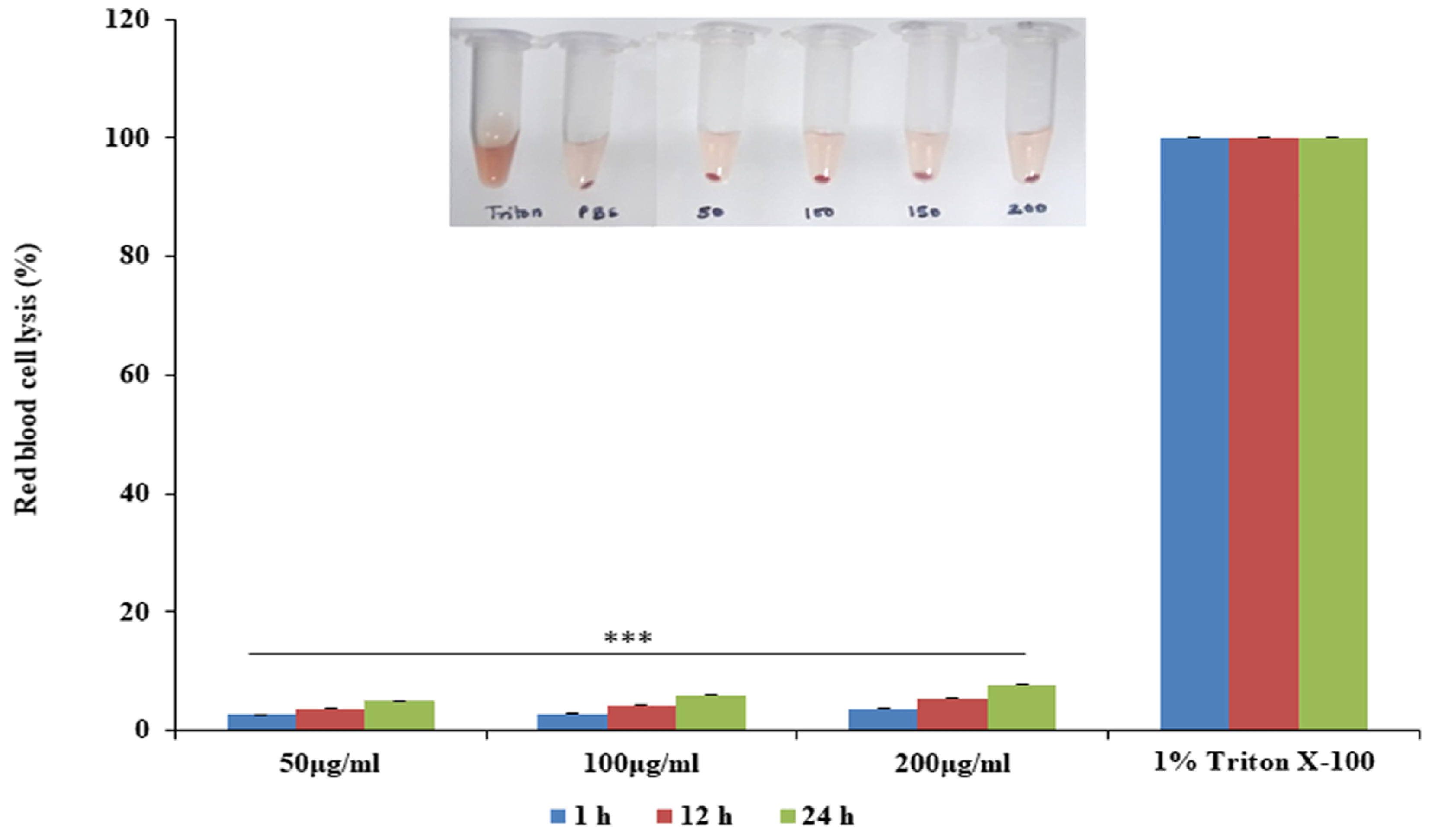
2.6. Hemolysis and Time-Kill Assay
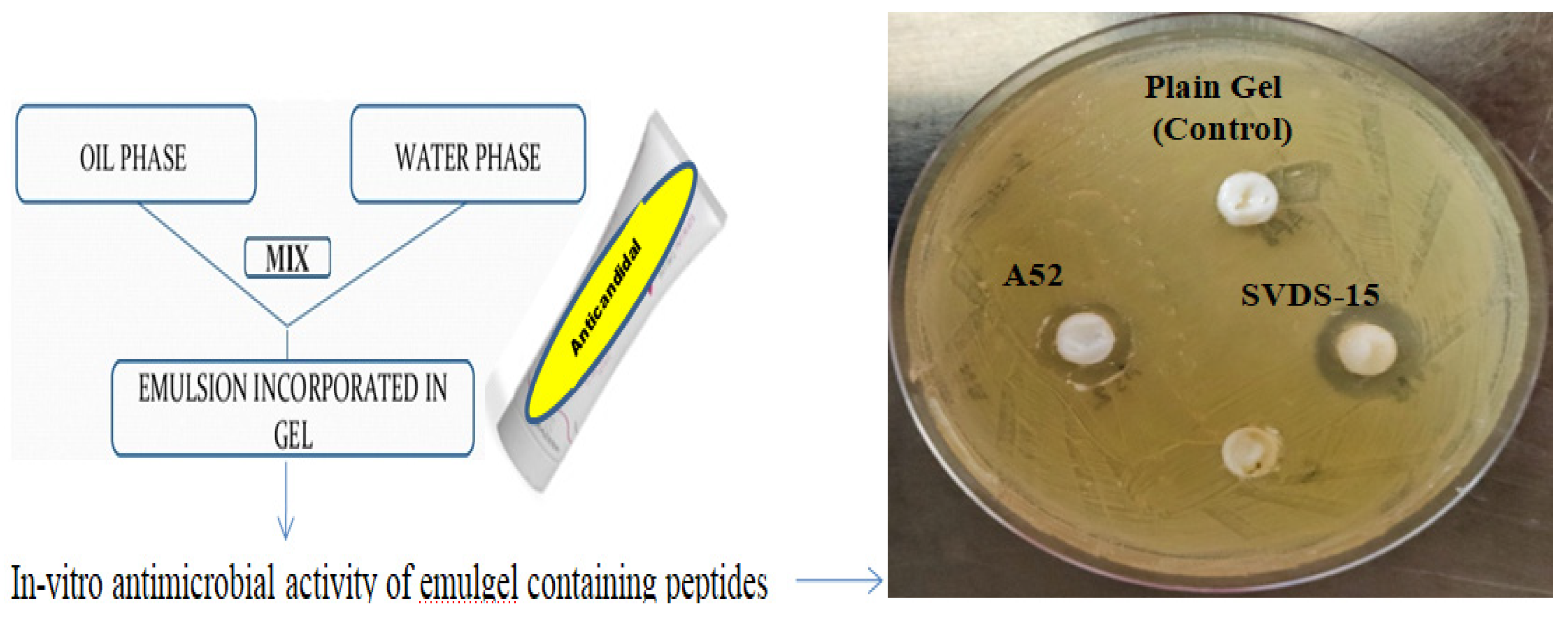
2.7. Emulgel Formulation and Skin Irritation Studies
3. Results and Discussion
3.1. Design and Synthesis of Peptides
3.2. Growth and Biofilm Optimization
3.3. Effect of Synthetic Peptides on Growth and Biofilm
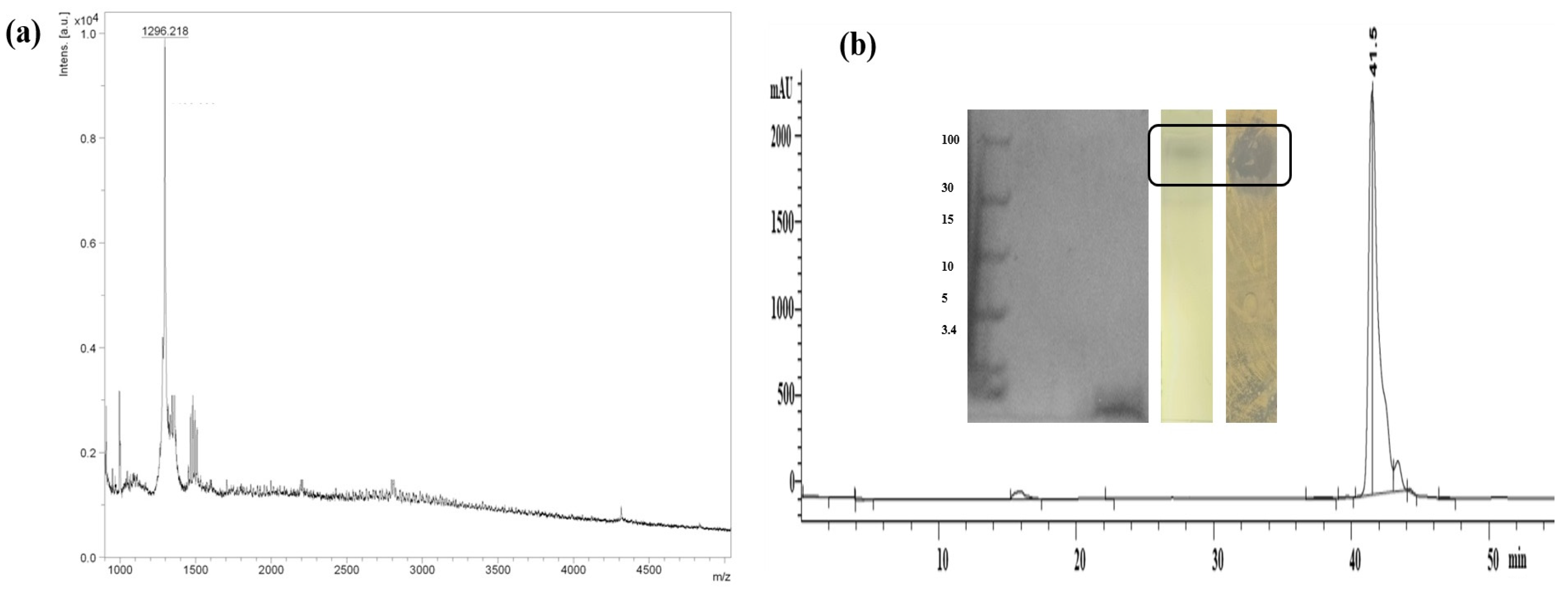
3.4. Characterization of SVDS-15 Peptide
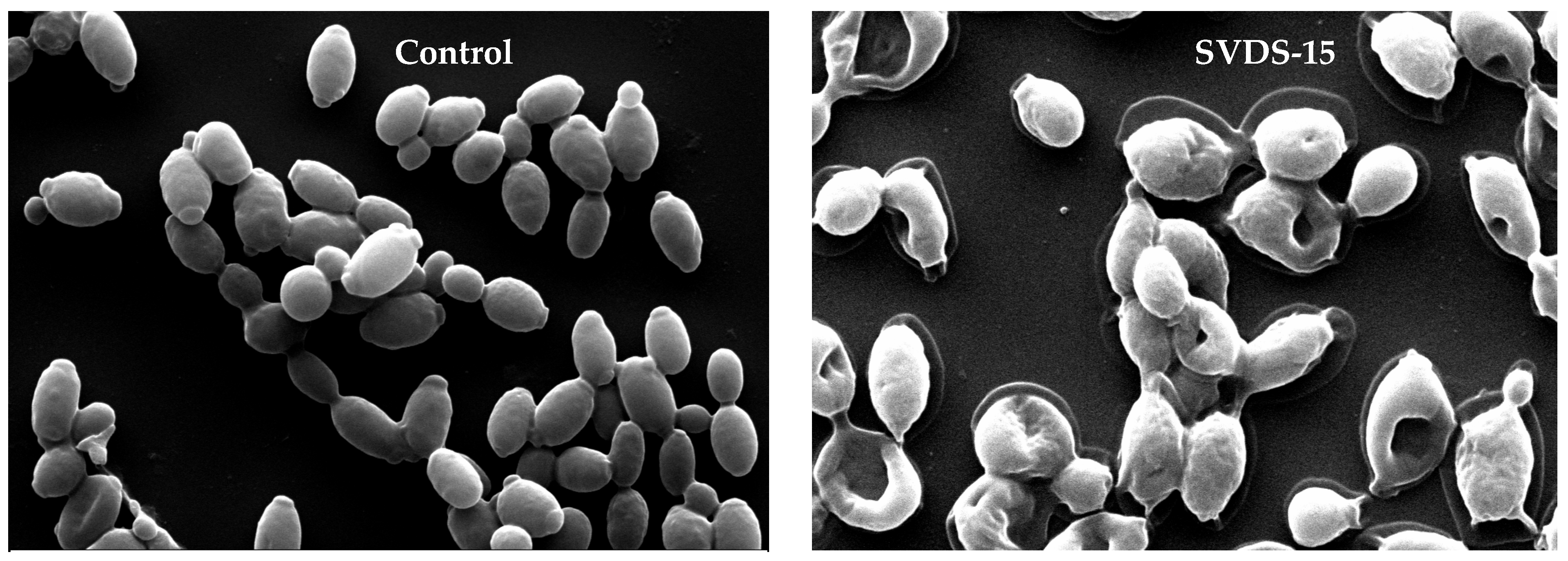
3.5. Scanning Electron Microscopy of SVDS-15 Peptide
3.6. Emulgel Formulation of Peptides
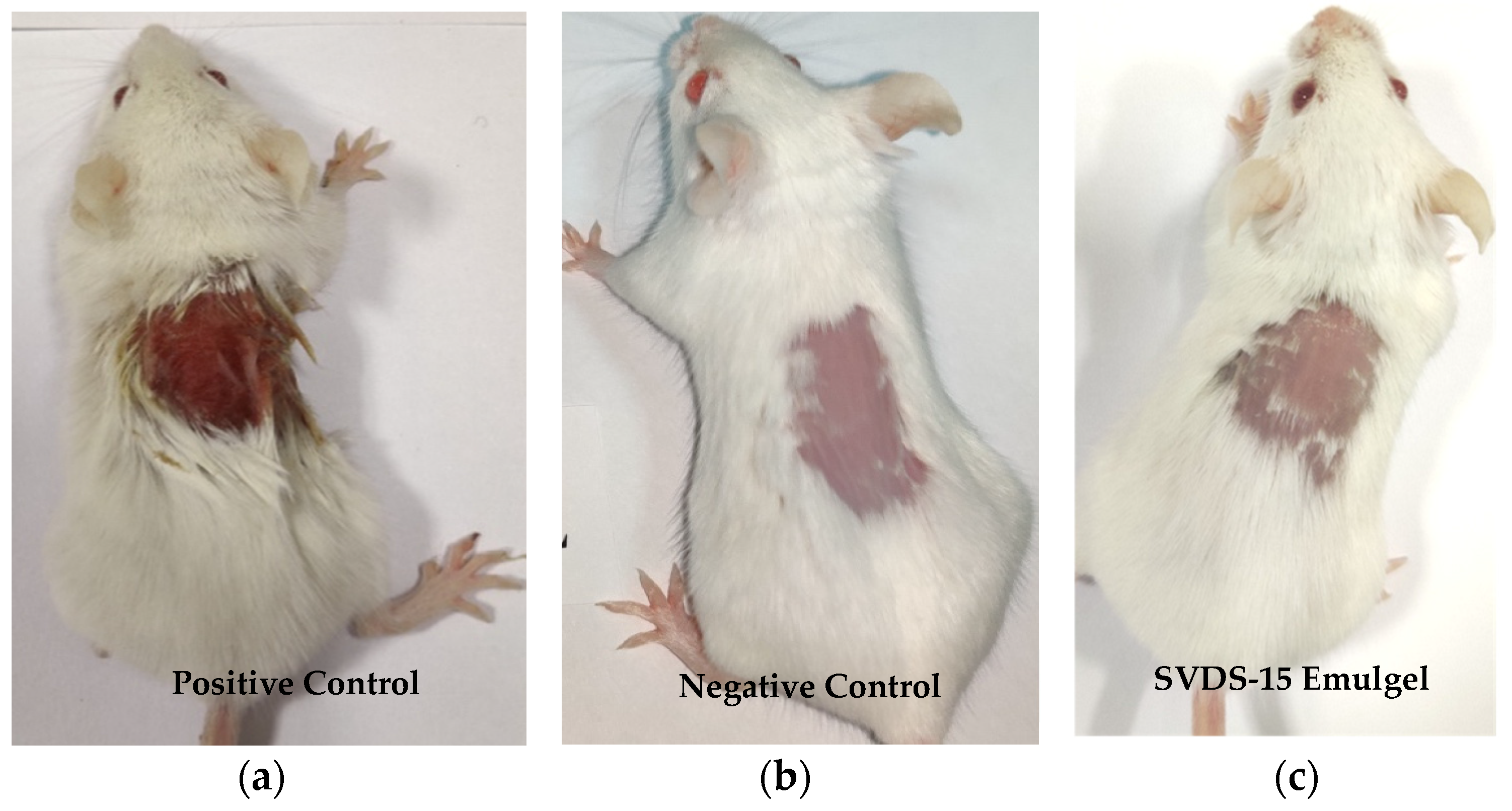
3.7. Skin Irritation Study of Emulgels
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naglik, J.R.; Moyes, D.L.; Wächtler, B.; Hube, B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011, 13, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Lum, K.; Tay, S.; Le, C.F.; Lee, V.S.; Sabri, N.H.; Velayuthan, R.D.; Hassan, H.; Sekaran, S.D. Activity of Novel Synthetic Peptides against Candida albicans. Sci. Rep. 2015, 5, 9657. [Google Scholar] [CrossRef] [PubMed]
- Eksi, F.; Gayyurhan, E.D.; Balci, I. In vitro susceptibility of Candida species to four antifungal agents assessed by the reference broth microdilution method. Sci. World J. 2013, 2013, 236903. [Google Scholar] [CrossRef] [PubMed]
- Ademe, M.; Girma, F. Candida auris: From multidrug resistance to pan-resistant strains. Infect Drug Resist. 2020, 13, 1287–1294. [Google Scholar]
- Singh, P.K.; Sharma, S.; Kumari, A.; Korpole, S. A non-pediocin low molecular weight antimicrobial peptide produced by Pediococcus pentosaceus strain IE-3 shows increased activity under reducing environment. BMC Microbial. 2014, 14, 226. [Google Scholar] [CrossRef]
- Freitas, C.G.; Felipe, M.S. Candida albicans and Antifungal Peptides. Infect. Dis. Ther. 2023, 12, 2631–2648. [Google Scholar] [CrossRef]
- Raman, N.; Lee, M.R.; Lynn, D.M.; Palecek, S.P. Antifungal activity of 14-helical β-peptides against planktonic cells and biofilms of Candida species. Pharmaceuticals 2015, 8, 483–503. [Google Scholar] [CrossRef]
- Lyu, Y.; Yang, Y.; Lyu, X.; Dong, N.; Shan, A. Antimicrobial activity, improved cell selectivity and mode of action of short PMAP-36-derived peptides against bacteria and Candida. Sci. Rep. 2016, 6, 27258. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Bhalla, S.; Chaudhary, K.; Kumar, R.; Sharma, M.; Raghava, G.P.S. In Silico Approach for Prediction of Antifungal Peptides. Front Microbiol. 2018, 9, 323. [Google Scholar] [CrossRef]
- Mól, A.R.; Castro, M.S.; Fontes, W. NetWheels: A web application to create high quality peptide helical wheel and net projections. bioRxiv 2018, 416347. [Google Scholar] [CrossRef]
- Baindara, P.; Singh, N.; Ranjan, M.; Nallabelli, N.; Chaudhry, V.; Pathania, G.L.; Sharma, N.; Kumar, A.; Patil, P.B.; Korpole, S. Laterosporulin10: A novel defensin like Class IId bacteriocin from Brevibacillus sp. strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology 2016, 162, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Singh, S.S.; Baindara, P.; Sharma, S.; Khatri, N.; Grover, V.; Korpole, S. Surfactin like broad spectrum antimicrobial lipopeptide co-produced with sublancin from Bacillus subtilis strain A52: Dual reservoir of bioactives. Front. Microbiol. 2020, 11, 1167. [Google Scholar] [CrossRef]
- Raje, M.; Dhiman, R.; Majumdar, S.; Dass, T.; Dikshit, K.L.; Kaur, R. Charged nylon membrane substrate for convenient and versatile high resolution microscopic analysis of Escherichia coli & mammalian cells in suspension culture. Cytotechnology 2006, 51, 111–117. [Google Scholar]
- Draize, J.H.; Woodard, G.; Calvery, H.O. Methods for the study of irritation and toxicity of substances applied topically to the skin and mucous membranes. J. Pharmacol. Exp. Ther. 1944, 82, 377–390. [Google Scholar]
- Gong, Z.; Karlsson, A.J. Translocation of cell-penetrating peptides into Candida fungal pathogens. Protein Sci. 2017, 26, 1714–1725. [Google Scholar] [CrossRef]
- Torres, R.; Barreto-Santamaría, A.; Arévalo-Pinzón, G.; Firacative, C.; Gómez, B.L.; Escandón, P.; Patarroyo, M.A.; Muñoz., J.E. In Vitro Antifungal Activity of Three Synthetic Peptides against Candida auris and Other Candida Species of Medical Importance. Antibiotics 2023, 12, 1234. [Google Scholar] [CrossRef] [PubMed]
- Gerard, J.M.; Haden, P.; Kelly, M.T.; Andersen, R.J. Loloatins A-D, cyclic decapeptide antibiotics produced in culture by a tropical marine bacterium. J. Nat. Prod. 1999, 62, 80–85. [Google Scholar] [CrossRef]
- Singh, S.S.; Sharma, D.; Baindara, P.; Choksket, S.; Harshvardhan; Mandal, S.M.; Grover, V.; Korpole, S. Characterization and antimicrobial studies of iturin-like and bogorol-like lipopeptides from Brevibacillus spp. strains GI9 and SKDU10. Front. Microbiol. 2021, 12, 729026. [Google Scholar] [CrossRef]
- Miller, M.A.; Zachary, J.F. Mechanisms and Morphology of Cellular Injury, Adaptation, and Death. Pathol. Basis Vet. Dis. 2017, 2, 43.e19. [Google Scholar]
- Neff, J.A.; Bayramov, D.F.; Patel, E.A.; Miao, J. Novel antimicrobial peptides formulated in chitosan matrices are effective against biofilms of multidrug-resistant wound pathogens. Mil. Med. 2020, 185, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Gordya, N.; Yakovlev, A.; Kruglikova, A.; Tulin, D.; Potolitsina, E.; Suborova, T.; Bordo, D.; Rosano, C.; Chernysh, S. Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: Calliphora vicina medicinal maggots. PLoS ONE 2017, 12, e0173559. [Google Scholar] [CrossRef] [PubMed]
| Peptide | Peptide Sequence | Hydrophobicity | Amphipathicity | Hydrophilicity | Charge | Mol. Wt. | Structure |
|---|---|---|---|---|---|---|---|
| SK01 | MACVNQCPKAIDRFIVK | −0.12 | 0.65 | −0.06 | 2 | 1937 | Defensin-like beta |
| SK02 | KQVYKACMNGKHLYC | −0.21 | 0.91 | −0.19 | 3.5 | 1786 | Defensin-like beta |
| SK03 | GIRWLVYRLRKV | −0.25 | 0.92 | −0.17 | 4 | 1559 | Helical |
| SK04 | HGLENKMYRHV | −0.31 | 0.94 | 0.12 | 2 | 1384 | Helical |
| SK05 | ATCHCSIHVSK | −0.09 | 0.6 | −0.33 | 2 | 1186 | Helical |
| SK06 | CMNGTQVYCR | −0.22 | 0.37 | −0.41 | 1 | 1175 | Helical |
| SK07 | KILKVARAWLAK | −0.13 | 1.12 | 0.02 | 4 | 1396.7 | Helical |
| SK08 | Fmoc-KILKVARAWLAK | −0.13 | 1.12 | 0.02 | 4 | 1655.4 | Helical |
| Pep1 | VKILAVALKWRAKR | −0.2 | 1.14 | 0.31 | 5 | 1652.3 | Helical |
| Pep2 | VIHKRHDGVKRI | −0.38 | 1.26 | 0.62 | 4 | 1457.9 | Helical |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, D. Novel Natural and Synthetic Anticandidal Therapeutic Peptides to Combat Drug-Resistant Infections. Med. Sci. Forum 2024, 24, 12. https://doi.org/10.3390/ECA2023-16385
Sharma D. Novel Natural and Synthetic Anticandidal Therapeutic Peptides to Combat Drug-Resistant Infections. Medical Sciences Forum. 2024; 24(1):12. https://doi.org/10.3390/ECA2023-16385
Chicago/Turabian StyleSharma, Deepika. 2024. "Novel Natural and Synthetic Anticandidal Therapeutic Peptides to Combat Drug-Resistant Infections" Medical Sciences Forum 24, no. 1: 12. https://doi.org/10.3390/ECA2023-16385
APA StyleSharma, D. (2024). Novel Natural and Synthetic Anticandidal Therapeutic Peptides to Combat Drug-Resistant Infections. Medical Sciences Forum, 24(1), 12. https://doi.org/10.3390/ECA2023-16385






